Abstract
The African trypanosome, Trypanosoma brucei, causes sleeping sickness in humans in sub-Saharan Africa. Here we report the sequence and analysis of the 1.1 Mb chromosome I, which encodes approximately 400 predicted genes organised into directional clusters, of which more than 100 are located in the largest cluster of 250 kb. A 160-kb region consists primarily of three gene families of unknown function, one of which contains a hotspot for retroelement insertion. We also identify five novel gene families. Indeed, almost 20% of predicted genes are members of families. In some cases, tandemly arrayed genes are 99–100% identical, suggesting an active process of amplification and gene conversion. One end of the chromosome consists of a putative bloodstream-form variant surface glycoprotein (VSG) gene expression site that appears truncated and degenerate. The other chromosome end carries VSG and expression site-associated genes and pseudogenes over 50 kb of subtelomeric sequence where, unusually, the telomere-proximal VSG gene is oriented away from the telomere. Our analysis includes the cataloguing of minor genetic variations between the chromosome I homologues and an estimate of crossing-over frequency during genetic exchange. Genetic polymorphisms are exceptionally rare in sequences located within and around the strand-switches between several gene clusters.
INTRODUCTION
African trypanosomes are eukaryotic parasites that replicate in the blood and tissue fluids of mammals, remaining extracellular throughout the lifecycle. Sleeping sickness in humans is caused by the Trypanosoma brucei species complex, which is transmitted by tsetse flies. The disease exists as a zoonosis in foci throughout sub-Saharan Africa. There are 300 000– 500 000 cases and over 2 million disability-adjusted life years (DALYs) are lost annually (1). Re-emergence can lead to epidemics, such as those reported in recent years (2). Infection of livestock with pathogenic trypanosomes is also a major economic restraint in much of sub-Saharan Africa (3). New chemotherapies are desperately required given the toxicity of available drugs and the rising prevalence of drug resistance (4,5).
The nuclear genome of T.brucei consists of three size classes of chromosome: 50–100 mini-chromosomes, a variable number of intermediate-sized chromosomes and 11 pairs of diploid megabase chromosomes (6). The latter contain the majority of genes and are being sequenced to completion in an international collaboration between the Sanger Institute and The Institute for Genomic Research (TIGR, USA). Large sections of the genome are transcribed polycistronically, and gene regulation is thought to be largely post-transcriptional (7). While the frequency of genetic exchange in the field remains controversial (8), a non-obligatory sexual cycle is observed and hybrid genotypes can be obtained (9).
Trypanosomes have a sophisticated method of immune avoidance. The mammal-infective (metacyclic) and bloodstream forms are covered in a variant surface glycoprotein (VSG) and serial expression of antigenically distinct VSGs leads to antigenic variation. The transcribed VSG gene is always located in one of several telomeric expression sites on a megabase or intermediate chromosome, together with multiple expression site-associated genes (10–13). The unexpressed VSG genes on the mini-chromosomes and at internal sites may be transposed to expression sites by duplicative transposition during antigenic switching.
We present here the complete DNA sequence and analysis of Tb927 chromosome I (chrI), including polymorphism analysis and genetic crossover frequency. Together with the accompanying research report from El-Sayed et al. (14), these data reveal for the first time the detailed organisation of trypanosome megabase chromosomes. Almost half of chromosome I is putative coding sequence (CDS: the open reading frame that forms the protein-coding region of a putative gene) and over half the predicted genes are novel to biology. The genes are organised into long directional clusters, which probably reflects the important role of polycistronic transcription. Several novel T.brucei gene families are described. Some show considerable sequence variation, while variable numbers of perfectly conserved tandemly arrayed genes suggests a role for gene amplification in control of protein levels in the parasite.
MATERIALS AND METHODS
Trypanosome stocks
The preparation of cloned T.brucei stocks of TREU (Trypanosomiasis Research, Edinburgh University) 927/4 (GPAL/KE/70/EATRO 1534) single VAT derivative GUTat 10.1 (Tb927), STIB (Swiss Tropical Institute, Basel) 247L (WA/TZ/71/STIB 247) (Tb247), 427 (Tb427), and laboratory hybrids have been described (6,15,16). TREU927/4GUTat 10.1 DNA is the sequencing substrate for the African trypanosome genome project and its growth and differentiation have been characterised (17).
Sequencing strategy and chromosome analysis
The DNA sequence of chromosome I was determined by whole chromosome shotgun (WCS). Briefly, the faster migrating homologue (chrIa) was excised and eluted from pulsed field gels (PFGs), sheared and cloned into a puc18 plasmid vector. Clones were selected randomly and end-sequenced to ∼10× coverage of the chromosome. Shotgun clones prepared from large-insert DNA cloned into bacteriophage P1 (18) and BAC vectors (selected by searching end-sequences at http://www.tigr.org/tdb/e2k1/tba1/gene.shtml) were also sequenced to ∼3× coverage, providing a framework on which to assemble the WCS. This method has been described previously (19). Sequence reads were assembled using PHRAP (P. Green, unpublished).
Annotation was performed using Artemis (20). CDS predictions were based on multiple lines of evidence including Glimmer (21), GC bias and sequence homology to known genes. Gene function was predicted using searches against sequences in public databases, Interpro hits and predicted domains (22,23), including signal peptide prediction [SignalP (24)] and trans-membrane domain searches [TMHMM2.0 (25)]. CDSs are labelled incrementally Tb927.1.10, Tb927.1.20, Tb927.1.30, etc. [nomenclature agreed with TIGR (14)]. Gene classification was based on the Gene Ontology system (26). Pseudogenes were identified where a region of DNA has significant BLASTX homology to known proteins but possible open reading frames were interrupted by stop codons or frameshifts. Repeated DNA was identified using Dotter (27) and BLASTN. Paralogous genes were initially identified using BLASTP searches against chromosome 1 predicted proteins and analysed using Clustal. Genes that were likely to have arisen by recent duplication from a common ancestor were defined as paralogous. Full details of CDS annotation may be viewed at http://www.genedb.org.
Nucleotide polymorphisms
The shotgun clone library of gel-eluted chrIa DNA is ∼80% pure and the remaining 20% derives mostly from the larger chrIb homologue (6). P1 and BAC clones derive from either homologue. Therefore, heterozygous DNA sequence polymorphisms between the chrI diploid homologues could be identified and catalogued manually during the finishing process. In the text, we use the term SSP (small scale polymorphism) to describe minor genetic variations including single nucleotide substitutions, single and small-scale multibase deletions and insertions (indels), and microsatellite repeat variations (as defined at http://www.ncbi.nlm.nih.gov/SNP). Identification of polymorphisms in DNA sequences found on multiple chromosomes is less reliable. Our method of SSP identification was verified by amplifying and sequencing 24 chrI regions of ∼500 bp from Tb927 genomic DNA, selected because they contained at least two putative SSPs. Heterozygous SSPs were identified in sequence traces derived from diploid amplification products and 97% of the annotated SSPs in these fragments were confirmed.
Repeated DNA length polymorphisms
To determine the length of large tandem arrays, restriction enzyme sites in unique sequence flanking tandem arrays, but not within array units, were identified for digestion of genomic DNA. Fragments were separated by pulsed field gel electrophoresis (PFGE) together with markers of known size, using optimal conditions for each fragment size range, then Southern-blotted and probed with a clone of the relevant repeat unit. Two different enzymes or combinations of enzymes were applied to show that length variation is not due to restriction site polymorphism. These enzymes were also combined with another cutting once inside the repeat unit, to check that expansion was not due to unique sequence insertions (data not shown). Tb927 chrI microsatellites (2–5 bp repeats, 70–100% identity) were identified using Tandem Repeats Finder (28). To facilitate comparison with the repeat lengths identified in other organisms (29), the analysis of number and length of perfect 2–4 bp repeat arrays (i.e. 100% identity between repeat units), includes only the number of perfect repeats to the left or right of the first or last point mutation, whichever is the larger. A subset of microsatellites was amplified by PCR. The primer sequences are available on request.
Genetic analysis
The generation of independent F1 progeny clones from crosses between cloned stocks Tb927 and Tb247 has been described previously (16,30). Heterozygous micro- and mini-satellite markers (>10 repeats in Tb927) were selected for polymorphism analysis. None was detected to the left of marker MS42. The genotypes of 38 progeny clones were determined for the 13 markers shown in Figure 2D. The two most frequently inherited haplotypes were presumed to be parental, and the less frequent haplotypes the result of crossovers. [See further details in El-Sayed et al. (14).] The proportion of crossovers between each pair of loci was calculated and expressed in centimorgans (cM). The physical size of the recombination unit is based on the Tb927 chrI sequence.
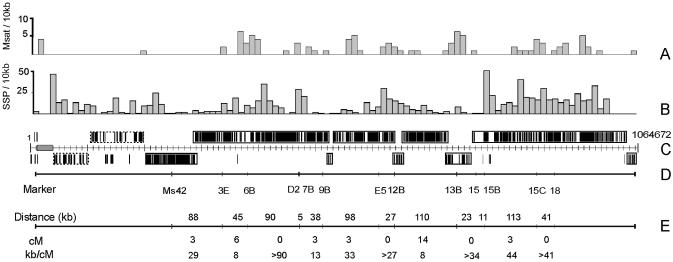
Figure 2.
Chromosome I structure with polymorphism and crossover frequency. The compressed chrI map in (C) shows the position and orientation of the gene clusters illustrated in greater detail in Figure 1. (A) Histogram showing the distribution of microsatellites containing >10 repeats in Tb927 chrI. (B) Histogram showing the distribution of SSPs. In (A) and (B), the width of each bar corresponds to 10 kb. (C) The distribution of directional gene clusters on chrI. Clusters are boxed to reveal those transcribed from left to right (above the line) and from right to left (below the line), and the strand-switch regions between them. Individual CDSs observed on the opposite strand to large clusters are either pseudogenes or annotated as hypothetical proteins, and may not be CDS (see Fig. 1). Clusters of RHS and LRRP CDSs are boxed with a dotted line and exclude two intact CDSs (LRRP1 and 1.380). (D) The position of polymorphic markers used to generate the genetic map. (E) A genetic crossover map of Tb927 chrI, created by analysing hybrid progeny from a cross between Tb927 and Tb247, showing the physical and genetic distances between markers (kb and cM respectively) and the physical size of the recombination unit (kb/cM).
RESULTS AND DISCUSSION
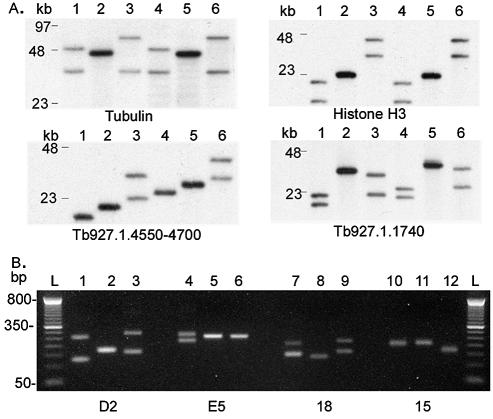
ChrIa has been resolved into three contigs, of which the largest is 1 056 003 bp. The remaining two contigs of 3373 bp and 5096 bp contain telomeric repeats (31) and were positioned by hybridisation to Southern-blotted PmeI genomic digests to the right and left ends of chrIa (18) respectively (results not shown). The gaps between contigs are bordered by degenerate 76 bp repeats (32). Tandemly arrayed CDSs with few sequence polymorphisms between copies could not be resolved by sequence alignment. The number of histone H3 genes was determined by comparison to a partial optical map (Schwartz et al., unpublished) and the numbers of α- and β-tubulin genes were estimated from mapping data, indicating 13–14 in one homologue and at least 9–10 in the other (producing HpaI/NcoI fragments of ∼50 and 36 kb respectively).
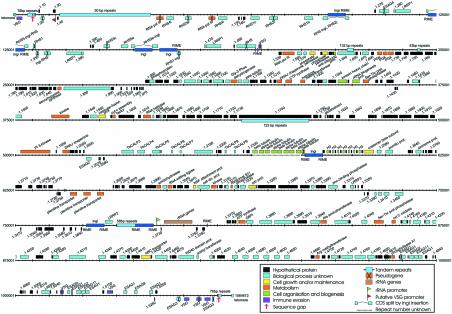
The megabase chromosomes of T.brucei are variable in size within and between isolates. Chromosome Ia of T.brucei stock TREU927 (Tb927 chrIa) is one of the smallest homologues observed to date (6,15). The total length of the DNA sequence presented here is 1 064 472 bp and manual annotation has revealed 534 putative CDSs. However, in the analysis presented here we do not include 139 short predicted CDSs (less than 150 codons) with no additional evidence of transcription, although these ‘unlikely CDSs’ are included in the available annotation and are listed in Table S1 (see Supplementary Material available at NAR Online). Therefore, Figure 1 shows 395 predicted CDSs, including 26 pseudogenes. Where it is possible to assign a (putative) function to the product of a CDS, these have been coloured according to the gene ontology (GO) categories listed (26,33). Some CDSs are assigned more than one term within these categories (details may be viewed at http://www.genedb.org). Figure 1 shows variation in CDS length from ∼100 bp to >21 kb, and considerable variation in CDS/kb in different regions (gene density). The means are given in Table 1. The G+C content of the left end of the chromosome (∼200 kb containing expression site-associated sequences, simple repeats and the RHS gene family, etc.) is lower than that of the rest of the chromosome, reflecting the lower gene density in this region. Table 2 summarises the numbers of genes or CDSs in each category shown in Figure 1. We have been unable to assign putative functions to the majority of annotated CDSs by sequence analysis, and many remain purely hypothetical (for definition, see Fig. 1). Sixty-two percent of CDSs did not have sufficient similarity to any genes sequenced in other organisms to allow homology assignments, a figure similar to that reported on completion of the Plasmodium falciparum genome sequence (34). This presumably reflects the large evolutionary distance between these parasites and other well-studied eukaryotes.
Figure 1.
Annotated features of the T.brucei chromosome I DNA sequence. The map shows the position and orientation of genes, putative CDSs, pseudogenes, DNA repeats and defined promoter sequences. All short CDSs (<150 codons) for which there is additional evidence of transcription (such as conservation in other species) are shown (see text). Coloured boxes above the central line represent genes oriented 5′→3′ left to right, and below in the reverse direction. The CDSs are coloured with respect to GO process categories: hypothetical proteins are predicted from DNA sequence alone, while proteins labelled ‘process unknown’ have homology to other proteins in T.brucei and/or other species but their biological process has not been defined.
Table 1. Features of T.brucei chromosome I DNA sequence.
| Total sequence length | 1 064 472 bp |
| G+C content | 45.2% |
| Expression site sequencesa | 37.9% |
| RHS region | 41.3% |
| CDS-rich regionb | 46.2% |
| Predicted CDS | 50.8% |
| Predicted non-CDS | 40.2% |
| Number of predicted CDSsc | 369 |
| Number of pseudogenes | 26 |
| Average CDS length | 1344 bp |
| Gene densityc | 1 per 2884 bp |
| Percentage CDS | 46.6% |
| Number of rRNA transcription units (18s, 5.8s, 28s) | 1 |
| Number of tRNA genes | 0 |
aFrom the 50-bp repeats to the first telomeric repeat.
bChromosome region containing directional gene clusters from 1.530 to 1.5260.
cNot including pseudogenes.
Table 2. Classification of putative CDSs in chromosome I.
| Total predicted protein-coding CDSs | 369 |
| Hypothetical proteins and proteins of unknown function | 277 |
| Proteins conserved in other species | 177 |
| Proteins conserved in T.brucei onlya | 42 |
| Proteins previously characterised in T.brucei | 37 |
| Proteins with annotated function | 55 |
| Gene ontology process assignmentsb | 156 |
| Cell organization and biogenesis | 18 |
| Metabolism | 8 |
| Other cell growth and/or maintenance | 4 |
| Immune-evasion | 3 |
| Interpro matchesc | 111 |
| Leucine-rich repeat, IPR001611 | 5 |
| G-protein βWD-40 repeat, IPR001680 | 3 |
| AAA ATPase, IPR003593 | 3 |
| SAM (and some other nucleotide) binding motif IPR000051 | 3 |
| Tubulin/FtsZ protein, IPR003008 | 3 |
| Zn-finger, RING, IPR001841 | 3 |
| CDSs with polymorphisms (chrIa versus chrIb) | 119 |
| Total number of non-conservative or complex polymorphismsd | 73 |
aFamilies 1–4, 7, 8, 13, 15–17 in Table 3, plus intact VSG genes.
bNumber of GO process terms assigned (see also Fig. 1), each category contains multiple terms, each CDS may have more than one assignment, GO process categories not assigned to chrI are not listed.
cSix most abundant domains (all domain annotations are available at http://www.genedb.org), non-redundant hits only, i.e. one hit of each interpro domain is counted per CDS and only one copy of repeated genes was used in the analysis.
dConservative amino acid substitutions were defined as those that score greater than zero using a BLOSUM 62 substitution matrix (56), complex polymorphisms include multiple substitutions and insertions or deletions but do not include insertions or deletions of three nucleotides.
Directional gene clusters
Figure 1 shows long arrays of annotated CDSs and genes on the same strand of the chromosome (directional gene clusters) separated by strand-switch regions (35). This organisation is more visible in Figure 2C, where the chromosome has been compressed to reveal the organisation of the clusters. There are eight possible strand-switches in the gene-rich region but >80% of the CDSs are located in just five large clusters, which may represent polycistronic transcription units (35,36). Mature mRNAs are derived from polycistronic molecules by splicing and addition of a 5′ cap (the spliced leader) and a poly(A) tail (7). It is known that mRNA-coding genes in trypanosomes are transcribed by an enzyme with the biochemical properties of a eukaryotic RNA polymerase II, although it is not clear whether there are discrete transcription initiation sites. A promoter element for transcription by RNA polymerase II has been identified only in association with the spliced leader RNA genes to date (37). We are unable to identify related sequence, unusual primary sequence features or physical properties in the strand-switch regions that might indicate common transcription start sites, as reported also by El-Sayed et al. (14) on chromosome II.
Synteny in coding regions and conservation of strand-switch regions are observed between several trypanosomatid parasites (38 and unpublished data), suggesting an evolutionary pressure to conserve the close association of certain genes within clusters and raising the question whether clusters encode proteins involved in related cellular processes, requiring co-regulation or similar expression levels. Since the majority of CDSs on chrI are currently of unknown function, precluding hypothesis, this discussion must await further experimentation.
The clusters contained within the dotted lines (CDS 1.60–1.530, left end) contain multiple members of the novel gene families, RHS and LRRP1 (13,39), and associated CDSs and mobile DNA elements. This region consists of only 17.8% predicted CDS, compared to 54% in the rest of the chromosome, and 44% of the predicted CDSs are pseudogenes. It is not clear that this region is organised, or likely to be transcribed, in the same manner as the gene-rich region, due to the low density of protein coding genes and because the intact genes do not fall into distinct directional gene clusters. In many organisms pseudogenes rarely occur in the central regions of chromosomes and mainly occur in genus- or species-specific gene families involved in adaptive functions, such as environmental response (40). Less conserved genes are also more commonly found in subtelomeric regions than in the chromosomal core. This organisation is thought to be related to the higher rate of recombination observed in these regions promoting amplification and subsequent sequence divergence, whilst conserving the housekeeping genes in the less recombinogenic central core (41,42). Therefore, the pseudogenes are not necessarily ‘dead’ (40). They may represent a reservoir of spare parts from which novel forms appear. The RHS region is located very close to the telomere in chromosomes that lack a bloodstream form VSG expression site (see below), as observed in chrIb and chrIIa (6,14). Larger homologues of chrI in other isolates contain many more RHS and associated sequences in subtelomeric locations and the considerable size variability of this region, between even closely related isolates (18), suggests a capacity for rapid divergence in these sequences. Interestingly, Tb927 chrII is much less size variable than chrI (6,15), yet has a greater number of RHS and associated sequences. However, fewer of these are pseudogenes, perhaps indicating a lower rate of recombination in this region on chrII.
VSG genes and VSG expression sites
Variant surface glycoprotein genes (VSG) are only expressed when inserted into an active expression site (ES). Transcribed VSG genes are invariably positioned adjacent to a telomere, with upstream ∼76 bp repeats. Both metacyclic and bloodstream-form trypomastigotes are covered in VSG protein and different expression sites are used at these stages (MES and BES respectively) (reviewed in 10). The left subtelomeric region of Tb927 chrI contains a sequence similar to that of previously described BES promoters. However, the degree of similarity to the chrI sequence is not as high as that between ES promoters that are known to be functional (normally >90%). The functional elements in BES promoters have been described (43,44) and the chromosome I sequence does not conform to a standard BES promoter using the criteria in either report. Downstream of this element are a single VSG and an ingi retrotransposon, but no complete copy of any expression site-associated genes (ESAGs). ESAGs 6 and 7 have been found in all functional BES characterised to date (13) and this putative BES appears truncated and degenerate compared with all others in the literature. It is not known if it is functional. Upstream of the promoter is an array of BES-associated 50 bp repeats and the entire region is haploid, in that no BES-associated sequences are present on chrIb (6,18).
At the other end of Tb927 chrI there are five VSG genes within 52 kb of the telomere (including one pseudogene), together with ESAG-like sequences interspersed with other CDSs, but no sequence similarity is detected to previously described MES or BES promoter sequences (10,44). Normally, telomeric VSG genes are transcribed towards the telomere. Unusually, the telomeric VSG gene at the right end of Tb927 chrI is positioned in a reverse orientation. Due to a single strand-switch in this region, the five genes adjacent to the telomere are on the reverse strand, such that the 76 bp repeats lie between the VSG gene and the telomere. Two VSG genes in this cluster lack the upstream 76 bp repeats that are usually involved in duplicative transposition (10). An inverted telomeric VSG gene has been described previously, at a position 24 kb from a telomere in T.brucei stock AnTat1.1 (45). This gene was shown to act exclusively as a donor in a transposition event that targeted it to a telomeric expression site elsewhere in the genome. Since this was a rare event that occurred only late in a chronic infection, the authors proposed that its inverse orientation with respect to the chromosome end reduced the efficiency of gene conversion by telomere pairing. It is possible, then, that the right end of Tb927 chrIa is not an active expression site but contains basic copy VSG genes that may be transposed to active sites during antigenic variation (10,14). In contrast to other reports (45 and unpublished data), we do not observe any ingi-like sequence adjacent to the VSGs oriented away from the telomere, or in the strand-switch region between VSGs.
Single nucleotide polymorphisms and other minor genetic variations between homologues
The distribution of minor genetic polymorphisms (referred to here as SSPs) is illustrated in Figure 2. We have identified 965 polymorphic sites, including 72% single nucleotide substitutions, 10% single nucleotide indels, 10% microsatellite variations and 3.5% dinucleotide indels. Such polymorphisms are observed at a mean frequency of ∼1 SSP/kb, ranging from 5 SSP/kb in some regions to 1 SSP/5 kb in one strand-switch region. It is more difficult to accurately define chrI SSPs in sequences found on multiple chromosomes (e.g. RHS, ingi and ribosomal RNA genes), and these should be treated with caution. Of the 965 polymorphic sites, ∼30% are found in putative CDS. Table 2 shows that 119 CDSs contain polymorphisms. Of these, 32 may be significantly altered in protein sequence.
One surprising observation is the low number of SSPs within and around several coding strand-switch regions (Fig. 2). In particular, 12 SSPs were detected in a 60 kb region (position 730 000–790 000 in Fig. 1) containing just 12 genes, of which eight code for relatively short hypothetical proteins, and three retroposon-like elements. While it is unlikely that evolutionary pressure is selecting against mutations over such a large region it is possible that other phenomena, such as increased gene conversion rates, could be acting to reduce accumulation of SSPs between homologues. This observation would be greatly strengthened if we were to observe a similar level of conservation when comparing multiple isolates.
Gene families
Table 3 summarises characteristics of the paralogous gene families identified on chromosome I. Remarkably, almost 20% of the annotated genes are closely related members of families. There are seven whose members have no homologues in other organisms and no characterised function (numbers 1–4, 7, 13 and 15 in Table 3), excluding the previously studied ESAGs and VSGs. Some families occur in alternating arrays, suggesting that they may require co-regulation as observed with α- and β-tubulin (46,47).
Table 3. Characteristics of gene families on chromosome Ia.
| Number | Product description | Distribution | % identity (protein) | % identity (nucleotide) | Conserved 3′ region | Comments | Gene identifiers (no. copies) |
|---|---|---|---|---|---|---|---|
| 1 | RHS | Clustered | 20–99% | 34–99% | Yes | Different 3′UTR associated with different subtypeb | Tb.1.70,120,180,220,420 (5) |
| 2 | LRRP1 | Clustered | 93–95% | 93–95% | Yes | Interspersed with 3 | Tb.1.290,370,480 (3) |
| 3 | Unknown function | Clustered | 75–95% | 93–95% | Yes | Interspersed with LRRP1 | Tb.1.280,380,470 (3) |
| 4 | Unknown function | Tandem | 23% | 45% | No | Tb.1.1470,1500 (2) | |
| 5 | Phosphate permease | Tandem | 97% | 97% | Yes | Tb.1.580,600 (2) | |
| 6 | PGK | Tandem | 65–89% | 63–91% | No | Tb.1.700,710,720 (3) | |
| 7 | Unknown function | Dispersed | 99% | 98% | Yes | Tb.1.1040,1650 (2) | |
| 8 | ESAG2 | Dispersed | 68–82% | 76–87% | Yes | 2 conserved 3′UTRs, 1 divergent | Tb.1.2040,4890,5100 (3) |
| 9 | Calpain-like | Clustered | 6–80% | NDc | No | Tb.1.2100,2110,2120,2150,2160,2230,2260 (7) | |
| 10 | β-tubulin | Tandem | >99% | >99% | Yes | Interspersed with α-tubulin | Tb.1.2330,2350,2370,2390 (ND) |
| 11 | α-tubulin | Tandem | >99% | >99% | Yes | Interspersed with β-tubulin | Tb.1.2340,2360,2380,2400 (ND) |
| 12 | Histone H3 | Tandem | 100% | >99% | Yes | Interspersed with 13 | Tb.1.2430,2450,2470,2490,2510,2530,2550 (7) |
| 13 | Unknown function | Tandem | 100% | >99% | Yes | Interspersed with histone-H3 | Tb.1.2440,2460,2480,2500,2520,2540,2560 (7) |
| 14 | Pteridine transporter | Tandem | 100% | >99% | Yes | Tb.1.2820,2850,2880 (3) | |
| 15 | Unknown function | Tandem | 51–99% | 60–99% | Yes | Last CDS of cluster is divergent in CDS and 3′UTR | Tb.1.4540,4560,4580,4600,4630,4650 (6) |
| 16 | ESAG1 | Clustered | 55–84% | 76–89% | No | 3 conserved 3′UTRs, 2 divergent | Tb.1.4870,4910,5120,5200,5240 (5) |
| 17 | ESAG11 | Clustered | 88% | 91% | Yes | Tb.1.4900,5110 (2) | |
| 18 | ESAG9, putative | Clustered | 30% | 48% | Yes | Tb.1.5080, 5220 (2) |
Clustered, confined to specific region of chrI but not arrayed in defined repeat units, genes may be on either strand, i.e. inverted relative to other copies. Tandem, within a defined repeat unit in close tandem arrays, repeat units never inverted relative to other copies. Dispersed, located in distant regions of the chromosome, may be inverted. ND, not determined.
aExcludes VSGs.
bRHS subtypes described in Bringaud et al. (39).
cNucleotide sequences too divergent for accurate alignment.
Differential expression of genes in T.brucei appears strongly influenced by their 3′ untranslated regions (UTRs) (7). There is no robust bioinformatic method of identifying the precise boundaries of 3′ UTRs, so we chose to compare 300 bp of sequence downstream of every intact CDS. Table 3 shows the results of this analysis for each gene family on chrI. We found that near-identical 3′ sequences (>90% identity over 100 bp or more) on chrI are always associated with gene families, although not all gene families have near-identical UTRs. As previously described, the differentially expressed members of the phosphoglycerate kinase (PGK) gene family have highly divergent 3′ sequences (48), while the α- and β-tubulin genes have identical 3′ sequences and are not differentially expressed (47). Family 15 has very conserved UTRs except the most 3′ family member, and this may be associated with a different expression pattern compared to the other family members. This may also be the explanation for the divergent UTRs of family 4 and the calpain-like proteases.
In some gene families, duplication and divergence serve to produce diversity in proteins within the family (for example, the RHS family discussed above). However, Table 3 shows that several tandem arrays contain multiple repeated CDSs that are almost identical. This is remarkable given that almost no polymorphisms, not even synonymous substitutions, are found on either chrIa or Ib. It appears likely that the level of cellular protein is altered by the amplification and deletion of identical gene copies within the array. It has been shown that there is a negative correlation between percentage identity and distance between repeats in eukaryotes and prokaryotes (49,50), also that the deletion/conversion rate is negatively correlated with the distance between repeats (51). Therefore, close direct repeats are more likely to be similar than dispersed repeats, either because they are more recent or are more subject to gene conversion. However, since the maintenance of repeat identity involves recombination, it is also observed that arrays of large repeats are most liable to deletion and are usually too unstable to persist in the absence of selection. For this reason, tandem repeats are usually smaller than dispersed repeats (49,50).
Several of the repeat units on chrI are very long by comparison with those discussed in other organisms (49,50) and they vary in number between trypanosome isolates. Figure 3A shows variation in the length of restriction fragments encompassing multiple tandemly arrayed copies of tubulin genes, histone H3 and CDSs of unknown function (families 10–13 and 15). Each of these arrays on Tb927 chrI contains CDSs that are 99–100% identical with no unique sequences between the repeat units. Since even third position nucleotide changes that would not alter amino acid sequence are not observed, we suggest that the process of amplification and deletion is an ongoing process and that gene conversion removes copy errors. It remains to be determined to what extent protein levels are altered and whether this is involved in adaptation to environmental factors, but the length of some of these arrays suggests that there is selective pressure to maintain them. Similarly, several long arrays of intergenic repeats (shown in Fig. 1) are also >98% identical. In Tb927 the open reading frame of CDS 1.1740 contains over 20 kb of almost perfect 123 bp repeats. Figure 3A shows that the array may expand to produce a putative CDS of >30 kb.
Figure 3.
Repeated DNA length polymorphism within and between cloned stocks of T.brucei. Columns 1, 4, 7, 10 = Tb927; columns 2, 5, 8, 11 = Tb247; columns 3, 6, 9, 12 = Tb427. L = 100 bp ladder. (A) Restriction fragments containing tandemly arrayed DNA sequences. Top left, tubulin gene array (unit size 3641 bp): lanes 1–3, HpaI/NcoI; lanes 4–6, SspI/XmnI, fragment lengths in Tb927chrIa sequence file 16 235 and 16 247 bp, respectively (but not represented in full, see Fig. 1), length polymorphism between stocks ∼35 kb. Top right, histone H3 gene array (unit size 1584 bp): lanes 1–3, Sau3AI; lanes 4–6, PstI/EcoRI, fragment lengths in sequence file 11 469 and 11 664 bp, respectively, length polymorphism ∼30 kb. Bottom left, Family 15 tandem array (unit size 3100 bp): lanes 1–3, SspI; lanes 4–6, BglII, fragment lengths in sequence file 18 171 and 26 134 bp, length polymorphism ∼15 kb. Bottom right, 123-bp repeat array in Tb927.1.1740: lanes 1–3, HindIII/SspI; lanes 4–6, XmnI, fragment lengths in sequence file 20 838 and 23 880 bp, length polymorphism ∼15 kb. (B) Length polymorphism of PCR amplification products containing microsatellite. Left to right: msat D2 in Tb.1.1780 and msats E5, 18 and 15 in intergenic sequence.
The sequences represented in Figure 3A contribute ∼100 kb to size variation between chrI homologues in Tb927 and Tb427 (Fig. 3A). In all cases, the smallest allele is observed in Tb927.
Polymorphic microsatellites and the genetic map
We have identified 90 perfect di-, tri- and tetra-nucleotide microsatellites of greater than or equal to five repeat units in chrI (excluding telomere and telomere-associated sequences). A comparative analysis of 1 Mb of sequence derived from each of human, mouse, fruit fly and yeast identified 278, 577, 235 and 120 microsatellites, respectively (29). The reduced counts in the lower eukaryotes presumably reflect the greater gene density. However, the remarkable feature of the T.brucei microsatellites is their median length. In 1 Mb of yeast sequence, 84% of perfect dinucleotide microsatellites contain <10 repeats, 99% contain <15 repeat units (total = 76) (29); the respective counts in T.brucei chrI are 40 and 86% (total = 54). Several studies have shown that variable length distribution of simple repeats in different species is most likely due to a difference in the frequency of polymerase template slippage during replication, i.e. that slippage rates vary taxonomically (29,52), and that this correlates with higher mutation frequencies in longer arrays. Analysis of repeat arrays of >70% identity in ChrI reveals even longer microsatellites, presumably because very long arrays are often interrupted by point mutations.
Figure 3B shows variation in the length of four chrI microsatellites in three T.brucei stocks. Many microsatellites in T.brucei may be conveniently scored across isolates on agarose rather than polyacrylamide gels (for example, D2 apparently varies by >170 bp between Tb927 and Tb427 in Fig. 3B). However, the apparently higher slippage rate in T.brucei may affect the usefulness of such genetic markers for comparison of diverse populations.
The distribution of microsatellites of >10 repeats (i.e. those most likely to show allelic polymorphism) and >70% identity is illustrated in Figure 2A. It is notable that very few exist in one-third of the chromosome, in the BES or the RHS region, despite the lower gene density. Application of a χ2 test does not detect any significant correlation in the distribution of the microsatellites and SSPs shown in Figure 2 (P > 0.05).
The map shown in Figure 2E was generated by analysis of the inheritance of a subset of the microsatellites [and one minisatellite, MS42 (53)] (Fig. 2D) in the progeny of Tb927 × Tb247 crosses (16). A degenerate 58-bp repeat that extends for 5.5 kb is located in a region where no crossovers were observed (13B/15B), within the SSP-poor strand-switch region described above. This region is conserved in Trypanosoma cruzi chromosome III where chromosome fragmentation experiments suggest that it is required for mitotic stability (J. Kelly, personal communication), suggesting it has centromeric properties. Centromeres are generally located in regions of low recombination frequency. Although this sequence is found only on chrI, centromeres are not always conserved in primary sequence (54).
Crossover frequencies range from ∼8 to >90 kb/cM, giving an average value for the physical size of the recombination unit of 22 kb/cM. This is similar to that determined for P.falciparum (55). A genetic map for chrII has also been constructed (14) and, although chrII is only 130 kb larger than chrI, a significantly larger number of crossovers was observed (χ2 = 10.3, P < 0.01).
The complete sequence and analysis of chromosome I of T.brucei stock Tb927 has revealed numerous novel CDSs. This chromosome is smaller than chromosome I homologues in almost all other trypanosome stocks studied to date. In some stocks, chrI is >3.5 Mb in size although no loss of linkage between cDNA markers has been observed (6,15). We propose that most, if not all, of this size variation is accounted for by amplification of DNA sequences contained within the chrI sequence presented here. Our data demonstrate that gene families are undergoing rapid expansion and contraction and we speculate that in some cases gene conversion events are acting to maintain a high level of sequence identity between copies. Analysis of SSPs in the chrI homologues reveals a lower density around strand-switch regions.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful for the close collaboration of the T.brucei group at TIGR, and especially the provision of end-sequences from large-insert DNA clones of the reference genome. We also thank numerous members of the T.brucei genome network for their support for the sequencing project and for comments and advice on annotation issues. This work was funded by the Wellcome Trust and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases. Correspondence and requests for materials should be addressed to N.H. (sequence data), S.E.M. (biological resources), A.T. or C.M.R.T. (genetic mapping).
REFERENCES
- 1.World Health Organisation (1998) Control and surveillance of African trypanosomiasis. Report of a WHO expert committee. Technical Report Series, 881. [PubMed]
- 2.Smith D.H., Pepin,J. and Stich,A.H.R. (1998) Human African trypanosomiasis: an emerging public health crisis. Brit. Med. Bull., 54, 341–355. [DOI] [PubMed] [Google Scholar]
- 3.Swallow B.M. (2000) Impacts of trypanosomiasis on African agriculture. PAAT Technical and Scientific Series, FAO, 2. [Google Scholar]
- 4.Barrett M.P. (2001) Veterinary link to drug resistance in human African trypanosomiasis? Lancet, 358, 603–604. [DOI] [PubMed] [Google Scholar]
- 5.Legros D., Ollivier,G., Gastellu-Etchegorry,M., Paquet,C., Burri,C., Jannin,J. and Büscher,P. (2002) Treatment of human African trypanosomiasis—present situation and needs for research and development. Lancet Infect. Dis., 2, 437–440. [DOI] [PubMed] [Google Scholar]
- 6.Melville S.E., Leech,V., Gerrard,C.S., Tait,A. and Blackwell,J.M. (1998) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasit., 94, 155–173. [DOI] [PubMed] [Google Scholar]
- 7.Clayton C.E. (2002) Life without transcriptional control? From fly to man and back again. EMBO J., 21, 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacLeod A., Tweedie,A., Welburn,S.C., Maudlin,I., Turner,C.M.R. and Tait,A. (2000) Minisatellite marker analysis of Trypanosoma brucei: Reconciliation of clonal, panmictic and epidemic population genetic structures. Proc. Natl Acad. Sci. USA, 97, 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson W. and Stevens,J. (1999) Genetic exchange in the trypanosomatidae. Adv. Parasit., 43, 1–46. [DOI] [PubMed] [Google Scholar]
- 10.Barry J.D. and McCulloch,R. (2001) Antigenic variation in trypanosomes: Enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol., 49, 1–70. [DOI] [PubMed] [Google Scholar]
- 11.LaCount D.J., El-Sayed,N.M., Kaul,S., Wanless,D., Turner,C.M.R. and Donelson,J.E. (2001) Analysis of a donor gene region for a variant surface glycoprotein and its expression site in African trypanosomes. Nucleic Acids Res., 29, 2012–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bringaud F., Biteau,N., Donelson,J.E. and Baltz,T. (2001) Conservation of metacyclic variant surface glycoprotein expression sites among different trypanosome isolates. Mol. Biochem. Parasit., 113, 67–78. [DOI] [PubMed] [Google Scholar]
- 13.Berriman M., Hall,N., Sheader,K., Bringaud,F.D., Tiwari,B., Isobe,T., Bowman,S., Corton,C., Clark,L., Cross,G.A.M. et al. (2002) The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasit., 122, 131–140. [DOI] [PubMed] [Google Scholar]
- 14.El-Sayed N.M.A., Ghedin,E., Song,J., MacLeod,A., Bringaud,F., Larkin,C., Wanless,D., Peterson,J., Hou,L., Taylor,S. et al. (2003) The sequence and analysis of Trypanosoma brucei chromosome II. Nucleic Acids Res., 31, 4856–4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melville S.E., Leech,V., Navarro,M. and Cross,G.A.M. (2000) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasit., 111, 261–273. [DOI] [PubMed] [Google Scholar]
- 16.Tait A., Masiga,D., Ouma,J., MacLeod,A., Sasse,J., Melville,S., Lindegard,G., McIntosh,A. and Turner,M. (2002) Genetic analysis of phenotype in Trypanosoma brucei: a classical approach to potentially complex traits. Phil. Trans. Royal Soc. B, 357, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Deursen F.J., Shahi,S.K., Turner,C.M.R., Hartmann,C., Guerra-Giraldez,C., Matthews,K.R. and Clayton,C.E. (2001) Characterisation of the growth and differentiation in vivo and in vitro of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasit., 112, 163–171. [DOI] [PubMed] [Google Scholar]
- 18.Melville S.E., Gerrard,C.S. and Blackwell,J.M. (1999) Multiple causes of size variation in the diploid megabase chromosomes of African trypanosomes. Chromosome Res., 7, 191–203. [DOI] [PubMed] [Google Scholar]
- 19.Bowman S., Lawson,D., Basham,D., Brown,D., Chillingworth,T., Churcher,C.M., Craig,A., Davies,R.M., Devlin,K., Feltwell,T. et al. (1999) The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature, 400, 532–538. [DOI] [PubMed] [Google Scholar]
- 20.Rutherford K., Parkhill,J., Crook,J., Horsnell,T., Rice,P., Rajandream,M.A. and Barrell,B. (2000) Artemis: sequence visualization and annotation. Bioinformatics, 16, 944–945. [DOI] [PubMed] [Google Scholar]
- 21.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,T., Corpet,F., Croning,M.D.R. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L.L. (2002) The Pfam Protein Families Database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen H., Brunak,S. and von Heijne,G. (1999) Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng., 12, 3–9. [DOI] [PubMed] [Google Scholar]
- 25.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene Ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnhammer E.L.L. and Durbin,R. (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein-sequence analysis. Gene-Combis, 167, 1–10. [DOI] [PubMed] [Google Scholar]
- 28.Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kruglyak S., Durrett,R.T., Schug,M.D. and Aquadro,C.F. (1998) Equilibrium distributions of microsatellite repeat length resulting from a balance between slippage events and point mutations. Proc. Natl Acad. Sci. USA, 95, 10774–10778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turner C.M.R., Sternberg,J., Buchanan,N., Smith,E., Hide,G. and Tait,A. (1990) Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology, 101, 377–386. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Jordan J.L., Cross,G.A.M., de Lange,T. and Griffith,J.D. (2001) T-loops at trypanosome telomeres. EMBO J., 20, 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell D.A., Vanbree,M.P. and Boothroyd,J.C. (1984) The 5′-limit of transposition and upstream barren region of a trypanosome VSG gene—tandem 76 base-pair repeats flanking (TAA)90. Nucleic Acids Res., 12, 2759–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berriman M., Aslett,M., Hall,N. and Ivens,A. (2001) Parasites are GO. Trends Parasitol., 17, 463–464. [DOI] [PubMed] [Google Scholar]
- 34.Gardner M.J., Hall,N., Fung,E., White,O., Berriman,M., Hyman,R.W., Carlton,J.M., Pain,A., Nelson,K.E., Bowman,S. et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Myler P.J., Audleman,L., DeVos,T., Hixson,G., Kiser,P., Lemley,C., Magness,C., Rickel,E., Sisk,E., Sunkin,S. et al. (1999) Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA, 96, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tschudi C. and Ullu,E. (1988) Polygene transcripts are precursors to calmodulin messenger RNAs in trypanosomes. EMBO J., 7, 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilinger G. and Bellofatto,V. (2001) Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res., 29, 1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bringaud F., Vedrenne,C., Cuvillier,A., Parzy,D., Baltz,D., Tetaud,E., Pays,E., Venegas,J., Merlin,G. and Baltz,T. (1998) Conserved organization of genes in trypanosomatids. Mol. Biochem. Parasit., 94, 249–264. [DOI] [PubMed] [Google Scholar]
- 39.Bringaud F., Biteau,N., Melville,S.E., Hez,S., El-Sayed,N.M., Leech,V., Berriman,M., Hall,N., Donelson,J.E. and Baltz,T. (2002) A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot. Cell, 1, 315–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison P.M. and Gerstein,M. (2002) Studying genomes through the aeons: protein families, pseudogenes and proteome evolution. J. Mol. Biol., 318, 1155–1174. [DOI] [PubMed] [Google Scholar]
- 41.Wilson R.K. (1999) How the worm was won—the C. elegans genome sequencing project. Trends Genet., 15, 51–58. [DOI] [PubMed] [Google Scholar]
- 42.Winzeler E.A., Lee,B., McCusker,J.H. and Davis,R.W. (1999) Whole genome genetic-typing in yeast using high-density oligonucleotide arrays. Parasitology, 118, 73–80. [DOI] [PubMed] [Google Scholar]
- 43.Pham V.P., Rothman,P.B. and Gottesdiener,K.M. (1997) Binding of trans-acting factors to the double-stranded variant surface glycoprotein (VSG) expression site promoter of Trypanosoma brucei. Mol. Biochem. Parasit., 89, 11–23. [DOI] [PubMed] [Google Scholar]
- 44.Ginger M.L., Blundell,P.A., Lewis,A.M., Browitt,A., Gunzl,A. and Barry,J.D. (2002) Ex vivo and in vitro identification of a consensus promoter for VSG genes expressed by metacyclic-stage trypanosomes in the tsetse fly. Eukaryot. Cell, 1, 1000–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Werf A., van Assel,S., Aerts,D., Steinert,M. and Pays,E. (1990) Telomere interactions may condition the programming of antigen expression in Trypanosoma brucei. EMBO J., 9, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinstein B. and Solomon,F. (1990) Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae—differences between alpha-tubulin and beta-tubulin. Mol. Cell Biol., 10, 5295–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matthews K.R., Tschudi,C. and Ullu,E. (1994) A common pyrimidine-rich motif governs transsplicing and polyadenylation of tubulin polycistronic pre-messenger-RNA in trypanosomes. Genes Dev., 8, 491–501. [DOI] [PubMed] [Google Scholar]
- 48.Blattner J. and Clayton,C.E. (1995) The 3′-untranslated regions from the Trypanosoma brucei phosphoglycerate kinase-encoding genes mediate developmental regulation. Gene, 162, 153–156. [DOI] [PubMed] [Google Scholar]
- 49.Achaz G., Netter,P. and Coissac,E. (2001) Study of intrachromosomal duplications among the eukaryote genomes. Mol. Biol. Evol., 18, 2280–2288. [DOI] [PubMed] [Google Scholar]
- 50.Achaz G., Rocha,E.P.C., Netter,P. and Coissac,E. (2002) Origin and fate of repeats in bacteria. Nucleic Acids Res., 30, 2987–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drouin G. (2002) Characterization of the gene conversions between the multigene family members of the yeast genome. J. Mol. Evol., 55, 14–23. [DOI] [PubMed] [Google Scholar]
- 52.Kruglyak S., Durrett,R., Schug,M.D. and Aquadro,C.F. (2000) Distribution and abundance of microsatellites in the yeast genome can be explained by a balance between slippage events and point mutations. Mol. Biol. Evol., 17, 1210–1219. [DOI] [PubMed] [Google Scholar]
- 53.Barrett M.P., MacLeod,A., Tovar,J., Sweetman,J.P., Tait,A., Le Page,R.W.F. and Melville,S.E. (1997) A single locus minisatellite sequence which distinguishes between Trypanosoma brucei isolates. Mol. Biochem. Parasit., 86, 95–99. [DOI] [PubMed] [Google Scholar]
- 54.Pluta A.F., Mackay,A.M., Ainsztein,A.M., Goldberg,I.G. and Earnshaw,W.C. (1995) Centromere—hub of chromosomal activities. Science, 270, 1591–1594. [DOI] [PubMed] [Google Scholar]
- 55.Su X.Z., Ferdig,M.T., Huang,Y.M., Huynh,C.Q., Liu,A., You,J.T., Wootton,J.C. and Wellems,T.E. (1999) A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science, 286, 1351–1353. [DOI] [PubMed] [Google Scholar]
- 56.Henikoff S. and Henikoff,J.G. (1993) Performance evaluation of amino acid substitution matrices. Proteins, 17, 49–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.