Abstract
The role of cysteine sulfhydryl residues on the RNA binding activity of human thymidylate synthase (TS) was investigated by mutating each cysteine residue on human TS to a corresponding alanine residue. Enzymatic activities of TS:C43A and TS:C210A mutant proteins were nearly identical to wild-type TS, while TS:C180A and TS:C199A mutants expressed >80% of wild-type enzyme activity. In contrast, TS:C195A was completely inactive. Mutant proteins, TS:C195A, TS:C199A and TS:C210A, retained RNA binding activity to nearly the same degree as wild-type human TS. RNA binding activity of TS:C43A was reduced by 30% when compared to wild-type TS, while TS:C180A was completely devoid of RNA binding activity. In vitro translation studies confirmed that mutant proteins TS:C43A, TS:C195A, TS:C199A and TS:C210A, significantly repressed human TS mRNA translation, while TS:C180A was unable to do so. To confirm the in vivo significance of the cysteine sulfhydryl residue, mutant proteins TS:C180A and TS:C195A were each expressed in human colon cancer HCT-C18:TS(–) cells that expressed a functionally inactive TS. A recombinant luciferase reporter gene under the control of a TS-response element was co-transfected into these same cells, and luciferase activity increased in the presence of the TS:C195A mutant TS protein to a level similar to that observed upon expression of wild-type TS protein. In contrast, luciferase activity remained unchanged in cells expressing the TS:C180A mutant protein. Taken together, these findings identify Cys-180 as a critical residue for the in vitro and in vivo translational regulatory effects of human TS.
INTRODUCTION
Thymidylate synthase (TS) is a folate-dependent enzyme that catalyzes the reductive methylation of 2′-deoxyuridine-5′-monophosphate (dUMP) by the reduced folate 5, 10-methylenetetrahydrofolate (CH2FH4) to yield thymidine-5′-monophosphate (dTMP, thymidylate) and dihydrofolate (1). Once synthesized, dTMP is phosphorylated within the cell by two successive enzymatic steps to dTTP, an essential precursor for DNA biosynthesis and DNA repair. Because this enzymatic reaction provides the sole intracellular de novo source of dTMP, TS is a critical therapeutic target in cancer chemotherapy (2,3).
In addition to its role in enzyme catalysis, TS also functions as an RNA binding protein (4–8). Specifically, translation of human TS mRNA is negatively regulated by direct binding of TS to two different cis-acting elements on its cognate mRNA. The first element is a 30-nt sequence contained within the 5′-untranslated region (UTR) and includes the translational start site in a stable stem–loop structure. The second binding site is a 70-nt sequence in the protein-coding region corresponding to nucleotides 480–550 (9). In vitro and in vivo studies have shown that each site can function independently of one another. However, both elements are required for the complete translational autoregulatory effects of TS.
There is growing evidence that the RNA binding activity of TS is affected by its state of ligand occupancy. When TS is ligand-free, maximal RNA binding activity is maintained, resulting in translational repression of TS mRNA. However, when TS is bound by either of its physiologic substrates, dUMP or CH2FH4, bound by the 5-FU metabolite FdUMP, or bound by an antifolate analog such as raltitrexed (ZD1694), RNA binding activity is markedly reduced. The net effect of ligand binding is abrogation of translational repression, resulting in synthesis of new TS protein.
Previous studies have shown that sulfhydryl group modification of TS significantly decreases enzymatic activity and its ability to form a stable ternary complex with the reduced folate CH2THF and FdUMP (10). Earlier work from our own laboratory showed that the redox state of the protein was a critical determinant of RNA binding (11). In the presence of reducing agents, such as 2-mercaptoethanol and/or dithiothreitol, the RNA binding activity of TS was significantly enhanced. In contrast, treatment with the oxidizing agent diamide or N-ethylmaleimide, significantly inhibited RNA binding activity. These studies suggested that the interaction between TS protein and its target TS mRNA was mediated by a reversible sulfhydryl switch mechanism and required the presence of at least one free sulfhydryl group.
In the present report, we investigated the role of the cysteine amino acid residues on the RNA binding activity of TS. For these studies, we used site-directed mutagenesis to express and purify mutant proteins corresponding to each cysteine moiety as well as RNA gel shift and in vitro translation assays to determine the RNA binding activity of these mutant proteins. The results presented herein demonstrate that Cys-180 of human TS plays a critical role in mediating RNA recognition.
MATERIALS AND METHODS
Cell culture
The human colon cancer HCT-C18 cell has been previously described (12,13) and was a kind gift of Dr Sondra Berger. HCT-C18 cells were maintained in RPMI 1640 growth medium containing 10% dialyzed fetal bovine serum and supplemented with 10 µM thymidine.
Synthesis of recombinant plasmid constructs
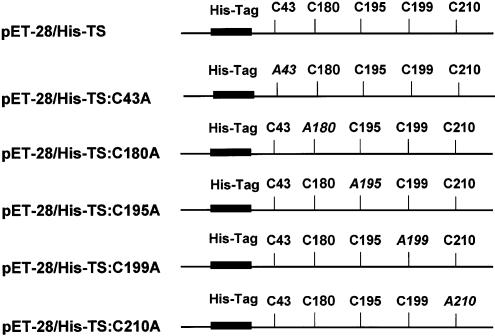
Full-length human TS cDNA was PCR-amplified and cloned into the HindIII and NdeI restriction sites of pET-28 to yield the His-Tag TS expression plasmid pET-28/His-TS. All point mutations were performed using the Quikchange site-directed mutagenesis kit (Strategene, La Jolla, CA) according to the manufacturer’s protocol. The five mutant TS expression plasmids constructed are presented in Figure 1, and each cysteine residue was mutated to a corresponding alanine. The mutant constructs are as follows: pET-28/His-TS:C43A, pET-28/His-TS:C180A, pET-28/His-TS:C195A, pET-28/His-TS: C199A and pET-28/His-TS:C210A. Recombinant plasmids pcDNA3.1(+):His-TS/C179A and pcDNA3.1(+):His-TS/C195A, which contain point mutations at Cys-180 and Cys-195, respectively, were also constructed using the site-directed mutagenesis technique. The sequences of the primers used are as follows (underlined bases represent the cysteine to alanine mutation): TS208–234 (sense), 5′-CAC ATC CTC CGC GCC GGC GTC AGG AAG-3′; TS234–208 (antisense), 5′-CTT CCT GAC GCC GGC GCG GAG GAT GTG-3′; TS619–645 (sense), 5′-AGA ATC ATC ATG GCC GCT TGG AAT CCA-3′; TS645–619 (antisense), 5′-TGG ATT CCA AGC GGC CAT GAT GAC TCT-3′; TS664–690 (sense), 5′-GCG CTG CCT CCA GCC CAT GCC CTC TGC-3′; TS-690–664 (antisense), 5′-GCA GAG GGC ATG GGC TGG AGG CAG CGC-3′; TS676–702 (sense), 5′-TGC CAT GCC CTC GCC CAG TTC TAT GTG-3′; TS702–676 (antisense), 5′-CAC ATA GAA CTG GGC GAG GGC ATC GCA-3′; TS709–735 (sense), 5′-AGT GAG CTG TCC GCC CAG CTG TAC CAG-3′; TS735–709 (antisense), 5′-CTG GTA CAG CTG GGC GGA CAG CTC ACT-3′
Figure 1.
Schematic diagram of expression plasmids of wild-type and mutant TS proteins. Full-length human TS cDNA was PCR-amplified, and the sequence was cloned in the HindIII and NdeI restriction site of pET-28 to yield the recombinant plasmid pET-28/His-Tag TS. Cysteine to alanine mutations of TS were constructed using the Stratagene Quikchange site- mutagenesis kit as detailed in the Materials and Methods. The amino acids in italics represent the cysteine to alanine mutation. The solid box indicates the His-Tag sequence at the N-terminal region of TS.
Expression and purification of wild-type and mutant human TS protein
Human recombinant His-Tag TS protein was purified using the Ni-NTA spin kit (Qiagen, Valencia, CA) according to previously described methods (14). In brief, Escherichia coli BL21 competent cells (Invitrogen, Carlsbad, CA) were transformed with recombinant wild-type or mutant human TS cDNA plasmids and grown overnight at 37°C. Cells were placed in fresh medium at a dilution of 1:10, grown to an absorbance of 0.6–0.8 at 600 nm and then induced with 1 mM IPTG at 30°C for 12 h. Cells were harvested by centrifugation at 30 000 g and cell pellets were suspended in a binding solution containing pH 8.0, 50 mM sodium phosphate, 10 mM imidazole and sonicated six times for 30 s using a Vibracell Model VC-600 sonicator (Sonics and Materials Inc., Danbury, CT). The sonicated cells were centrifuged for 20 min at 43 000 g at 4°C. The supernatant was then loaded onto a Ni-NTA spin column, and the column was washed with 20 mM imidazole, and 50 mM sodium phosphate buffer three times. His-Tag TS proteins were eluted with a buffer of 300 mM imidazole, 50 mM sodium phosphate, pH 8.0, and protein samples were dialyzed in 100 mM, Tris–HCl, pH 7.4.
In vitro transcription
Full-length human TS mRNA (TS1-1524) was synthesized in vitro with SP6 RNA polymerase using linearized pcEHTS plasmid as template. In vitro transcription was performed using the in vitro transcription kit (Ambion, Austin, TX), according to previously described methods (9). Once synthesized, all RNA transcripts were resolved on a 15% polyacrylamide/8 M urea gel. The concentration of RNA was then determined by UV absorbance at 260 nm.
32P-radiolabeled human TS mRNA was synthesized in vitro using the Promega in vitro transcription kit as previously described (4,5). The radiolabeled RNA was resolved on a 15% polyacrylamide/8 M urea gel and subsequently gel-purified to ensure integrity and purity.
RNA gel-mobility shift assay
The RNA gel-mobility assay was performed according to previously published methods (4,5). In brief, 32P-radiolabeled human TS mRNA (100 000 c.p.m., 1–2 fmol) was incubated with human TS protein in a reaction mixture containing 10 mM HEPES, pH 7.4, 40 mM KCI, 3 mM MgCI2, 5% glycerol, 250 mM 2-mercaptoethanol and 2 U of Prime RNase inhibitor, for 15 min at room temperature. RNase T1 (12 U, Eppendorf) was then added for 10 min, followed by incubation with heparin sulfate (5 mg/ml, Sigma) for an additional 10 min at room temperature. The entire reaction mixture was then resolved on a 5% non-denaturing polyacrylamide gel (acrylamide/methylenebisacrylamide weight ratio, 60:1), transferred to Whatman filter paper, dried, and then visualized by autoradiography. Quantitation was performed using a Hewlett Packard ScanJet 4P Plus Scanner and NIH Image 1.51 software. The apparent dissociation constants (Kd) were determined by Scatchard analysis as previously described (11,15).
In vitro translation
In vitro translation reactions were performed using a rabbit reticulocyte lysate system (Promega, Madison, MI) as previously outlined (4,9). Reaction mixtures (final volume, 20 µl) containing rabbit reticulocyte lysate, amino acid mixture without methionine (0.4 µl), RNase inhibitor (0.4 µl), and 12 µCi of 35S-methionine were incubated with the respective mRNA transcript at 30°C for 1 h. Translation products were analyzed by SDS–PAGE (15% acrylamide) according to the method of Laemmli (16) and gels were processed as previously described (5). After drying for 2 h, the translation products were visualized by autoradiography.
Transient transfection and luciferase assay
Transient transfection experiments were performed as previously described (17). In brief, cells were plated at 2 × 105/ 60 mm dish and grown to ∼50% confluence. In each transfection experiment, cells were incubated with either 5 µg of pcDNA3.1(+):His-TS, pcDNA3.1(+):His-TS/C195A or pcDNA3.1(+):His-TS/C180A along with 5 µg of p644 or p644/TS:N25. The p644 and p644/TS:N25 recombinant plasmids have been previously characterized (18). Cells were co-transfected with 0.05 µg of ppRL-SV40 plasmid DNA. This plasmid encodes renilla luciferase, and was included to provide an internal control for transfection efficiency. After incubation at 37°C for 24 h, cells were washed twice with serum-free RPMI 1640 medium, and then incubated in 2 ml RPMI 1640 containing 10% dialyzed fetal bovine serum for an additional 48 h at 37°C. Cells were harvested using reagents from the dual luciferase assay kit (Promega, Madison, MI), as previously described (9).
TS catalytic assay
The catalytic activity of wild-type and mutant TS proteins was determined as previously outlined (19). This assay was performed in a total volume of 200 µl containing 10–5 M [5-3H] dUMP (specific activity, 20 Ci/mmol), 100 mM 2-mercaptoethanol, 50 mM KH2PO4, pH 7.2, 150 µM CH2THF and 0.5 µg TS protein. Each reaction mixture was incubated at 37°C for 30 min. The reaction was terminated upon addition of 100 µl of ice-cold 20% trichloroacetic acid. Residual [5-3H] dUMP was removed by adding 200 µl of an albumin-coated activated charcoal solution. Samples were vortexed and allowed to stand at room temperature for 10 min. The charcoal was removed by centrifugation at 10 000 g for 30 min. A 250 µl sample of the supernatant was then assayed for [3H]H2O radioactivity by liquid scintillation counting.
RESULTS
As a first step towards investigating the potential effect of the sulfhydryl group on the RNA binding activity of TS, each cysteine residue on TS protein was mutated to a corresponding alanine residue. Wild-type and mutant TS proteins were then expressed and purified to homogeneity. We first determined whether mutation of any one of the cysteine residues resulted in loss of catalytic activity, using the well-characterized TS catalytic assay. As seen in Table 1, mutant proteins TS:C43A and TS:C210A expressed completely intact enzymatic activity when compared to wild-type TS protein. TS:C180A and TS:C199A mutants expressed >80% of wild-type enzyme activity. In contrast, TS:C195A was catalytically inactive. These studies confirmed that the cysteine amino acid residue at position 195, which resides in the nucleotide-binding domain (20), is essential for TS catalytic activity.
Table 1. Enzyme activity of wild-type and mutant human TS proteins.
| Protein | Specific activity (U/mg) |
|---|---|
| Wild-type TS | 0.50 |
| TS:C43A | 0.50 |
| TS:C180A | 0.40 |
| TS:C195A | 0.00 |
| TS:C199A | 0.45 |
| TS:C210A | 0.50 |
The catalytic activity of wild-type and mutant human TS proteins was determined using the radioenzymatic assay as outlined in the Materials and Methods section. One unit of activity is defined as 1 µmol of thymidylate formed per min at 37°C under the conditions of the assay.
We next performed a series of RNA gel-mobility shift experiments to determine the effect of the respective C→A mutations on RNA binding activity. RNA binding experiments confirmed that three of the mutant proteins, TS:C43A (Fig. 2, lane 3), TS:C195A (Fig. 2, lane 5) and TS:C210A (Fig. 2, lane 6), interacted with human TS mRNA probe to the same degree as wild-type TS (Fig. 2, lane 2). The binding activity of TS:C199A was reduced by nearly 30% when compared with wild-type TS (Fig. 2, lane 6). Of note, the RNA binding activity of the TS:C180A protein was completely abrogated (Fig. 2, lane 4).
Figure 2.
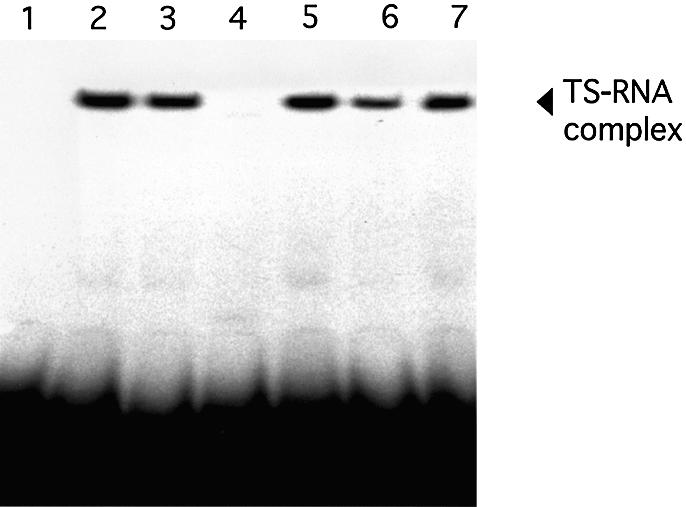
RNA binding activity of wild-type and mutant human TS proteins. RNA gel mobility-shift assays were performed as described in the Materials and Methods. Samples were resolved on a 5% non-denaturing acrylamide gel. 32P-radiolabeled full-length human TS mRNA probe was incubated in the absence (lane 1) or presence of wild-type TS (lane 2), TS:C43A (lane 3), TS:C180A (lane 4), TS:C195A (lane 5), TS:C199A (lane 6) and TS:C210A (lane 7). The arrow indicates the position of the RNP complex as visualized by autoradiography.
The binding affinities of wild-type and mutant TS proteins, as measured by the apparent dissociation constants (Kd), were then determined using the well-established Scatchard analysis. As seen in Table 2, mutant proteins TS:C43A (Kd, 2.8 nM), TS:C195A (Kd, 3.0 nM) and TS:C210A (Kd, 2.9 nM) bound to TS mRNA with an affinity nearly identical to that of wild-type TS (Kd, 2.9 nM). The TS:C199A mutant protein displayed partially decreased RNA binding affinity by ∼30% when compared to wild-type TS (Kd, 4.3 nM). In contrast, the binding affinity of TS:C180A was dramatically reduced by >100-fold (Kd, > 300 nM).
Table 2. RNA binding activity of wild-type and mutant TS proteins.
| Protein | Kd (nM) |
|---|---|
| Wild-type TS | 2.9 ± 0.50 |
| TS:C43A | 2.8 ± 0.44 |
| TS:C180A | >300 |
| TS:C195A | 3.0 ± 0.45 |
| TS:C199A | 4.3 ± 0.65 |
| TS:C210A | 2.9 ± 0.49 |
The dissociation constant (Kd) of wild-type and mutant human TS proteins was determined using the Scatchard analysis as outlined in the Materials and Methods. Each value represents the mean ± SE of three to five experiments.
To begin to determine the functional consequences of mutations of the cysteine residues, we employed a rabbit reticulocyte lysate in vitro translation system to investigate their effects on the translational repressive activity of TS. When human TS mRNA was incubated in the lysate reaction mixture, the corresponding in vitro translation products were observed at 37 kDa for TS (Fig. 3, lane 1). In the presence of exogenous wild-type TS, dose-dependent inhibition of TS mRNA translation was observed (Fig. 3, wild-type TS). The same level of translational repression was also observed when human TS mRNA was incubated with mutant proteins, TS:C43A, TS:C195A, TS:C199A and TS:C210A (Fig. 3). In contrast, the translational inhibitory effect was nearly completely abrogated when TS:C180A was included in the rabbit lysate reaction (Fig. 3). These studies suggest that Cys-180 is required to maintain the translational repressive effects of TS protein in vitro.
Figure 3.
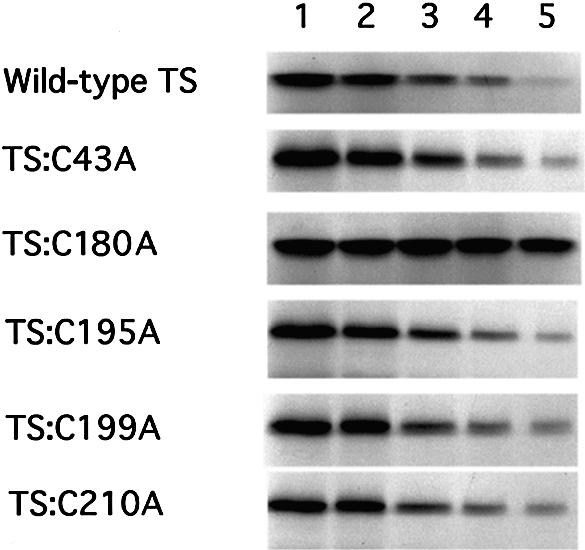
Inhibition of human TS mRNA translation in vitro by wild-type and mutant human TS proteins. In vitro translation reactions containing rabbit lysate were incubated with human TS mRNA (0.4 pmol, lanes 1–5) as outlined in the Materials and Methods. Varying amounts of TS protein, lanes 1–5: 0, 10, 20, 30 and 40 pmol, respectively, were included in each reaction. After incubation at 30°C for 1 h, samples were resolved on 12.5% SDS–PAGE, and protein products were visualized by autoradiography.
We next investigated the potential in vivo biological significance of the cysteine residue. For this series of experiments, we used human colon cancer HCT-C18 cells which express a functionally inactive TS and the p644/TS:N25 luciferase reporter construct. In previously published studies, we had used an in vitro RNA selection method in which a completely degenerate, linear RNA pool of 25 nt was incubated with human recombinant TS protein. The TS:N25 sequence was isolated after 10 rounds of selection and amplification, and in vitro RNA binding studies revealed that this selected sequence bound human recombinant TS protein with nearly 20-fold higher affinity than native, wild-type TS RNA sequences (18). The cDNA sequence corresponding to TS:N25 RNA was then cloned onto the upstream of luciferase gene to yield the heterologous luciferase reporter plasmid, p644/TS:N25. In vivo transfection experiments confirmed that the TS:N25 sequence functioned as a positive enhancer element in that luciferase activity was significantly increased by nearly 3-fold in human colon cancer HCT-C18-TS(+) cells which overexpress human TS, but not in HCT-C18 cells, which express a functionally inactive TS. RNase protection assays revealed that the significant changes in luciferase activity was not associated with corresponding changes in luciferase mRNA levels. These findings suggested that the biological effects of the selected TS:N25 sequence were mediated at the translational level.
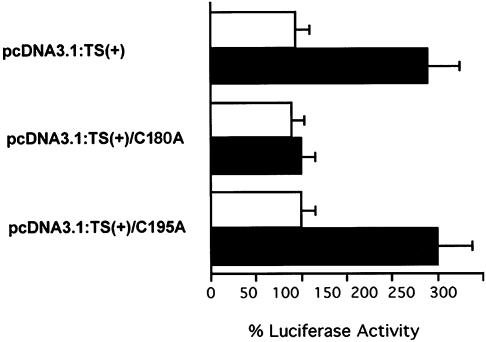
In the present study, we co-transfected HCT-C18 cells with either parent luciferase plasmid p644 or the p644/TS:N25 heterologous plasmid along with a plasmid expressing either wild-type TS [pcDNA3.1(+):His-TS] or plasmids expressing mutant TS:C180A [pcDNA3.1(+):His-TS/C180A] or TS: C195A TS proteins [pcDNA3.1(+):His-TS/C195A]. As seen in Figure 4, luciferase activity was increased by nearly 2.5-fold in cells expressing either wild-type TS or TS:C195A mutant protein. In contrast, luciferase activity remained unchanged in cells expressing the TS:C180A mutant TS protein following transfection with p644/TS:N25 when compared to transfection with p644.
Figure 4.
Effect of cysteine mutation on the luciferase activity mediated by the enhancer TS-response element. Human colon cancer HCT-C18 cells were transiently transfected with p644 (open bar) or p644/TS:N25 (solid bar). Three different plasmids, including pcDNA3.1:TS(+), pcDNA3.1: TS(+)/C180A or pcDNA3.1:TS(+)/C195A were co-transfected with p644 or p644/TS:N25. Luciferase activity was measured 48 h post-transfection using the dual luciferase assay system as described in the Materials and Methods. The activity in cells transfected with p644 was defined as 100%.
DISCUSSION
In the present study, we performed a series of experiments to identify the amino acid residues on human TS protein that are required for RNA recognition. For this work, we used a site-directed mutagenesis strategy to express and purify mutant TS proteins. In this initial series of experiments, we focused our efforts on expressing mutant proteins with mutations at the critical cysteine residues, of which there are five located at amino acid residues 43, 180, 195, 199 and 210. The rationale for focusing on these particular residues was based on earlier work from our laboratory that demonstrated that the redox state of the protein was a critical determinant of RNA binding (11). In the presence of a reducing environment with either 2-ME or DTT, the RNA binding activity of human TS was significantly enhanced. However, treatment of human TS with either of the oxidizing agents diamide or N-ethylmaleimide, significantly repressed RNA binding activity. These studies suggested that the interaction between TS protein and its target TS mRNA was mediated by a reversible sulfhydryl switch mechanism and involved at least one cysteine amino acid residue. However, this initial work was unable to identify the specific cysteine residue responsible for this effect.
Our findings demonstrate that mutations at cysteine residues 43, 195, 199 and 210 do not impair the RNA binding activity of TS nor do these mutations appear to significantly alter the translational repressive effects of the protein. In contrast, the TS:C180A mutant protein, which retains nearly complete catalytic activity when compared to wild-type TS, is completely devoid of RNA binding activity and is unable to inhibit TS mRNA translation. The fact that a catalytically active TS, such as TS:C180A, has lost RNA binding function and that a catalytically inactive TS, such as TS:C195A, is able to maintain RNA binding activity provides additional evidence that the functions of RNA binding and enzyme catalysis are not mediated by the same domain(s) on the TS protein. To provide further support for the biological effects of this specific mutation, in vivo transfection experiments were performed. As seen in Figure 4, transient transfection experiments using a reporter luciferase construct, under the control of an enhancer TS-response element, revealed a marked increase in luciferase activity in cells expressing wild-type TS or TS:C195A mutant protein. In contrast, luciferase activity remained unchanged in cells expressing the TS:C180A mutant TS protein. Taken together, these findings highlight the critical role of Cys-180 in mediating the translational regulatory function of human TS in vitro and in vivo.
Several studies have focused on characterizing the molecular elements that mediate the interaction between RNA binding proteins and their target RNA. The R17 bacteriophage coat protein (21–23), aminoacyl-tRNA synthetase (24,25) and iron-responsive factor (26,27) represent three well-characterized RNA binding proteins. In each instance, a free cysteine sulfhydryl group(s) on the RNA binding protein forms a covalent Michael adduct with the C-6 position on the corresponding uracil ring of the target RNA. In the present case, it is clear that the sulfhydryl residue at Cys-180 is critical for RNA binding. However, the precise mechanism by which this specific amino acid moiety mediates its effects on RNA binding remains unclear. There are, at least, three potential mechanisms by which it might be exerting its effects. First, the cysteine sulfhydryl may form a direct Michael adduct with the C-6 position of a uracil ring on TS mRNA. Second, occupation of the cysteine residue may result in altered RNA binding through a steric hindrance mechanism. Finally, this cysteine may be critical in maintaining TS in a certain conformational structure that then allows the actual domain on the protein to be readily accessible for RNA binding.
Previous work from this laboratory revealed that E.coli TS proteins with point mutations in the nucleotide-binding domain were able to retain their RNA binding function (11). The only mutant protein that lost RNA binding activity was the one at Cys-146 (C146S). Of note, for the E.coli species this represents the active site cysteine residue. In contrast, RNA binding activity was completely abrogated in mutant proteins with point mutations in the folate-binding region of E.coli TS, including the C50F mutant protein (11). These initial studies suggested that the active site cysteine sulfyhydryl and the folate-binding domain may play an important role in the RNA binding activity of E.coli TS. Our findings with the human TS species differ somewhat from those obtained with the E.coli protein, in that a mutation at the active site cysteine of human TS did not impair the ability of the mutant protein to interact with its own cognate TS mRNA. In addition, a mutation in the cysteine residue of human TS at position 43, which is located in the folate-binding pocket, did not alter RNA binding function. The crystal structures of E.coli (28–30) and human TS have been resolved (31,32), and the evidence, to date, suggests that the structures of these two species of TS may indeed be different. Taken together, our RNA binding studies would suggest that the main effect of the cysteine sulfhydryl residue is on maintaining the TS protein in a proper conformation that then allows for RNA binding to occur. However, to more directly address this issue, studies are underway in our laboratory to resolve the crystal structure of the human TS protein–human TS mRNA complex.
Further studies are needed to more carefully elucidate the key molecular elements underlying the translational autoregulation of TS and the interaction between TS protein and its target mRNA. Our laboratory is presently dissecting the critical cis-elements on TS mRNA that are required for in vitro and in vivo RNA recognition. Studies are also in progress to determine the precise intracellular localization of the TS RNP complex in intact human colon cancer cells. Such work may help to further elucidate the biological significance of this particular RNP complex. In addition, mutant proteins with point mutations in the folate- and nucleotide-binding domain of human TS are being evaluated to determine their relative RNA binding activity. These studies should provide further insights as to the critical domain(s) on human TS that are required for the process of RNA binding. However, the work presented herein provides additional insights into the determinants that mediate the TS translational autoregulation. Finally, as the process of translational autoregulation is becoming an increasingly recognized mechanism for the control of gene expression, these studies may provide new insights that can be applied to the regulation of other critical cellular genes.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank the Yale Cancer Center and the VACT Cancer Center for their continued support of this research. This work was supported in part by grants from the National Cancer Institute (CA75712 and CA16359) to E.C.
REFERENCES
- 1.Hardy L.W., Finer-Moore,J.S., Montfort,W.R., Jones,M.O., Santi,D.V. and Stroud,R.M. (1987) Atomic structure of thymidylate synthase: target for rational drug design. Science, 235, 448–455. [DOI] [PubMed] [Google Scholar]
- 2.Danenberg P.V. (1977) Thymidylate synthase: a target enzyme in cancer chemotherapy. Biochim. Biophys Acta, 473, 73–79. [DOI] [PubMed] [Google Scholar]
- 3.Carreras C. and Santi,D.V. (1995) The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem., 64, 721–762. [DOI] [PubMed] [Google Scholar]
- 4.Chu E., Koeller,D.M., Casey,J.L., Drake,J.C., Chabner,B.A., Elwood,P.C., Zinn,S. and Allegra,C.J. (1991) Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl Acad. Sci. USA, 88, 8977–8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu E., Voeller,D., Koeller,D.M., Drake,J.C., Takimoto,C.H., Maley,G.F., Maley,F. and Allegra,C.J. (1993) Identification of an RNA binding site for human thymidylate synthase. Proc. Natl Acad. Sci. USA, 90, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu E., Voeller,D.M., Jones,K.L., Takechi,T., Maley,G.F., Maley,F., Segal,S. and Allegra,C.J. (1994) Identification of a thymidylate synthase ribonucleoprotein complex in human colon cancer cells. Mol. Cell. Biol., 14, 207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voeller D.M., Changchien,L.M., Maley,G.F., Maley,F., Takechi,T., Turner,R.E., Montfort,W.R., Allegra,C.J. and Chu,E. (1995) Characterization of a specific interaction between Escherichia coli thymidylate synthase and Escherichia coli thymidylate synthase. Nucleic Acids Res., 23, 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu E. and Allegra,C.J. (1996) The role of thymidylate synthase as an RNA binding protein. BioEssays, 18, 191–198. [DOI] [PubMed] [Google Scholar]
- 9.Lin X., Voeller,D.M., Allegra,C.J., Maley,F. and Chu,E. (2000) Characterization of a cis-acting regulatory element in the protein coding region of thymidylate synthase mRNA. Nucleic Acids Res., 28, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plese P.C. and Dunlap,R.B. (1977) Sulfhydryl group modification of thymidylate synthetase and its effect on activity and ternary complex formation. J. Biol. Chem., 252, 6139–6144. [PubMed] [Google Scholar]
- 11.Chu E., Voeller,D.M., Morrison,P.F., Jones,K.L., Takechi,T., Maley,G.F., Maley,M.F. and Allegra,C.J. (1994) The effect of reducing reagents on binding of thymidylate synthase protein to thymidylate synthase messenger RNA. J. Biol. Chem., 269, 20289–20293. [PubMed] [Google Scholar]
- 12.Hoganson D.K., Williams,A.W. and Berger,S.H. (1999) Isolation and characterization of a thymidylate synthase-deficient human colon tumor cell line. Biochem. Pharmacol., 58, 1529–1537. [DOI] [PubMed] [Google Scholar]
- 13.Ju J.F., Pedersen-lane,J., Maley F. and Chu,E. (1999) Regulation of p53 expression by thymidylate synthase. Proc. Natl Acad. Sci. USA, 96, 3769–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedersen-Lane J., Maley,G.F., Chu,E. and Maley,F. (1997) High-level expression of human thymidylate synthase. Protein Expr. Purif., 10, 256–262. [DOI] [PubMed] [Google Scholar]
- 15.Liebold E. and Munro,H.N. (1988) Structural requirements of iron-responsive elements for binding of the protein involved in both transferrin receptor and ferritin mRNA post-transcriptional regulation. Proc. Natl Acad. Sci. USA, 85, 2171–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriphage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 17.Felgner P.L., Gadek,T.R., Holm,M., Roman,R., Chan,H.W., Wenz,M., Northrop,J.P., Ringold,G.M. and Danielsen,M. (1987) Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl Acad. Sci. USA, 84, 7413–7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin X., Mizunuma,N., Chen,T.M., Copur,S.M., Maley,G.F., Liu,J., Maley,F. and Chu E. (2000) In vitro selection of an RNA sequence that interacts with high affinity with thymidylate synthase. Nucleic Acids Res., 28, 1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu E., Lai,G.-M., Zinn,S. and Allegra,C.J. (1990) Resistance of a human ovarian cancer line to 5-fluorouracil associated with decreased levels of 5-fluorouracil in DNA. Mol. Pharmacol., 38, 410–417. [PubMed] [Google Scholar]
- 20.Dunlap R.B., Harding,N.G.L. and Huennekens,F.M. (1971) Thymidylate synthase and its relationship to dihydrofolate reductase. Ann. N. Y. Acad. Sci., 186, 153–165. [DOI] [PubMed] [Google Scholar]
- 21.Bernardi A. and Spahr,P.F. (1972) Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc. Natl Acad. Sci. USA, 69, 3033–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carey J. and Uhlenbeck,O.C. (1983) Sequence-specific interaction of R17 coat protein-ribonucleic acid interaction. Biochemistry, 22, 2601–2615. [DOI] [PubMed] [Google Scholar]
- 23.Romaniuk P.J. and Uhlenbeck,O.C. (1985) Nucleoside and nucleotide inactivation of the R-17 coat protein: evidence for the transient covalent RNA-protein bond. Biochemistry, 24, 4239–4244. [DOI] [PubMed] [Google Scholar]
- 24.Koontz S.W. and Schimmel,P.R. (1979) Aminoacyl-tRNA synthetase-catalyzed cleavage of the glycosidic bond of 5-halogenated uridines. J. Biol. Chem., 254, 12277–12284. [PubMed] [Google Scholar]
- 25.Starzyk R.M., Koontz,S.W. and Schimmel,P.R. (1982) A covalent adduct between the uracil ring and the active site of an aminoacyl tRNA synthetase. Nature, 298, 136–140. [DOI] [PubMed] [Google Scholar]
- 26.Klausner R.D., Rouault,T.A. and Harford,J.B. (1993) Regulating the fate of mRNA: the control of cellular iron metabolism. Cell, 72, 19–28. [DOI] [PubMed] [Google Scholar]
- 27.Hentze M.W., Rouault,T.A., Harford,J.B. and Klausner,R.D. (1989) Oxidation-reduction and the molecular mechanism of a regulatory RNA-protein interaction. Science, 244, 357–359. [DOI] [PubMed] [Google Scholar]
- 28.Matthews D.A., Appelt,K., Oatley,S.J. and Xuong,N.H. (1990) Crystal structure of Escherichia coli thymidylate synthase containing bound 5-fluoro-2′-deoxyuridylate and 10-propargyl-5, 8,-dideazafolate. J. Mol. Biol., 214, 937–948. [DOI] [PubMed] [Google Scholar]
- 29.Montfort W.R., Perry,K.M., Fauman,E.B., Finer-Moore,J.S., Maley,G.F., Hardy,L., Maley,F. and Stroud,R.M. (1990) Structure, multiple site binding and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate. Biochemistry, 29, 6964–6977. [DOI] [PubMed] [Google Scholar]
- 30.Hyatt D.C., Maley,F. and Montfort,W.R. (1977) Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate. Biochemistry, 36, 4584–4594. [DOI] [PubMed] [Google Scholar]
- 31.Phan J., Koli,S., Minor,W., Dunlap,R.B., Berger,S.H. and Lebioda,L.(2001) Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry, 40, 1897–1902. [DOI] [PubMed] [Google Scholar]
- 32.Almog R., Waddling,C.A., Maley,F., Maley,G.F. and Van Roey,P. (2001) Crystal structure of a deletion mutant of human thymidylate synthase delta(7–29) and its ternary complex with tomudex and dUMP. Protein Sci., 10, 988–996. [DOI] [PMC free article] [PubMed] [Google Scholar]