Abstract
The spontaneous rate of G·C→A·T mutations and a hotspot T·A→G·C transversion are known to increase with the frequency of transcription—increases that have been ascribed primarily to processes that affect only these specific mutations. To investigate how transcription induces other spontaneous point mutations, we tested for its effects in repair-proficient Salmonella enterica using reversion assays of chromosomally inserted alleles. Our results indicate that transcription increases rates of all tested point mutations in the induced gene: induction significantly increased the individual rates of an A·T→T·A transversion, an A·T→G·C transition and the pooled rates of the three other point mutations assayed. Although the S.enterica genome is thought to have a mutational bias towards G·C base pairs, transitions creating A·T pairs were approximately 10 times more frequent than the reverse mutation, resulting in an overall mutation pressure to lower G+C contents. Transitions occurred at roughly twice the rate of transversions, similar to results from sequence comparisons; however, several individual transversions are more frequent than the least common transition.
INTRODUCTION
Spontaneous mutations, like those induced by mutagens, should become more frequent with increased transcription (1–3) because DNA becomes locally exposed during synthesis of RNA transcripts. In initial support of this supposition, reversions of an Escherichia coli trpA strain were found to be more common in a constitutive mutant (4). However, a similar transcription effect was not observed in a different strain, suggesting that the result was an artifact of the use of potentially misleading statistic—average mutants per culture—rather than more reliable alternatives. More robust studies have since shown that elevating transcription levels can influence varying types of mutations: for example, as transcription rates increase, frameshift reversions in yeast are more frequent (5,6), whereas chromosomal rearrangements and deletions in a human cell lines are less common (7).
Transcription is known to increase the spontaneous rates of certain point mutations in E.coli: C→T reversions in both plasmid-borne and chromosomal genes (8,9), and a T·A→G·C forward mutation at a mutation hotspot in an alternate plasmid-borne gene (10). In addition, transcription promotes the spontaneous formation of three forward mutations due to non-C→T point substitutions in E.coli cells situated in different sequence contexts (11). Although rates of point mutations were elevated by transcription in each of these cases, other mutations might be reduced or unaffected, because each substitution rate is set by mutagenic and anti-mutagenic processes that do not necessarily affect other substitutions. For example, the elevated C→T transition rates are attributable to the increased susceptibility of cytosines on single-stranded DNA to deamination (8,12), a process that does not significantly affect other mutations. If the rate of the predominant mutagenic processes for a particular mutation is not significantly altered by transcription, the antimutagenic effect of transcription-coupled repair (13,14) could overwhelm them and reduce mutations with increased expression (15), as appears to be the case in mammalian cells (7,16).
To investigate whether there are general effects of transcription on spontaneous base substitutions, we measured the rates of several point mutations under both induced and non-induced conditions. Our findings suggest that heightened transcription increases mutagenesis for all substitutions tested and that for this class of mutations, this effect always overrides that of transcription-coupled repair in bacteria.
Differences among point mutation rates can also affect the overall G+C content of a genome. For example, when C→T transitions occur at a higher rate than T→C transitions, A·T pairs will accumulate at neutral sites, such as non-coding DNA, and degenerate codon positions. Experimental data suggest a mutational bias towards A·T pairs in E.coli (17–21) making it difficult to explain the observed preponderance towards G·C base pairs at third codon positions (22). By measuring mutation rates for all possible point mutations, we found a highly significant bias towards the formation of A.T pairs among transitions, but no significant bias among transversions, which is consistent with experimental rather than comparative sequence data.
MATERIALS AND METHODS
Bacterial strains
Strains used in mutation assays contain mutant lacZ alleles inserted into the tre locus of the Salmonella enterica sv. Typhimurium LT2 chromosome (Table 1); reversion rates of these alleles at this locus have been determined previously to be similar to those of the same alleles inserted into other loci (23). Strains were constructed by moving lacZ alleles of E.coli strains CC101 through CC106 (24) into S.enterica TR10000, as described (23). To confirm that strains possessed the appropriate mutations, the central portion of each lacZ allele was amplified by the polymerase chain reaction and sequenced. To test whether the lac operons of these strains were inducible, revertants were isolated and plated with and without isopropyl-β-d-thiogalactoside (IPTG) on minimal glucose media (25) supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-gal).
Table 1. Effect of inducer on reversion rates.
| Strain | Reverts to G1A2G3 bya | Reversion rateb No IPTG | 250 µM IPTG | % increase (P value) |
|---|---|---|---|---|
| UB589 | G2·C→A2·T | 2.110 × 10–10 | 3.416 × 10–10 | 62% (0.26) |
| TT22914 | A1·T→G1·C | 0.149 × 10–10 | 0.384 × 10–10 | 156% (***) |
| UB609 | C2·G→A2·T | 0.954 × 10–10 | 1.181 × 10–10 | 24% (>0.50) |
| TT22910 | T1·A→G1·C | 0.878 × 10–10 | 1.292 × 10–10 | 47% (>0.50) |
| UB631 | T2·A→A2·T | 0.409 × 10–10 | 2.005 × 10–10 | 390% (****) |
| UB569 | C1·G→G1·C | 0.017 × 10–10 | n.a. | n.a. |
| Transitions | 1.130 × 10–10 | 1.900 × 10–10 | 68% (****)c | |
| Transversions testedd | 0.747 × 10–10 | 1.492 × 10–10 | 100% (****)c | |
| Totald | 0.900 × 10–10 | 1.656 × 10–10 | 86% (****)c |
aSequence on coding strand.
bEstimates of exponential-phase mutation rates in the presence and absence of IPTG. Statistical comparisons made by analyzing log-transformed data using the Tukey–Kramer test for a posteriori comparisons of means for each strain, and two-factor ANOVA of ln (µ) versus mutation type and IPTG presence/absence for groups of strains. ***, P < 0·001; ****, P < 0·0001; n.a., data not available because mutation rates were too low for accurate determination.
cSignificance of P values due primarily to TT22914 and/or UB631.
dEstimate does not include data for strain UB569.
Mutation assays
Mutation rates for all strains except UB569 were determined at two IPTG concentrations, 0 and 250 µM, for four or five batches of 10 to 52 cultures. The mutation rate for UB569 was found to be very low and therefore was not assayed in the presence of IPTG. For all strains, larger batches were used when a significant proportion of the cultures did not produce mutants. To make the estimates determined for different batches statistically independent, each batch was derived from a different colony. Cultures were initiated by adding ∼100 cells (20 µl of a 1:106 dilution from an overnight culture) to 20 ml of LB media and incubated at 37°C for 22 h with moderate shaking (160 r.p.m.). To enumerate total cells, serial dilutions of two to six cultures per batch were plated and counted on LB; to enumerate revertants, the cells of each culture were washed with 5 ml of M9 salts, resuspended into 0.4 ml of the same, and then plated onto minimal lactose (0.2%) plates supplemented with 100 µM IPTG and 20 µg/ml X-gal. Plates were pretreated with resuspensions of ∼109 cells of scavengers, S.enterica serovar Typhimurium strain 14028s, to reduce the survival of non-revertants. Blue colonies visible after 2 days incubation at 37°C were scored as revertants.
To test for differences in overall mutation rates among strains, we assayed for mutations resistant to 100 µg/ml rifampicin, using four batches per treatment of eighteen 0.3 ml cultures. Each culture was inoculated with ∼106 cells and incubated for 6.5 h at 37°C with shaking (200 r.p.m.). Three cultures per batch were serially diluted and plated for total cells, and the rest were spread directly onto LB–rifampicin plates.
Calculation of mutation rates
Mutants arising from exponential-phase mutations are expected to follow a Luria–Delbrück (LD) distribution (26), whereas those arising in stationary phase and after plating are expected to be Poisson distributed. Because we anticipate both exponential and post-exponential-phase mutations, we investigated the fit of combined Poisson–LD distributions to the data of each batch. Such distributions are characterized by two parameters: m, the overall number of mutations per culture, and θ, the fraction of mutations following the Poisson rather than the LD distribution. When compared to pure LD distributions, combined distributions can fit the data better even if no post-exponential-phase mutations actually occur because they employ an additional parameter. Therefore, our default assumption was that there were no post-exponential-phase mutations (θ = 0). However, if the best-fitting combined distribution had a likelihood 10-fold greater than that of the pure LD distribution, we applied its value of θ. To determine θ for each treatment—a given strain grown under a particular IPTG concentration—we averaged estimates derived from batches containing >36 cultures. (Estimates from smaller batches were ignored because they yielded much less accurate estimates.) The values of θ thus determined were then used in generating maximum likelihood estimates of overall (m) and exponential-phase (mLD = [1 – θ] × m) mutations per culture for each batch. Overall and exponential-phase mutation rates for each treatment were calculated using the following formula:
where mi and ni are the estimates for number of mutations and total cells per culture of batch i, respectively, N is the total number of batches and mi is replaced with mLD,i when calculating µLD.treatment. This equation was derived assuming, as justified previously, that both m and n follow a log-normal distribution with respect to batches, which yields a log-normal distribution for µ (24).
RESULTS
Effects of transcription on mutation rates
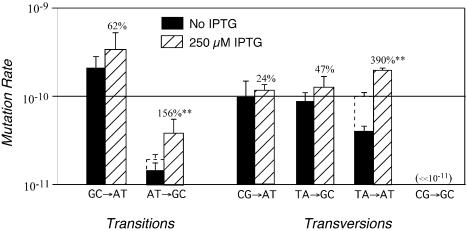
To determine the extent to which transcription affects particular point mutations, reversion rates were calculated for mutant lacZ alleles in the presence and absence of inducer (Fig. 1, Table 1). Exponential-phase and overall mutation rates were significantly higher in the presence of IPTG for the T·A→A·T transversion and the A·T→G·C transition. When the assumption that some mutations occurred after exponential-phase was relaxed, that is when θ was required to equal zero, some increases with induction were less pronounced: the rate of the T·A→A·T transversion rose 1.9- rather than 2.6-fold, and that of the A·T→G·C transition increased 2.0 rather than 4.9 times. However, both of the results remained statistically significant whether or not we included post-exponential-phase mutations. The three remaining mutations—the G·C→A·T, C·G→A·T and T·A→G·C reversions—were unaffected by the requirement that θ = 0. For these mutations, exponential-phase and overall rates were higher in the presence of IPTG, although individually none reached statistical significance. However, there is a statistically significant overall effect of induction in an analysis restricted to these mutations (P < 0·01, two-factor ANOVA).
Figure 1.
Exponential-phase reversion rates in the presence (hatched bars) or absence (solid bars) of IPTG, calculated under the assumptions that post-plating mutations did or did not occur. Rates calculated under these two assumptions were identical for all strains in the presence of IPTG but differed for strains TT22914 and UB631 in its absence. In these two cases, the gray extended regions of the solid bars reflect rates that include post-plating mutations.
To test whether IPTG, in addition to inducing lac expression, might cause a genome-wide increase in mutation rates, we also assayed mutations to rifampicin resistance in the presence and absence of IPTG (Table 2). None of the strains tested has a significantly different mutation rate to rifampicin resistance in the presence of IPTG nor was there a significant effect when the data from different strains were pooled, or subjected to an analysis of variance (P = 0·52) (Table 2).
Table 2. Effect of inducer on rates of mutations to rifampicin resistance.
| Strain | Mutation rate (n) No IPTG | 250 µM IPTG | ln (µ)a No IPTG | 250 µM IPTG | Change with IPTGb |
|---|---|---|---|---|---|
| TT22910 | 3.08 × 10–9 (4) | 3.55 × 10–9 (2) | –19.60 ± 0·39 | –19.46 ± 0·08 | +15% (0·99) |
| UB569 | 3.60 × 10–9 (5) | 3.14 × 10–9 (3) | –19.44 ± 0·14 | –19.58 ± 0·27 | –13% (0·99) |
| UB631 | 3.51 × 10–9 (4) | 3.62 × 10–9 (4) | –19.47 ± 0·30 | –19.44 ± 0·41 | +3% (0·99) |
| Pooled | 3.41 × 10–9 (13) | 3.44 × 10–9 (9) | –19.50 ± 0·29 | –19.49 ± 0·32 | +1% (0·90) |
aMean ± 95% confidence interval of ln (µ).
bComparisons between ln (mutation rate) in presence and absence of IPTG using Tukey–Kramer test for a posteriori comparisons of means for individual strains, and Student’s t-test for pooled data. P values in parentheses.
In the presence and absence of inducer, the relative frequencies of mutations (except T·A→A·T) were the same for both the exponential-phase and overall mutation rates: µG·C→A·T > µC·G→A·T ≈ µT·A→G·C > µA·T→G·C, where all differences were significant (Tukey–Kramer test for a posteriori comparisons of means, P < 0·05). The rate of the T·A→A·T exponential-phase mutation in the absence of inducer was significantly lower than those of the C·G→A·T and T·A→G·C mutations; however, in the presence of inducer, the rates of these three mutations were indistinguishable statistically. The C·G→G·C mutation rate was virtually undetectable in our system and, due to its extremely low frequency, was only measured under non-induced conditions.
DISCUSSION
The previously observed increases in spontaneous reversion rates of C→T transitions and a particular T·A→G·C transversion with transcription have both been attributed to specific mutagenic processes: cytosine deamination in the first case (8,12) and a mechanism involving local six-base pseudo-inverted repeats in the second (10). We have found that transcription also causes a highly significant increase in the rate of the T·A→A·T transversion and the A·T→G·C transition, mutations that cannot be attributed to deamination or to the presence of local sequence features. Our findings complement and extend previous work showing that spontaneous mutation rates of ble and kan gene constructs that mutate by the G·C→T·A transversion (and other non-C→T base substitutions) are elevated by transcription (11). Different spontaneous mutations were primarily affected in our system, and we were able to statistically test the effects of transcription on individual mutations. Moreover, by assaying chromosomal mutations in repair-proficient strains (as opposed to plasmid-borne mutations in uracil-DNA-glycosylase-deficient strain), we were able to investigate mutation rates that were lower, and more representative, of natural spontaneous mutations.
To test for a general effect of transcription on the remaining substitutions, we combined the reversion rates of the G·C→A·T, T·A→G·C and T·A→G·C mutations into a single ANOVA. Whereas the increases in the rates of these mutations did not reach statistical significance when analyzed individually, there is a statistically significant effect when these data are pooled. Cumulatively, this outcome, along with the results for other mutations in this and other studies, suggests that there are general mechanisms by which transcription augments rates of spontaneous point mutations.
The increase in mutation rates with gene expression is observed whether or not post-plating mutations were estimated to have occurred. In all but two of the cases, the frequency of post-plating mutations (θ) was calculated to be zero, arguing against the possibility that substantial numbers of mutants arose from plated cells sustained by nutrients derived from dying bacteria. However, the frequency of non-induced T·A→A·T transversions had a non-zero value of θ: the likelihood of the distribution assuming θ = 0.77 was over 700 times greater than that of the distribution where θ = 0, thus indicating that post-exponential-phase mutations were detectable. In this case, the presence of Poisson-distributed mutations likely reflects stationary-phase rather than post-plating mutations because the growth rate (under non-induced conditions) of the strain harboring this allele was among the highest of all strains tested, and cultures had entered stationary phase prior to plating.
In addition to inducing transcription of the lac operon, the addition of IPTG reduced the total number of cells by 23%; but it is not likely that this decrease contributes to the increased lacZ reversion rates for three reasons. (i) Control mutation rates tested at a non-induced locus did not increase (Table 2). (ii) Spontaneous reversion rates of lacZ alleles are not affected when S.enterica cells are stressed or starved (27). (iii) There is no significant negative correlation between total cell counts and mutation rates among batches of the same strain (r = –0.06, N = 44, P = 0.71).
The rates of both transitions investigated, the A·T→G·C and G·C→A·T mutations, were elevated by transcription, and in both cases, the pyrimidine, the base where the mutation was likely initiated, was situated on the transcribed strand. Although the transcribed strand spends less time than its counterpart in the vulnerable single-stranded state [and is therefore considered to be less prone to damage (8)], these results suggest that it also exhibits higher rates of damage during transcription. The most plausible explanation for the observed increase of multiple types of spontaneous substitutions is an elevation in rates of a variety of damages with transcription on both DNA strands, and not just those caused by deamination or errors generated by local repeats. For example, the increase in the forward mutation rates of the ble and kan gene constructs (11) are mainly attributable to more frequent G→T transversions on the non-transcribed strand caused by either oxidative damage or the hydrolysis of the glycosidic linkage of guanosine residues. Because neither of these processes, nor those already mentioned, can account for the significant increase in the rate of the T·A→A·T transversion detected in our system, this mutation must be due to yet another transcription-sensitive damage process. These results therefore imply that the increased susceptibility to damage due to the disassociation of the DNA strands normally overrides any reduction attributable to transcription-coupled repair.
Both chromosomal location and immediate sequence context are also known to affect rates of point mutations (23,28). In particular, substitutions at specific hot spots occur at significantly higher rates than those at other positions and can dominate mutational spectra (28). However, most mutable sites are not hotspots and have spontaneous mutation rates of similar magnitude, as observed for the 91 of 93 detectable sites in the rpsL gene of E.coli (28). This phenomenon, coupled with the observation that the reversion rates that we estimated are much lower than those typical of a mutational hotspot, suggests that the single-base substitutions we assayed are characteristic of other mutations in this class.
In our system, there was a significant cumulative bias towards the formation of A·T pairs, which would elevate the A+T content of the genome. This is due to the G·C→A·T transition, which occurs at 10-fold higher than the reverse mutation. This trend among transitions towards A·T base pairs has previously been observed in reversion-assay studies of the same alleles when borne on episomes (18,25,29) and on the E.coli chromosome (30). In contrast, there is no compositional bias among transversions because the rate of C·G→A·T is approximately equal to that of the reverse mutation, and the other transversions do not alter base composition.
It is also possible to deduce mutational biases from the spectrum of point mutations that inactivate a particular gene. Such studies provide complementary information to reversion assays because the same point mutation can be observed in numerous sequence positions and contexts. Although it is difficult to statistically compare mutation rates derived from such studies (because computing mutation rates would require the identification of all potential target sites), the trends detected among spectra of spontaneous mutations match those detected by reversion assays. For example, in studies employing the lacI, supF, rpsL and tonB genes of E.coli, C→T transitions occur at higher per-target frequencies and are more common than T→C transitions (Table 3). A similar A·T bias has also been observed in studies of human and yeast cells, and in a coliphage (31–34).
Table 3. Mutational spectra of enterobacterial genesa.
| Mutation | lacI M | T | M/T | rpsL M | T | M/T | supF M | T | M/T | tonB M | T | M/T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C·G→T·A | 182 | 78 | 2.3 | 111 | 27 | 4.1 | 6 | 2 | 3.0 | 8 | 3 | 2.7 |
| T·A→C·G | 43 | 29 | 1.5 | 61 | 21 | 2.9 | 1 | 1 | 1.0 | 0 | 0 | un |
| G·C→A·T | 36 | 74 | 0.5 | 89 | 22 | 4.0 | 19 | 8 | 2.4 | 0 | 3 | 0.0 |
| A·T→C·G | 55 | 43 | 1.3 | 12 | 13 | 0.9 | 2 | 1 | 2.0 | 3 | 3 | 1.0 |
| T·A→A·T | 42 | 53 | 0.8 | 17 | 13 | 1.3 | 3 | 1 | 3.0 | 6 | 4 | 1.5 |
| C·G→G·C | 10 | 50 | 0.2 | 31 | 13 | 2.4 | 14 | 5 | 2.8 | 0 | 0 | un |
| Transitions | 225 | 107 | 2.1 | 172 | 48 | 3.6 | 7 | 3 | 2.3 | 8 | 3 | 2.7 |
| Transversions | 148 | 220 | 0.7 | 149 | 61 | 2.4 | 38 | 15 | 2.5 | 9 | 10 | 0.9 |
| Total | 373 | 321 | 45 | 17 |
aColumns present numbers of mutants (M) isolated for the specified mutation, numbers of target sites (T) for each mutation and the fraction of mutants per target sites (M/T). Mutants were determined using wild type strains and target sites were identified using both mutant and wild type strains (although some targets may still escape detection). un, values undefined. Data compiled from: Schaaper and Dunn (19), Halliday and Glickman (20 and references therein) (for lacI), Mo et al. (38), Timms et al. (39), Yoshiyama et al. (40), Yoshiyama and Maki (10) (for rpsL), Akasaka et al. (21), Obata et al. (22) (for supF), Yamamura et al. (41) (for tonB).
We detected no significant bias for (or against) A·T pairs among transversions. Previous reversion assay studies yield similar results (18,25,29) (Table 3); however, the evidence from mutational spectra is contradictory; in the supF and rpsL genes, there appears to be a bias for A·T pairs, but in the lacI gene the opposite bias is observed (Table 3).
The spontaneous transitions assayed in this study occur at nearly twice the rate of transversions (Table 1). As observed previously for these lacZ alleles (18,25,29), ranges of the transition and transversion rates overlap. The C·G→G·C transversion occurs at a significantly lower rate than all other reversions examined and is also the least common point mutation detected in two of four mutational spectra (Table 3).
We have documented a general mutagenic effect of transcription on spontaneous point mutations in the induced gene and a mutational bias towards A·T pairs among spontaneous transitions. Though such results reflect and extend many of the trends observed in other experimental studies, both results conflict with previously held views about the mutational process in enteric bacteria based on the analysis of sequence data in two ways:
First, comparative analyses of E.coli and S.enterica homologs suggest that transcription reduces spontaneous point mutation rates (35,36) because synonymous substitution rates, after accounting for the effects of adaptive codon bias, decrease as indicators of transcription frequency increase. In contrast, our experimental data indicate that the rates of potentially all point mutations increase with transcription. Our results suggest that theoretical studies have overestimated the degree of selection acting on synonymous sites in highly expressed genes, thereby underestimating their mutation rates.
Second, the sequence compositions of E.coli and S.enterica genes suggest an intrinsic mutational bias against A·T pairs (23,37) because third codon positions, where most sites are under no selective constraints and should reflect the underlying mutational process, are G+C rich. It is possible that the A·T mutational bias observed in rapidly growing, aerobic, experimental populations is not representative of the mutational process in nature. Alternatively, these enteric species could be undergoing a shift in base composition that is not apparent from the cumulative measures of G+C contents at codon positions. Changes in the patterns of mutations within a species can be exposed by reconstructing the substitutions in a phylogenetic context, and such studies, as well as the analysis of mutational patterns under more natural growth conditions, will help reconcile the differences between the experimental and genomic measures of mutational bias.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank John Roth for support, advice and bacterial strains, and Frank Stewart and Phil Gerrish for providing computer programs. This research was supported by NIH grants GM55535 and GM56120 (to H.O.), and U.B. was supported by NIH grant GM27068 (to J.R.Roth).
REFERENCES
- 1.Brock R.D. (1971) Differential mutation of beta-galactosidase gene of Escherichia coli. Mutat. Res., 11, 181–186. [PubMed] [Google Scholar]
- 2.Savic D.J. and Kanazir,D.T. (1972) Effect of a histidine operator constitutive mutation on UV-induced mutability within histidine operon of Salmonella typhimurium. Mol. Gen. Genet., 118, 45–50. [DOI] [PubMed] [Google Scholar]
- 3.Davis B. (1989) Transcriptional bias: A non-Lamarckian mechanism for substrate-induced mutations. Proc. Natl Acad. Sci. USA, 86, 5005–5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balbinder E., Callahan,R., McCann,P.P., Cordaro,J.C., Weber,A.R., Smith,A.M. and Angelosanto,F. (1970) Regulatory mutants of the tryptophan operon of Salmonella typhimurium. Genetics, 66, 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Datta A. and Jinks-Robertson,S. (1995) Association of increased spontaneous mutation rates with high levels of transcription in yeast. Science, 268, 1616–1619. [DOI] [PubMed] [Google Scholar]
- 6.Morey N.J., Greene,C.N. and Jinks-Robertson,S. (2000) Genetic analysis of transcription-associated mutations in Saccharomyces cerevisiae. Genetics, 154, 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippert M.J., Chen,Q. and Liber,H.L. (1998) Increased transcription decreases the spontaneous mutation rate at the thymidine kinase locus in human cells. Mutat. Res., 401, 1–10. [DOI] [PubMed] [Google Scholar]
- 8.Beletskii A. and Bhagwat,A.S. (1996) Transcription-induced mutations: Increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc. Natl Acad. Sci. USA, 93, 13919–13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright B.E., Longacre,A. and Reimers,J.M. (1999) Hypermutation in derepressed operons of Escherichia coli K12. Proc. Natl Acad. Sci. USA, 96, 5089–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshiyama K. and Maki,H. (2003) Spontaneous hotspot mutations resistant to mismatch correction in Escherichia coli: Transcription-dependent mutagenesis involving template-switching mechanisms. J. Mol. Biol., 327, 7–18. [DOI] [PubMed] [Google Scholar]
- 11.Klapacz J. and Bhagwat,A.S. (2002) Transcription-dependent increase in multiple classes of base substitution mutations in Escherichia coli. J. Bacteriol., 184, 6866–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beletskii A. and Bhagwat,A.S. (1998) Correlation between transcription and C to T mutations in the non-transcribed DNA strand. J. Biol. Chem., 379, 549–551. [PubMed] [Google Scholar]
- 13.Selby C.P. and Sancar,A. (1993) Transcription-repair coupling and mutation frequency decline. J. Bacteriol., 175, 7509–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leadon S.A. (1999) Transcription-coupled repair of DNA damage: unanticipated players, unexpected complexities. Am. J. Hum. Genet., 64, 1259–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunala S. and Brash,D.E. (1992) Excision repair at individual bases of the Escherichia coli lacI gene: relation to mutation hot-spots and transcription coupling activity. Proc. Natl Acad. Sci. USA, 89, 11031–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vrieling H., van Zeeland,A.A. and Mullenders,L.H.F. (1998) Transcription coupled repair and its impact on mutagenesis. Mutat. Res., 400, 135–142. [DOI] [PubMed] [Google Scholar]
- 17.Hall B.G. (1991) Spectrum of mutations that occur under selective and non-selective conditions in E. coli. Genetica, 84, 73–76. [DOI] [PubMed] [Google Scholar]
- 18.Schaaper R.M. and Dunn,R.L. (1991) Spontaneous mutation in the Escherichia coli lacI gene. Genetics, 129, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halliday J.A. and Glickman,B.W. (1991) Mechanisms of spontaneous mutation in DNA repair-proficient Escherichia coli. Mutat. Res., 250, 55–71. [DOI] [PubMed] [Google Scholar]
- 20.Akasaka S., Takimoto,K. and Yamamoto,K. (1992) G:C→T:A and G:C→C:G mutations in the Escherichia coli supF gene: an improved lacZ(am) E. coli host designed for assaying pZ189 supF mutational specificity. Mol. Gen. Genet., 235, 173–178. [DOI] [PubMed] [Google Scholar]
- 21.Obata F., Nunoshiba,T., Hashimoto-Gotoh,T. and Yamamoto,K. (1998) An improved system for selection of forward mutations in an Escherichia coli supF gene carried by plasmids. J. Rad. Res., 39, 263–270. [DOI] [PubMed] [Google Scholar]
- 22.Muto A. and Osawa,S. (1987) The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl Acad. Sci. USA, 84, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hudson R.E., Bergthorsson,U., Roth,J.R. and Ochman,H. (2002) Effect of chromosome location on bacterial mutation rate. Mol. Biol. Evol., 19, 85–92. [DOI] [PubMed] [Google Scholar]
- 24.Cupples C.G. and Miller,J.H. (1989) A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA, 86, 5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J.H. (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 26.Luria S.E. and Delbrück,M. (1943) Mutations of bacteria from virus sensitivity to virus resistance. Genetics, 28, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes D. and Andersson,D.I. (1997) Carbon starvation of Salmonella typhimurium does not cause a general increase of mutation rates. J. Bacteriol., 179, 6688–6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maki H. (2002) Origins of spontaneous mutations: specificity and directionality of base-substitutions, frameshifts, and sequence-substitution mutageneses. Annu. Rev. Genet., 36, 279–303. [DOI] [PubMed] [Google Scholar]
- 29.MacKay W.J., Han,S. and Samson,L.D. (1994) DNA alkylation repair limits spontaneous base substitution mutations in Escherichia coli. J. Bacteriol., 176, 3224–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fijalkowska I.J., Jonczyk,P., Tkaczyk,M.M., Bialoskorska,M. and Schaaper,R.M. (1998) Unequal fidelity of leading strand and lagging strand DNA replication on the Escherichia coli chromosome. Proc. Natl Acad. Sci. USA, 95, 10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lichtenauer-Kaligis E.G.R., Thijssen,J., de Dulk,H., van de Putte,P., Tasseron-de Jong,J. and Giphart-Gassler,M. (1993) Genome wide spontaneous mutation in human cells determined by the spectrum of mutations in hprt cDNA genes. Mutagenesis, 8, 207–220. [DOI] [PubMed] [Google Scholar]
- 32.DeBoer J.G., Erfle,H., Walsh,D., Holcroft,J., Provost,J.S., Rogers,B., Tindall,K.R. and Glickman,B.W. (1997) Spectrum of spontaneous mutations in liver tissue of lacI transgenic mice. Environ. Mol. Mutagen., 30, 273–286. [PubMed] [Google Scholar]
- 33.Kunz B.A., Ramchandran,K. and Vonarx,E.J. (1998) DNA sequence analysis of spontaneous mutagenesis in Saccharomyces cerevisiae. Genetics, 148, 1491–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yatagi F. and Glickman,B.W. (1990) Specificity of spontaneous mutation in the lacI gene cloned into bacteriophage M13. Mutat. Res., 243, 21–28. [DOI] [PubMed] [Google Scholar]
- 35.Berg O.G. and Martelius,M. (1995) Synonymous substititon-rate constants in Escherichia coli and Salmonella typhimurium and their relationship to gene expression and selection pressure. J. Mol. Evol., 41, 449–456. [DOI] [PubMed] [Google Scholar]
- 36.Eyre-Walker A. and Bulmer,M. (1995) Synonymous substitution rates in enterobacteria. Genetics, 140, 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence J.G. and Ochman,H. (1997) Amelioration of bacterial genomics: rates of change and exchange. J. Mol. Evol., 44, 383–397. [DOI] [PubMed] [Google Scholar]
- 38.Mo J.-Y., Maki,H. and Sekiguchi,M. (1991) Mutational specificity of the dnaE173 mutator associated with a defect in the catalytic subunit of DNA Polymerase III of Escherichia coli. J. Mol. Biol., 222, 925–936. [DOI] [PubMed] [Google Scholar]
- 39.Timms A.R., Steingrimsdottir,H., Lehmann,A.R. and. Bridges,B.A. (1992) Mutant sequences in the rpsL gene of Escherichia coli B/r: mechanistic implications for spontaneous and ultraviolet light mutagenesis. Mol. Gen. Genet., 232, 89–96. [DOI] [PubMed] [Google Scholar]
- 40.Yoshiyama K., Higuchi,K., Matsumara,H. and Maki,H. (2001) Directionality of DNA replication fork movement strongly affects the generation of spontaneous mutations in Escherichia coli. J. Mol. Biol., 307, 1195–1206. [DOI] [PubMed] [Google Scholar]
- 41.Yamamura E., Nunoshiba,T., Kawata,M. and Yamamoto,K. (2000) Characterization of spontaneous mutation in the oxyR strain of Escherichia coli. Biochem. Biophys. Res. Commun., 279, 427–432. [DOI] [PubMed] [Google Scholar]