Abstract
In the archaeal RNA polymerase and the eukaryotic RNA polymerase II, two subunits (E/F and RPB4/RPB7, respectively) form a heterodimer that reversibly associates with the core of the enzyme. Recently it has emerged that this heterodimer also has a counterpart in the other eukaryotic RNA polymerases: in particular two subunits of RNA polymerase I (A14 and A43) display genetic and biochemical characteristics that are similar to those of the RPB4 and RPB7 subunits, despite the fact that only A43 shows some sequence homology to RPB7. We demonstrate that the sequence of A14 strongly suggests the presence of a HRDC domain, a motif that is found at the C-terminus of a number of helicases and RNases. The same motif is also seen in the structure of the F subunit, suggesting a structural link between A14 and the RPB4/C17/subunit F family, even in the absence of direct sequence homology. We show that it is possible to co-express and co-purify large amounts of the recombinant A14/A43 heterodimer, indicating a tight and specific interaction between the two subunits. To shed light on the function of the heterodimer, we performed gel mobility shift assays and showed that the A14/A43 heterodimer binds single-stranded RNA in a similar way to the archaeal E/F complex.
INTRODUCTION
While bacteria and archaea have a single RNA polymerase (RNAP), eukaryotic nuclei contain three different types of RNAPs (I, II and III), each specialising in the transcription of a different class of RNA (rRNA, mRNA and tRNA, respectively). Most of the research on eukaryotic polymerases has focussed on the RNAPII from Saccharomyces cerevisiae, a large complex consisting of 12 polypeptides (RPB1-12), organised as a compact core of 10 subunits and a more loosely associated heterodimer (RPB4 and RPB7). Crystals of yeast RNAPII have been obtained only for the 10-subunit core, since the heterogeneity in the polymerase preparations caused by the variable stoichiometry of the RPB4/RPB7 heterodimer interfered with crystallisation. The three-dimensional structure of RNAPII therefore did not include these subunits (1,2).
Archaeal cells contain a single RNAP made up of about 12 subunits which display considerable homology to the eukaryotic RNAPII subunits. The RPB4 and RPB7 homologues are termed subunits F and E, but whereas the RPB7 homologue is reasonably well conserved, the similarity between the eukaryotic RPB4 and the archaeal F subunit is barely detectable. The three-dimensional structure of the complex between subunits E and F has been determined by crystallography at high resolution (3). Subunit E (the RPB7 homologue) has an elongated two-domain structure containing two potential single-stranded RNA (ssRNA) binding motifs: a truncated RNP motif at the N-terminus and an S1 motif at the C-terminus. The S1 motif folds into a five-stranded β-barrel known as OB fold. The smaller F subunit (the RPB4 homologue) wraps around one side of subunit E at the interface between the two domains.
Far less is known about the other two eukaryotic RNAPs: RNAPI that transcribes the 18S-5.8S-28S ribosomal RNA precursor and RNAPIII that is involved in the production of tRNAs and a variety of other small RNAs. There is a high level of homology between the large subunits of all three RNAPs (A190/RPB1/C160 and A135/RPB2/C128), which contain the main catalytic centre. Of the small subunits, five are universal and are present in all three enzymes (RPB5, RPB6, RPB8, RPB10, RPB12), a few are homologous and some are clearly type-specific subunits. Such subunits are likely to play important roles in ensuring the type-specific assembly of the different RNAPs in vivo and in facilitating their communication with a range of specialised transcription factors.
Two subunits of RNAPIII (C25 and C17) show significant sequence homology to RPB7 and RPB4. Less clear is the situation for RNAPI where a polypeptide (A43) shows similarity to RPB7 but no obvious subunit with sequence homologue of RPB4 can be found. However the RNAPI subunits A14 and A43 display a biochemical behaviour that is reminiscent of RPB4 and RPB7. It has therefore been suggested that A14 and A43 are the counterparts of the RPB4/RPB7 heterodimer in RNAPI (4). Recently, genetic evidence was presented to support this hypothesis, showing an interaction between A14, A43 and RPB6 (5).
While a number of the smaller RNAP subunits have essentially a structural role in the RNAP architecture, the presence of an S1 motif in the amino acid sequence of the archaeal E subunit already indicated that the likely role of this subunit is to bind ssRNA. Indeed, gel mobility shift assays have confirmed that the yeast RPB4/RPB7 heterodimer binds to single-stranded nucleic acid (6).
Here we show that the sequence of A14 strongly suggests the presence of a structural motif that is also seen in the three-dimensional structure of the archaeal F subunit. Based on this structural similarity, we propose a revised model for the relationship between RPB4/F subunit and A14. We show that it is possible to co-express and purify the A14/A43 heterodimer in milligram amounts and we analyse the effect of A43 deletion mutants on strength of the interaction. Furthermore, we show that the archaeal E/F heterodimer binds ssRNA in a non-sequence-specific manner, confirming and extending the results obtained for the RPB4/RPB7 heterodimer. We also show that the A14/A43 heterodimer binds ssRNA with a similar affinity to the archaeal E/F complex, and analyse the effect of A43 deletion mutants on the nucleic acid binding properties.
MATERIALS AND METHODS
Sequence and structural alignments
The structure of the Methanococcus jannaschii RNAP F subunit was submitted to the Dali server (http://www2.embl-ebi.ac.uk/dali) to be compared with the entire Protein Data Bank. Only structural similarities with a Z-score >3.0 were further analysed, and they were considered only if the regions of structural similarity did not contain large insertions. The Sgs1-HRDC (helicase and RNaseD C-terminal) domain was the top match, with a Z-score of 3.7 and a r.m.s. deviation of 3.1 Å for 61/80 α-carbons. The PDB accession numbers are 1go3 for the E/F heterodimer and 1d8b for the yeast Sgs1-HRDC domain.
The sequence database was searched using the BLAST-PSI program (7). Multiple sequence alignments were generated with the ClustalW program (8) and manually modified to account for the positioning of the secondary structure elements. The colour code for residue conservation shown in Figure 1 was based on the alignment of the amino acid sequences of 19 archaeal E subunits, 13 RNAPII RPB7 subunits, 7 RNAPIII C25 subunits and 7 RNAPI A43 subunits. The full alignment including all the 46 sequences is available as Supplementary Material.
Figure 1.
Amino acid sequence alignment for the RPB7 family. An alignment of the A43 homologues from different organisms (accession numbers: RPA43_YEAST, NP_014985; RPA43_SCHPO, NP_596665; RPA43_DROME, AAF52451; RPA43_ANOGA, EAA00625; RPA43_HUMAN, XP_166508; RPA43_ARATH, NP_565115) with the M.jannaschii E subunit (RPOE_METJA, NP_247371) and the S.cerevisiae RPB7 (RPB7_YEAST, NP_010692.1) and C25 (RPC25_YEAST, NP_012778.1) subunits. The position of the secondary structure elements for the E subunit (3) is shown below the sequence; the secondary structure elements that form the canonical S1 motif/OB fold are indicated in light blue. The N- and C-terminal extensions of the A43 sequences are not explicitly shown. Highlighted in red are the residues that are conserved in all the four subfamilies (A43, RPB7, C25, E subunits); in yellow are the amino acids conserved in at least four out of six A43 sequences; in blue are the amino acids conserved in five out of seven eukaryotic C25 subunits; in green are the residues conserved in 10 out of 13 RPB7 sequences; in violet are the residues conserved in 15 out of 19 archaeal homologues (the following groups of amino acid residues have been considered similar: Ile/Val/Leu/Met, Phe/Tyr/Trp, Asp/Glu, Ser/Thr, Arg/Lys, Gly/Ala). Red asterisks indicate the position of the N- and C-terminal deletions for the yeast A43 subunit. A full sequence alignment including 46 homologues is presented as Supplementary Material.
The alignment in Figure 2 is based on the phylogenetic classification of proteins as reported in the NCBI-COG00514 (Clusters of Orthologous Groups of proteins: http://www.ncbi.nlm.nih.gov/COG/), representing the superfamily of DNA helicases (including the RecQ helicases) sharing the HRDC domain. The alignment of A14 with the Escherichia coli and Vibrio cholera RecQ helicase was based on a BLAST result, while the alignment between subunit F and the Sgs1-HRDC domain was based on the previously described Dali result. The alignment between S.cerevisiae RPB4 and C17 subunits (RPB4_YEAST and C17_YEAST, respectively) is the one reported in the SMART classification of protein domains (http://smart.embl-heidelberg.de) and corresponds to the family SM00657.
Figure 2.
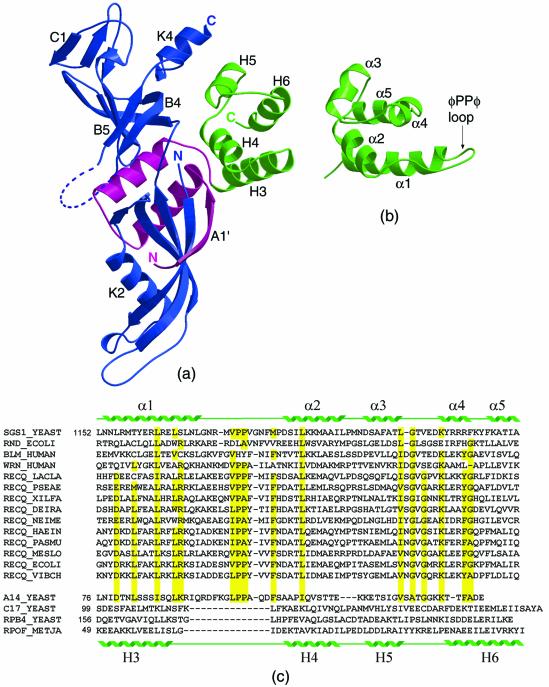
(Opposite) Both the F subunit and A14 contain a HRDC domain. (a) A ribbon representation of the E/F heterodimer. The E subunit (shown in blue) is an elongated molecule that folds into two domains: a β-sheet wrapped around a helix (K2) forms the N-terminal domain (at the bottom of the molecule) while the S1 motif in the C-terminal half of the protein folds into an antiparallel β-barrel with an OB-fold topology. The disordered loop (residues 152–158) between strands B4 and B5 in the OB fold has been modelled for the sake of clarity and is shown as a dashed line. The F subunit (shown in magenta and green) folds into a series of helices that pack against one side of the E subunit at the interface between the two domains, with the N-terminus contributing one strand to the N-terminal β-sheet of the E subunit. The last four helices (H3–H6, shown in green) form a globular cluster that is structurally similar to a HRDC domain. (b) A ribbon representation of the HRDC domain from the S.cerevisiae helicase Sgs1 (14). The structure consists of five helices (α1–α5); between helices α1 and α2 is a conserved loop containing the φPPφ sequence. (c) An alignment of the HRDC domains from a variety of helicases and RNases from prokaryotic and eukaryotic sources. The first sequence corresponds to the C-terminal domain of the yeast Sgs1 helicase, whose structure is shown in (b). The position of the secondary structure elements is shown above the sequence. The sequence of yeast A14 is aligned with the HRDC domains based on sequence similarity; residues that are conserved in the HRDC motif and are present in the A14 subunit are highlighted in yellow. The sequence of subunit F is shown at the bottom of the table, with the position of the secondary structure elements shown below the sequence. Subunit F has been aligned with the HRDC motif based on the structural homology with the Sgs1-HRDC domain. A tentative alignment with S.cerevisiae RPB4 and C17 is shown, although the low level of sequence homology leaves a number of uncertainties as to the correct match. The alignment between A14 and RPB4/subunit F is therefore not based on direct sequence homology, but is the results of a three-step process: (i) the detection of the presence of a HRDC motif in the sequence of A14 and the subsequent alignment of A14 with members of the RecQ family; (ii) the sequence alignment between the subunit F, RPB4 and C17 family members; (iii) the alignment of the structurally equivalent residues of subunit F and the S.cerevisiae RecQ helicase Sgs1.
Protein expression and purification
Co-expression of the recombinant S.cerevisiae A14/A43 complex in E.coli cells was achieved using a two-plasmid strategy. The gene for the A14 subunit was amplified by polymerase chain reaction and cloned into the NcoI and BamHI sites of the kanamycin-resistant pETM-30 vector (EMBL). The vector was designed to express the A14 protein fused to a His6-glutathione-S-transferase tag, cleavable by the tobacco etch virus (TEV) protease. Subunit A43 and its deletion mutants were cloned into the NheI and BamHI sites of the ampicillin-resistant pET21-a vector (Novagen), for expression as untagged proteins.
Escherichia coli BL21(DE3) cells were transformed with both plasmids, under the control of kanamycin and ampicillin and, after induction, cell cultures were grown at 30°C overnight. The cell pellet was thawed and resuspended in 20 mM Tris–HCl, pH 8.0, 300 mM potassium acetate, 7 mM magnesium acetate, 10% glycerol (buffer P300), supplemented with lysozyme at 1 mg/ml, benzonase (0.1 U/ml) and 1 mM phenylmethylsulfonylfluoride (PMSF). Cell lysis was performed by sonication and the resulting lysate clarified by centrifugation and filtered.
All the chromatographic steps were performed using an ÄKTA FPLC apparatus (Amersham Biosciences). The crude extract was loaded onto a GSTrap chelating column (Amersham Biosciences); the column was extensively washed with buffer P300 and the protein eluted with 10 mM glutathione. After dialysis (20 mM Tris–HCl pH 7.9, 500 mM NaCl, 10% glycerol), recombinant His-tagged TEV protease was added to the sample and the digestion carried out for 24 h. Removal of the His-tagged protease and of the cleaved His-GST tag were performed using a HiTrap chelating column (Amersham Biosciences) previously charged with Ni2+ ions. The final yields were of the order of 1–3 mg of pure complex per litre of cell culture.
RNA binding experiments
The probe template was a 63-ribonucleotide fragment of the Methanococcus tuberculosis rrn leader region in pGEM3Z (Promega). The RNA probe was produced by in vitro transcription using SP6 RNA polymerase and 32P[UTP] according to standard procedures and purified on an RNeasy column (Qiagen) after DNase digestion. Probe at a concentration of 2 nM was used in a 10 µl reaction containing 10 mM HEPES pH 7.5, 0.2 mM EDTA, 10 mM ammonium acetate, 1 mM DTT, 200 mM potassium acetate, 10 mM magnesium acetate, 0.5 µg/ml tRNA, 10 µg/ml BSA, 10% glycerol, 40 U of RNasin (Promega) and varying amounts of protein. The samples were incubated at room temperature for 15 min before loading onto a native 8% acrylamide, 0.5× TBE gel. The gel was electrophoresed at 25 mA for 2.5 h at 4°C before exposing to autoradiography. Increasing protein concentrations were 0.14, 0.7 and 3.5 µM.
RESULTS
A43 displays sequence similarity to the RPB7 family
The sequence similarity between the archaeal RNAP E subunit, the RNAPII subunit RPB7 and the RNAPIII subunit C25 is clear and unambiguous (Fig. 1; a full sequence alignment including 46 homologues is presented in the Supplementary Material). Within each subgroup (E subunit, RPB7, C25), the percentage of identity between different species is generally >30%, while across the subgroups the level of identity is ∼20%.
The homology extends to most of the secondary structure elements that are seen in the archaeal E subunit (3) with the exception of the C-terminal helix (K4), which is almost certainly unique to the archaeal homologues. The homology is higher in the truncated RNP motif and the OB fold, and less pronounced for the small β-sheet (C1–C3) which is inserted between strands B3 and B4 of the β-barrel. However, a few key residues are conserved in strands C1 and C2, indicating that the β-sheet is likely to be conserved. In the C25 subunits, the loop between B4 and B5 is longer and unusually rich in polar and charged amino-acid residues. In a number of OB folds the corresponding loop has been shown to play a key role in nucleic acid binding by closing over the nucleotide strand, with both polar and charged side-chains interacting with the sugar-phosphate backbone. In the structure of the M.jannaschii E subunit this loop is disordered, consistent with the possibility that it may become ordered upon nucleic-acid binding.
It is possible to detect a weak but significant sequence homology between A43 and the E/RPB7/C25 family of proteins (9) (Fig. 1). The A43 subunits are generally larger than the RPB7 homologues (up to 340 amino acid residues) and have diverged far more than the other members of the family, with a level of identity as low as 10% between A43 subunits from different species. The sequence similarity is more pronounced in the first half of the polypeptide, while the C-terminal regions tend to be rather different and unusually rich in charged residues and/or repetitive sequences. An exception is represented by the sequences of the Schizosaccharomyces pombe and Arabidopsis thaliana A43, which are shorter (173 and 196 amino acids, respectively) and lack both the N-terminal and the low complexity C-terminal extensions. Beyond strand B4 the relationship between the A43 subunits and the other orthologues is uncertain. The alignment shown in Figure 1 is based on the positioning of the shorter S.pombe and A.thaliana A43 sequences to obtain a complete five-stranded OB-fold barrel; this places the low complexity sequences of the mammalian and insect A43 subunits between strands B4 and B5, which corresponds to the position of similar insertions in the C25 subunits. Moreover, a deletion mutant of A43 lacking the last 87 amino acid residues is viable in S.cerevisiae (10) and our data show that the corresponding A43 fragment can be expressed in large amounts, suggesting that this fragment has a complete and independent fold.
When comparing the similarities across the subgroups, whereas the sequence identity between E/RPB7/C25 is generally ∼20%, the level of identity between A43 and other members is only between 7 and 14%, a level that is below the accepted significance threshold. However, a clear pattern emerges when all the sequences are aligned and particularly when the sequence alignment is compared with the 3D structure of the archaeal E subunit, showing conservation of key structural residues in the core of the protein.
In some of the A43 homologues, the loop between B4 and B5 [involved in ssRNA/ssDNA binding in the canonical OB fold (11)] includes long extensions. Such extensions often consist of long stretches of negatively and positively charged amino acid residues (particularly lysines), consistent with the hypothesis of an involvement in the binding of the sugar-phosphate backbone of the nucleic acid. However it has to be stressed that the sequence homology between A43 and the other proteins is rather poor beyond strand B3 and therefore there is a serious ambiguity in the exact positioning of strands B4 and B5 along the A43 sequence.
RPB4 and A14 share a HRDC motif at the C-terminus
No significant sequence homology could be detected between the A14 subunit of RNAPI (consisting of 137 amino acid residues) and any other RNA polymerase subunit. In particular no similarity could be detected between A14 and either the archaeal F subunit, RPB4 (RNAPII) or C17 (RNAPIII). However, a structural connection can be established between the A14 subunit and subunit F.
The structure of archaeal RNAP subunit F consist of a short β-strand (A′) and two helices (H1, H2) wrapping around the E subunit, followed by a more globular cluster of four helices (H3, H4, H5 and H6) at the C-terminus (3). A comparison of the structure with known protein folds identified a structural similarity between these four helices and the HRDC domain (Fig. 2a). This is a small domain (consisting of about 80 residues) found in RecQ helicases and RNaseD from various organisms (12). Mutations in the HRDC domain have been shown to play a role in a number of human diseases, such as the Werner and Bloom syndromes (13).
The NMR structure of the C-terminal domain of the S.cerevisiae RecQ helicase Sgs1 (Fig. 2b) shows that the HRDC domain folds as a cluster of five helices (α1–α5), where the last two helices (α4 and α5) can be described as a single helix with a kink (14). A similar fold is found in auxiliary domains of DNA and RNA polymerases, DExx box helicases and site-specific recombinases. A number of the HRDC sequences have a conserved hydrophobic insertion including the φPPφ consensus sequence (where the first hydrophobic residue φ is generally V/L/I and the second is F/Y/A). The region is located in an exposed loop between helices α1 and α2 and is therefore a likely candidate for protein–protein interactions. Despite evidence that the Sgs1-HRDC domain weakly binds ssDNA, none of the residues that were shown to interact with the nucleic acid is conserved within the HRDC family, nor is the positive electrostatic charge distribution on the surface conserved. This suggests that the domain is not associated with a specific function, but may simply represent a common structural motif in proteins involved in nucleic acid binding.
When the C-terminal domain of the F subunit and the Sgs1-HRDC domain are superposed, the largest difference is the deletion in the F subunit of the α1–α2 loop which includes the φPPφ motif. None of the residues involved in DNA binding are conserved and the overall electrostatic character of the molecular surface is very different (positively charged in HRDC and negatively charged in subunit F), making it unlikely that the F subunit shares the nucleic acid binding properties of the Sgs1-HRDC domain.
The amino acid sequence of A14 suggests the presence of a HRDC motif at the C-terminus (Fig. 2c). A significant number of residues conserved in most proteins belonging to the HRDC family are present in A14, including the loop between α1 and α2 which contains the sequence LPPA. The level of sequence identity is higher with the RecQ helicases. Given the level of sequence identity, it is likely that the C-terminus of A14 folds into a similar helical cluster, although the fact that the A14 sequence is slightly shorter suggests the presence of only four helices. Of the residues that confer a positively charged character to the Sgs1-HRDC domain surface, and that have been proposed to be involved in DNA binding, only one (Lys 131) is present on A14.
The presence of a structurally equivalent domain in subunit F, despite the lack of a detectable signal for a HRDC motif in the sequence, clearly suggests a link between A14 and F, even in the absence of direct sequence homology between the proteins. The proposed similarity between A14 and the subunit F/RPB4/C17 does not involve a direct sequence homology, but it is demonstrated by the fact that they share a common structural domain, namely the HRDC motif. The presence of the HRDC signature sequence can be detected in the A14 sequence, while it is directly observed in the structure of subunit F, which is a prototype for the RPB4 and C17 structures. Considering the low level of conservation of the subunit F/RPB4/C17 family, in sharp contrast to the obvious similarities between most of the other RNAP subunits, the fact that the sequences of the RPB4 and A14 may have diverged so as to lose any resemblance is not surprising. But our results indicate that the overall fold of the molecule is likely to be conserved.
An interesting observation is that in the E/F complex the loop between helices H3 and H4 (the structural equivalent of helices α1 and α2 in the HRDC motif) is close to the N-terminus of subunit E (Fig. 2a). The N-terminal extension of S.cerevisiae A43 can therefore be a possible candidate for interaction with the LPPA loop of A14.
Recombinant A43 and A14 can be co-expressed and form a stable heterodimer
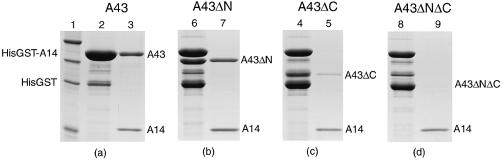
In order to confirm that A14 and A43 form a stable heterodimer, we used a two-plasmid system to co-express the two yeast subunits in E.coli cells. Full-length A14 was cloned into a vector conferring kanamycin resistance and expressing the target protein with a His-GST tag at the N-terminus, while A43 was cloned into an ampicillin-resistant vector. Co-expression of His-GST-A14 and untagged A43 resulted in the formation of a stable heterodimeric complex that could be purified on a glutathione sepharose matrix (Fig. 3a). The possibility that A43 alone could bind to the matrix was ruled out by expression of this subunit alone (data not shown). Cleavage of the His-GST tag and a further metal affinity chromatography step, followed by size exclusion chromatography, were used to purify the heterodimer, clearly demonstrating a tight and specific interaction between the two subunits.
Figure 3.
Co-expression of the A14/A43 heterodimer. (a) Co-expression of His-GST-A14 and full-length A43. After each purification step protein samples were subjected to SDS–PAGE. Lane 1 shows the molecular mass markers (50, 35, 25 and 15 kDa, respectively); lane 2 is the eluate from a glutathione column (where A43 and the His-GST-A14 fusion have the same mobility); lane 3 shows the binary complex after cleavage of the His-GST tag and further purification with metal affinity chromatography to remove the tag. (b–d) Similarly, the first lane is the eluate from the glutathione column and the second lane the result of cleavage of the tag and further purification. (b) Co-expression of His-GST-A14 and A43 lacking the first 32 amino acids. Lane 7 shows the presence of a stable stoichiometric heterodimer. (c) Co-expression of His-GST-A14 and A43 lacking the last 87 amino acids. After digestion and further purification (lane 5) only a small amount of A43ΔC is present. (d) Co-expression of His-GST-A14 and A43 lacking the first 32 amino acids and the last 87 amino acids. There is no evidence of a band corresponding to the A43ΔNΔC, suggesting that the formation of the complex is impaired by the presence of both deletions.
Based on the sequence alignment in Figure 1, a number of deletion mutants of A43 were designed and tested for their capacity to interact with A14. Most of the A43 sequences have an N-terminal extension with respect to the other members of the RPB7 family. We chose to delete the first 32 amino acid residues based on the alignment of the S.pombe and A.thaliana A43 homologues. A much longer tail is present at the C-terminus of some of the A43 subunits; not only is this extension an obvious addition to the OB fold seen in the archaeal E subunit but it is also very rich in charged residues. Again, such a long tail is not present in the more compact S.pombe and A.thaliana homologues. Furthermore, it has been shown that the last 87 amino acids of S.cerevisiae A43 are not essential for its function in vivo, despite conferring to the cell temperature sensitivity (10).
We therefore prepared constructs encoding for A43ΔN (including residues 33–326), A43ΔC (1–239) and A43ΔNΔC (33–239) and used them in co-expression experiments together with the plasmid expressing His-GST-A14. The results of these experiments are shown in Figure 3. The interaction with A14 is clearly maintained for A43ΔN, showing that the first 32 amino acid residues are not essential for the formation of the heterodimer (Fig. 3b). A weaker interaction is observed for the C-terminal deletion mutant (Fig. 3c), while it is abolished for the double mutant lacking both the N- and C-termini (Fig. 3d). This can be explained by postulating that the deletion at the N-terminus causes a minor destabilisation of the complex, which has a synergic effect with the much more severe impairment caused by the C-terminal deletion.
However, the difficulty of generating a reliable alignment with the E subunit beyond strand B3 makes the design of C-terminal deletion mutants uncertain and it is possible that the mutant we have chosen to construct may lack an essential structural element necessary for the integrity of the OB fold. Therefore we cannot rule out that the weaker interaction of A43ΔC with A14 is simply the result of a structural defect rather than reflecting an active role of the C-terminal tail in the formation of the heterodimer.
Similar experiments to try to identify the interactions between A43 and A14 have been recently reported in which in vitro synthesised A14 was incubated with truncated mutants of 35S-labelled A43 fused with an HA epitope (5). Contrary to our data, results from these experiments show no binding for both a ΔN36 and a ΔC87 deletion mutant, while we were able to purify a stable heterodimer for mutant ΔN33 and observed a weaker, but significant interaction for ΔC87. We suggest that the discrepancy may be due to the fact that we co-express both proteins in the same cells, and therefore minimise the folding problems these subunits are likely to encounter when produced in the absence of the correct partner. Indeed, in a two-hybrid assay the first 36 amino acids of A43 were found dispensable for the interaction (5), consistent with our co-expression experiments.
Both the E/F and A43/A14 heterodimers bind single-stranded RNA
The presence of an S1 motif in the sequence of the archaeal E subunit strongly suggests that its role is to bind single-stranded nucleic acids. Indeed, the eukaryotic RPB4/RPB7 complex has been shown to bind to both ssDNA and ssRNA with comparable affinities (6).
To test for ssRNA binding activity, we performed electrophoretic mobility shift assays with a radiolabelled RNA probe. The assay was done using the archaeal E/F heterodimer, the full-length yeast RNAPI heterodimer (A14/A43) and the two deletion mutants which maintained the integrity of the complex (A14/A43ΔN, A14/A43ΔC; Fig. 4). Both the E/F complex and the A14/A43 heterodimers bound to RNA. Protein–RNA complexes with comparable affinities were obtained in the presence of various RNA probes (data not shown) indicating that the binding is sequence-non-specific. Neither of the deletion mutants bound to the probe, suggesting that the N- and C-terminal extensions play a role in interacting with the nucleic acid. We also carried out RNA binding experiments with the A14 and A43 subunits in the absence of a partner (data not shown); while we detected no binding in the presence of A14 alone, there was evidence of an interaction between A43 and the RNA probe.
Figure 4.
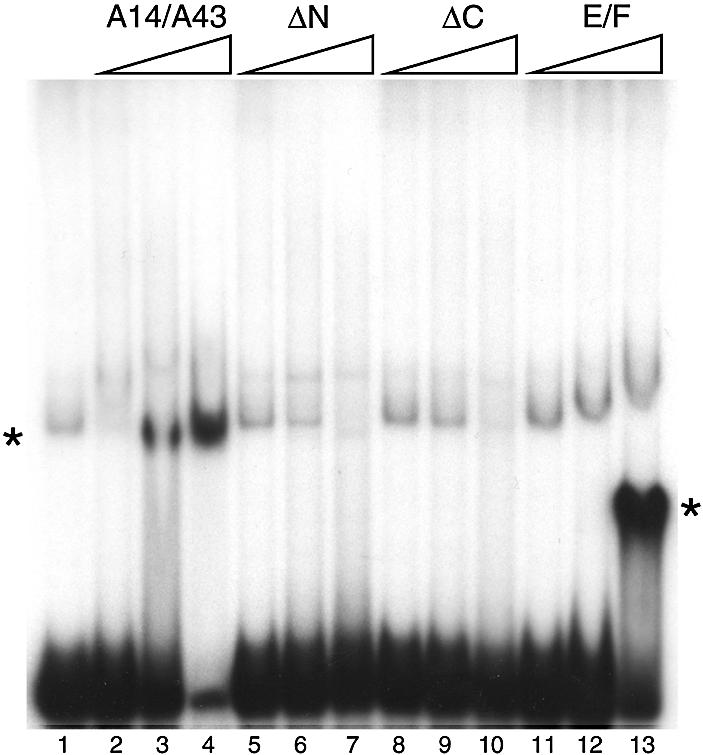
Nucleic acid binding properties of the A14/A43 complex. A radiolabelled RNA probe was incubated with increasing amounts of protein and the resulting complexes resolved from unbound RNA by electrophoresis in a non-denaturating polyacrylamide gel. Bands corresponding to the free probe and the RNA–protein complexes were detected by autoradiography. The protein concentrations were 0.14, 0.7 and 3.5 µM. Lane 1, free probe; lanes 2–4, full length A14/A43 heterodimer; lanes 5–7, A14/A43ΔN; lanes 8–10, A14/A43ΔC; lanes 11–13, E/F. Asterisks indicate the positions of the protein–RNA complexes. A weak band, running at a position similar to that of A14/A43–RNA complex, is present also in lane 1 (free probe) and is likely to be due to the formation of secondary structure in the RNA probe. Both the full-length A14/A43 and the E/F heterodimer bind the RNA probe (as clearly indicated by the formation of a strong band corresponding to the complex and confirmed by the depletion of the free probe), while the formation of the complex was not detected in the presence of the deletion mutants.
DISCUSSION
While deletion of RPB7 in yeast is lethal (15,16), RPB4 is not essential under optimal growth conditions. However, rpb4Δ cells are heat and cold sensitive (17,18) and are unable to enter the stationary phase when encountering nutrient depletion (19). RNAPII purified from the rpb4Δ strain lacks both RPB4 and RPB7, but RPB7 can interact with RNAPII independently of RPB4 and overexpression of RPB7 partially suppresses the stress phenotypes of the rpb4Δ strain.
The RNAPI subunits A14 and A43 display biochemical behaviour that is similar to RPB4 and RPB7. It is possible to purify an inactive form of S.cerevisiae RNAPI that does not contain the subunits A14, RPB6 (a subunit present in all three eukaryotic RNAPs, also known as ABC23) and A43 (20). Deletion of RPA43 is lethal (21), while RPA14 is not essential for growth, although its absence results in a slight but detectable reduction in the synthesis of rRNA, growth defects at temperatures >30°C and a marked instability of the entire RNAPI (20). Polymerase purified from cells in which the gene for A14 has been deleted also lacks A43 and RPB6, indicating that the absence of A14 causes a major instability of two other essential subunits. Although A43 is essential in vivo, this subunit is not necessary for enzyme activity in a non-specific transcription assay in vitro (22). This behaviour is similar to that of RPB4 and RPB7, suggesting a functional similarity between RPB4/RPB7 and A14/A43.
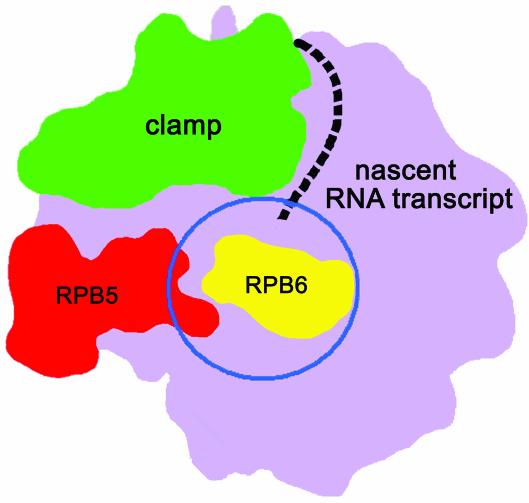
An obvious difference between RNAPI and RNAPII is the fact that the absence of A14 not only weakens the interaction between the core and A43, but also affects RPB6. Given that the position of RPB6 in the RNAP core is adjacent to the proposed location of the RPB4/RPB7 heterodimer (3), on the right-hand side of RPB5, underneath the clamp (Fig. 5), it is possible that A43, being considerably more bulky than RPB7, interacts more tightly with RPB6 in the RNAPI structure and therefore affects its stability as well. Indeed, a direct interaction between RPB6 and A43 has been shown by co-expression in E.coli of His-tagged RPB6 and A43, followed by co-purification by metal affinity chromatography (5). This model is also consistent with the position of the A14 and A43 subunits as seen by electron microscopy (23).
Figure 5.
A model for the interaction of A14/A43 with the RNA polymerase core. A schematic representation of the RNAPII 10-subunit core is shown [viewed in a similar orientation as the side view in Figs 3 and 6D by Cramer et al. (31)], with RPB5 shown in red, RPB6 shown in yellow and the mobile clamp, which closes onto the active side cleft upon nucleic acid binding, shown in green. The predicted exit path for the nascent RNA transcript is shown as a dashed line, and the proposed location of the RPB4/RPB7 heterodimer (3) (Fig. 4) is indicated by a blue circle. The position of RPB6 in the RNAPII is exactly underneath the proposed position for the RPB4/RPB7 heterodimer, making it possible that in RNAPI the absence of the A14/A43 complex may also affect the stability of RPB6.
Sequence homology between A43 and the RPB7 family can be easily detected (9), while no sequence relationship could be found between A14 and the subunit F/RPB4/C17 family. A model has been proposed in which the structural similarity between A14 and the F subunit is limited to the N-terminal half of the protein, including strand A1′ and helices H1 and H2 (5), while the C-terminal domain (corresponding to the 4-helix bundle) has been hypothesised to have a different fold. However such speculation is supported neither by sequence homology nor by structural similarity. Here we present evidence for a clear relationship between the C-terminal domain of the F subunit and A14; the amino-acid sequence of A14 suggests the presence of a HRDC motif at the C-terminus, a structural unit also seen in the F subunit (Fig. 2). We therefore propose a very different structural model from Peyroche et al. (5); in our model the region between residues 76–137 of A14 folds into a helical bundle analogous to helices H3, H4, H5, H6 in the F subunit structure.
Before discussing the functional role of the A14/A43 heterodimer, it is interesting to note that A14 seems to be unique to S.cerevisiae and has no clear homologue in any other organisms. It is possible that the sequence is so divergent that the similarity cannot be recognised; this would be consistent with both the relatively low level of conservation shown by the members of the subunit F/RPB4 family, and the far lower degree of conservation shown by the RNAPI A43 subunits compared to the RPB7/C25/E subunits. However, no unidentified polypeptide that could correspond to an A14 homologue has been observed in preparations of RNAPI from different species. It is conceivable that the loss of this subunit is due to a labile interaction that does not survive the biochemical purification procedures needed to isolate the polymerase core, but it may also suggest that A14, a non-essential subunit in yeast, has not been conserved in evolution. This would be in agreement with the genetic, biochemical and structural data indicating that RPB4 has simply a structural role in stabilising RPB7 and its interaction with the RNAP 10-subunit core, a role that could be dispensable. A 163 amino acid reading frame from the Candida albicans genome sequence displaying some sequence similarity to yeast A14 has been reported (5). Although a short stretch of 15 amino acid residues is almost identical, the overall similarity between the two sequences is low and there is no evidence that the C.albicans protein is part of the RNAPI. When the sequence homology between the S.cerevisiae A14 and C.albicans putative homologue is used to build a profile to carry out further searches in the sequence database by BLAST-PSI (NCBI), no other sequence that matches this profile can be found.
While a number of the smaller RNAP subunits have essentially a structural role in the RNAP architecture, the consistent presence of this structurally independent binary complex in all the eukaryotic and archaeal RNA polymerases indicates a critical functional role for these subunits. The presence of an S1 motif in the amino acid sequence of the archaeal E subunit has already indicated that the likely role of this subunit is to bind ssRNA. Although no trace of the characteristic S1 sequence pattern can be found in any of the eukaryotic homologues, based on the crystal structure of the E subunit it is possible to identify a number of alternative residues that may play a similar role to the canonical S1 motif in all the eukaryotic subunits. Indeed, gel mobility shift assays have confirmed that the yeast RPB4/RPB7 heterodimer binds to single-stranded nucleic acid (6).
Although proteins containing an OB fold can bind either ssRNA or ssDNA, the particular subgroup containing the sequence signature of the S1 motif binds ssRNA. For example, the proposed function for the cold-shock proteins is to bind to nascent mRNA during bacterial transcription, thereby preventing formation of intramolecular hydrogen bonds at low temperatures like an ‘RNA chaperone’ (24). In a similar manner the ribosomal S1 protein binds to a subset of mRNAs, particularly those containing long 5′ untranslated sequences, preventing the formation of secondary structures that would interfere with efficient translation (25). The bacterial transcription anti-terminator factor NusA has been proposed to bind the nascent mRNA directly after its emergence from the RNAP exit tunnel (26); the transcription terminator Rho is thought to translocate along the RNA (27); translation initiation factor eIF1A is employed in scanning mRNA for the initiation codon (28). The emerging common theme is, therefore, ssRNA binding without sequence specificity. We therefore proposed that the OB fold of RPB7 interacts with the nascent RNA transcript, possibly assisted by the truncated RNP domain (3).
We have now measured binding to ssRNA for both the archaeal E/F complex and the eukaryotic A14/A43 complex and shown that the two complexes bind nucleic acid with comparable affinities (Fig. 4). This indicates not only that the structure of the heterodimer is likely to be conserved but also that the RNA binding function has remained constant in the evolution of the archaeal and eukaryotic RNA polymerases. On the other hand, while it is likely that the S1 motif of the E/F heterodimer plays a major role in nucleic acid binding, the N- and C-terminal extensions of A43 seem to play a significant role in RNA binding, since their deletion abolished the formation of a protein–nucleic acid complex.
An additional role for the heterodimer, although not as well understood, may involve the initiation of transcription at specific promoters. RPB4 and RPB7 have been implicated in promoter-specific transcription initiation in vitro (6,29) and in promoter-activated transcription in vivo (30). No obvious mechanism has been proposed, nor has the structure of the archaeal homologue shed any light on a possible role in transcription initiation. A possibility is that the heterodimer simply triggers or facilitates the closure of the clamp as the nascent transcript emerges from the active site, assisting the polymerase conformational changes that are essential for transcription initiation (31).
Another possibility is that the heterodimer may be directly involved in recruitment of activators. There is some evidence that human RPB7 interacts with the N-terminal moiety of transcription factor EWS (32), the highly homologous hTAFII68 (33), as well with the ligand-free retinoic acid receptor (34), but it difficult to determine whether these few examples are physiologically relevant and whether they are representative of a more general function of RPB7.
A far clearer picture of the participation of A43 in the formation of the pre-initiation complex is emerging, showing that A43 plays a key role in RNAPI recruitment onto rRNA promoters by directly interacting with transcription factor RRN3 (10,35). RRN3, also known as TIF-IA in mammalian cells, is an essential transcription factor which associates with the polymerase also in the absence of DNA and defines a distinct population of transcriptionally active and initiation-competent RNAPI (36). Furthermore the RNAPIII subunit C17 (the homologue of RPB4) from S.cerevisiae has been shown to interact with the general transcription factor TFIIIC, and possibly have a role in transcription initiation (37).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a Wellcome Trust Grant to S.O. and P.B.
NOTE ADDED IN PROOF
Recently the structure of the S.cerevisiae RNAPIII 12-subunit complex has been determined by two groups [Armache,K.J., Kettenberger,H. and Cramer,P. (2003) Proc. Natl Acad. Sci. USA, 100, 6964–6968 and Bushnell,D.A. and Kornberg,R.D. (2003) Proc. Natl Acad. Sci. USA, 100, 6969–6973]. The position of the RPB4/RPB7 heterodimer in the crystal structure is in agreement with the model presented in Figure 5.
REFERENCES
- 1.Cramer P., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science, 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- 2.Gnatt A.L., Cramer,P., Fu,J., Bushnell,D.A. and Kornberg,R.D. (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science, 292, 1876–1882. [DOI] [PubMed] [Google Scholar]
- 3.Todone F., Brick,P., Werner,F., Weinzierl,R.O.J. and Onesti,S. (2001) Structure of an archaeal homologue of the eukaryotic RNA polymerase II RPB4/RPB7 complex. Mol. Cell, 8, 1137–1143. [DOI] [PubMed] [Google Scholar]
- 4.Cramer P. (2002) Multisubunit RNA polymerases. Curr. Opin. Struct. Biol., 12, 89–97. [DOI] [PubMed] [Google Scholar]
- 5.Peyroche G., Levillain,E., Siaut,M., Callebaut,I., Schultz,P., Sentenac,A., Riva,M. and Carles,C. (2002) The A14-A43 heterodimer subunit in yeast RNA pol I and their relationship to Rpb4-Rpb7 pol II subunits. Proc. Natl Acad. Sci. USA, 99, 14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlicky S.M., Trans,P.T., Sayre,M.H. and Edwards,A.M. (2001) Dissociable Rpb4-Rpb7 subassembly of RNA polymerase II binds to single-stranded nucleic acid and mediates a post-recruitment step in transcription initiation. J. Biol. Chem., 276, 10097–10102. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 8.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4867–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shpakovski G.V. and Shematorova,E.K. (1999) Characterization of the rpa43+ cDNA of Schizosaccharomyces pombe: structural similarity of subunit RPA43 of RNA polymerase I and subunit RPC25 of RNA polymerase III. Bioorg. Khim., 25, 791–796. [PubMed] [Google Scholar]
- 10.Peyroche G., Milkereit,P., Bischler,N., Tschochner,H., Schultz,P., Sentenac,A., Carles,C. and Riva,M. (2000) The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J., 19, 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theobald D.L., Mitton-Fry,R.M. and Wuttke,D.S. (2003) Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys Biomol. Struct., 32, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hickson I.D. (2003) RecQ helicases: caretakers of the genome. Nature Rev. Cancer, 3, 169–178. [DOI] [PubMed] [Google Scholar]
- 13.Morozov V., Mushegian,A.R., Koonin,E.V. and Bork,P. (1997) A putative nucleic acid-binding domain in Bloom’s and Werner’s syndrome helicases. Trends Biochem. Sci., 22, 417–418. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z., Macias,M.J., Bottomley,M.J., Sier,G., Linge,J.P., Nilges,M., Bork,P. and Sattler,M. (1999) The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Struct. Fold. Des., 7, 1557–1566. [DOI] [PubMed] [Google Scholar]
- 15.McKune K., Moore,P.A., Hull,M.W. and Woychik,N.A. (1995) Six human RNA polymerase subunits functionally substitute for their yeast counterparts. Mol. Cell. Biol., 15, 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young R.A. (1991) RNA polymerase II. Annu. Rev. Biochem., 60, 689–715. [DOI] [PubMed] [Google Scholar]
- 17.Rosenheck S. and Choder,M. (1998) Rpb4, a subunit of RNA polymerase II, enables the enzyme to transcribe at temperature extremes in vitro. J. Bacteriol., 180, 6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woychik N.A. and Young,R.A. (1989) RNA polymerase II subunit RPB4 is essential for high- and low-temperature yeast cell growth. Mol. Cell. Biol., 9, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choder M. (1993) A growth rate-limiting process in the last growth phase of the yeast life-cycle involves RPB4, a subunit of RNA polymerase II. J. Bacteriol., 175, 6358–6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smid A., Riva,M., Bouet,F., Sentenac,A. and Carles,C. (1995) The association of three subunits with yeast RNA polymerase is stabilised by A14. J. Biol. Chem., 270, 13534–13540. [DOI] [PubMed] [Google Scholar]
- 21.Thuriaux P., Mariotte,S., Buhler,J.-M. and Sentenac,A. (1995) Gene RPA43 in Saccharomyces cerevisiae encodes an essential subunit of RNA Polymerase I. J. Biol. Chem., 270, 24252–24257. [DOI] [PubMed] [Google Scholar]
- 22.Hager G.L., Holland,M.J. and Rutter,W.J. (1977) Isolation of ribonucleic acid polymerases I, II and III from Saccharomyces cerevisiae. Biochemistry, 16, 1–8. [DOI] [PubMed] [Google Scholar]
- 23.Bischler N., Brino,L., Carles,C., Riva,M., Tschochner,H., Mallouh,V. and Schultz,P. (2002) Localization of the yeast RNA polymerase I-specific subunits. EMBO J., 21, 4136–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graumann P.L. and Marahiel,M.M. (1998) A superfamily of proteins that contain the cold-shock domain. Trends Biochem. Sci., 20, 286–290. [DOI] [PubMed] [Google Scholar]
- 25.Subramanian A.R. (1983) Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol., 28, 101–142. [DOI] [PubMed] [Google Scholar]
- 26.Worbs M., Bourenkov,G.P., Bartunik,H.D., Huber,R. and Wahl,M.C. (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol. Cell, 7, 1177–1189. [DOI] [PubMed] [Google Scholar]
- 27.Bodgen C.E., Fass,D., Bergman,N., Nichols,M.D. and Berger,J.M. (1999) The structural basis for terminator recognition by the Rho transcription termination factor. Mol. Cell, 3, 487–493. [DOI] [PubMed] [Google Scholar]
- 28.Battiste J.L., Pestova,T.V., Hellen,C.U.T. and Wagner,G. (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell, 5, 109–119. [DOI] [PubMed] [Google Scholar]
- 29.Edwards A.M., Kane,C.M., Young,R.A. and Kornberg,R.D. (1991) Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro. J. Biol. Chem., 266, 71–75. [PubMed] [Google Scholar]
- 30.Pillai B., Sampath,V., Sharma,N. and Sadhale,P. (2001) Rpb4, a non-essential subunit of core RNA polymerase II of Saccharomyces cerevisiae is important for activated transcription of a subset of genes. J. Biol. Chem., 276, 30641–30647. [DOI] [PubMed] [Google Scholar]
- 31.Cramer P., Bushnell,D.A., Fu,J., Gnatt,A.L., Maier-Davis,B., Thompson,N.E., Burgess,R.R., Edwards,A.M., David,P.R. and Kornberg,R.D. (2000) Architecture of RNA polymerase II and implication for the transcription mechanism. Science, 288, 640–649. [DOI] [PubMed] [Google Scholar]
- 32.Petermann R., Mossier,B.M., Aryee,D.N., Khazak,V., Golemis,E.A. and Kovar,H. (1998) Oncogenic EWS-Fli1 interacts with hsRPB7, a subunit of human RNA polymerase II. Oncogene, 17, 603–610. [DOI] [PubMed] [Google Scholar]
- 33.Bertolotti A., Melot,T., Acker,J., Vigneron,M., Delattre,O. and Tora,L. (1998) EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68 and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol., 18, 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen X.Q., Bubulya,A., Zhou,X.F., Khazak,V., Golemis,E.A. and Shemshedini,L. (1999) Ligand-free RAR can interact with the RNA polymerase II subunit hsRPB7 and repress transcription. Endocrine, 10, 281–289. [DOI] [PubMed] [Google Scholar]
- 35.Imazawa Y., Hisatake,K., Nakagawa,K., Muramatsu,M. and Nogi,Y. (2002) The fission yeast RPA21 subunit of RNA polymerase I: an evolutionarily conserved subunit interacting with ribosomal DNA (rDNA) transcription factor Rrn3p for recruitment to rDNA promoter. Genes Genet. Syst., 77, 147–157. [DOI] [PubMed] [Google Scholar]
- 36.Miller G., Panov,K.I., Friedrich,J.K., Trinkle-Mulcahy,L., Lamond,A.I. and Zomerdijk,J.C. (2001) hRRN3 is essential in the SL1-mediated recruitment of RNA Polymerase I to rRNA gene promoters. EMBO J., 20, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferri M.L., Peyroche,G., Siaut,M., Lefebvre,O., Carles,C., Conesa,C. and Sentenac,A. (2000) A novel subunit of yeast RNA polymerase III interacts with the TFIIB-related domain of TFIIIB70. Mol. Cell. Biol., 20, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.