Abstract
Sequencing errors in combination with repeated regions cause major problems in shotgun sequencing, mainly due to the failure of assembly programs to distinguish single base differences between repeat copies from erroneous base calls. In this paper, a new strategy designed to correct errors in shotgun sequence data using defined nucleotide positions, DNPs, is presented. The method distinguishes single base differences from sequencing errors by analyzing multiple alignments consisting of a read and all its overlaps with other reads. The construction of multiple alignments is performed using a novel pattern matching algorithm, which takes advantage of the symmetry between indices that can be computed for similar words of the same length. This allows for rapid construction of multiple alignments, with no previous pair-wise matching of sequence reads required. Results from a C++ implementation of this method show that up to 99% of sequencing errors can be corrected, while up to 87% of the single base differences remain and up to 80% of the corrected reads contain at most one error. The results also show that the method outperforms the error correction method used in the EULER assembler. The prototype software, MisEd, is freely available from the authors for academic use.
INTRODUCTION
Genome sequencing is important for the study and comparison of organisms and has generated a wealth of new biological knowledge. However, as more sequence is continuously produced for different organisms, increasing amounts of complex repeated regions are encountered. These regions often contain important biological information (1), and it is therefore important to be able to efficiently determine their sequences.
The shotgun sequencing method is today the strategy of choice for large scale genome sequencing projects. The method is relatively cost effective and easy to automate and, in addition, the redundant sequencing increases the accuracy of the finished sequence. Problems in this approach are mainly caused by the limited quality of primary sequence data and the presence of repetitive sequences. Sequencing errors, especially combined with repeats, often cause problems in the sequence assembly step due to the inability of assemblers to distinguish between sequencing errors and single base differences between repeats. These problems make finishing a time consuming task, if at all possible. Although several eukaryote genomes have been published, none of them is complete in all regions.
One way to simplify and improve shotgun fragment assembly results is to correct sequencing errors. For example the EULER (2) and Arachne (3) assembly programs contain integrated error correction steps. These are, however, not ideal and further improvements are needed.
A statistical method presented in a previous paper (4) was developed in order to identify single base differences between repeat copies for the purpose of correct assembly of repeats. The differences between repeats are located by constructing and analyzing multiple alignments consisting of all shotgun reads in a dataset that may sample several repeat copies. Detected differences occurring at a certain rate are labeled as defined nucleotide positions, DNPs. In this paper, we present an algorithm to correct sequencing errors in shotgun sequence reads, using the DNP method. By performing error correction prior to actual shotgun fragment assembly, the complexity of the task can be reduced (2).
The DNP method uses multiple alignments consisting of a read and all its overlaps with other reads. The construction of multiple alignments is computationally demanding, while large scale sequencing projects require reliable software that is able to handle large numbers of sequences more and more efficiently. For this reason, we also describe an algorithm for finding overlaps between shotgun fragments that can be used for rapid construction of multiple alignments. Previous methods commonly use exact matches as seeds for finding alignments followed by dynamic programming, e.g. TRAP (5), Phrap (http://www.phrap.org) and ARACHNE (3). These methods are fast, at a cost of low sensitivity. Our method uses a novel algorithm for finding approximate matches that allows for fast overlap detection while maintaining high sensitivity. The multiple alignments are directly constructed from q-grams, i.e. words of length q, that match a pattern with a maximum number of substitutions.
These methods have been implemented in a prototype program, MisEd, that is available from the authors at no cost for academic and non-profit users. We also present the results of a comparison of the performances of MisEd with the error correction algorithm used in the EULER assembler. The results show that MisEd outperforms EULER.
MATERIALS AND METHODS
The DNP method presented in (4) discriminates sequencing errors from real differences between repeat copies, making it a suitable tool for error correction. This is achieved by constructing multiple alignments that contain reads sampling the same region in different repeat copies. The input to the DNP method is an optimized multiple alignment consisting of a read and all its overlaps with other reads in the dataset. This includes true overlaps as well as apparent overlaps, i.e. overlaps with reads from similar repeat copies.
Our error correction method can be divided into the following parts: trimming, construction of multiple alignments and error correction. The sections below describe each part in detail.
Trimming
The purpose of the trimming step is to remove unusable ends of sequences from long runs. It is desirable to trim the reads as little as possible, since the purpose of the method is to correct errors rather than trim them out of the dataset. A longer mean read length leads to higher shotgun coverage in the subsequent assembly step. Furthermore, the amount of DNPs detected drops significantly with stringent trimming conditions. In our previous investigation, an increase in mean read quality from 95.7 to 97.4%, due to more stringent trimming, resulted in a decrease in shotgun coverage of 22% while the number of detected DNPs decreased by 29% (4).
The trimming is performed using Phred quality values (6). A window of length lw is slid along the read from 3′ to 5′ and from 5′ to 3′ with step size sw, until nw consecutive windows with a mean error rate below a threshold εmax have been found. The starting position of the first window is marked as the beginning, or end, of the analyzable sequence. A minimum length of high quality region lminhq is required in order to keep the read in the database.
Construction of multiple alignments
The construction of multiple alignments consists of two steps: construction of raw multiple alignments, and optimization.
Construction of raw multiple alignments
The objective of the first step in multiple alignment construction is to rapidly construct a multiple alignment of all reads sampling the same region in different repeat copies. This is accomplished by choosing a starting read and locating all reads that truly or apparently overlap this read. True and apparent overlaps between the starting read and a candidate read begin in one of the reads and end in one of the reads. This fact can be used for rapid verification of candidate overlaps for the multiple alignment. We create the multiple alignment by locating short approximate matches to the starting read and determining whether there are matches in the beginnings and ends of the potential overlaps (Fig. 1). The result is a raw multiple alignment that has to be locally optimized. The optimization step is computationally expensive. For this reason, the raw multiple alignment should contain as few false, i.e. neither true nor apparent, overlaps as possible.
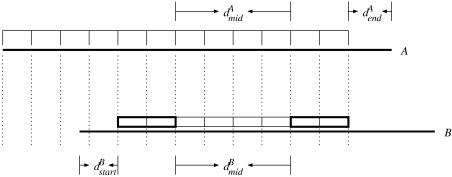
Figure 1.
Principles of raw multiple alignment construction. Horizontal bars indicate reads. Boxes indicate v-neighbors of the starting read A that are present in the candidate read B. Bold boxes indicate patterns that confirm the overlap, if the following criteria are met: Δd = |dAmid – dBmid| ≤ µmid; dBstart ≤ µend; dAend ≤ µend.
A sequence S′ is said to be a v-neighbor of a sequence S, if S′ differs in at most v positions compared to S, i.e. the Hamming distance (7) between S′ and S is at most v. A pattern P = {q1, q2,…, qn} is defined as ngrams concatenated q-grams, words of length q. Thus, the pattern length |P| = q · ngrams. A read A is said to overlap with a read B if there exists two patterns PA1 and PA2 in A and two patterns PB1 and PB2 in B, fulfilling the following criteria: (i) each qi in PA1 and the corresponding qi in PB1 are v-neighbors, and analogously in PA2 and PB2. (ii) The difference in distances Δd = |dAmid – dBmid| ≤ µmid, where dAmid is the distance between the first index of PA1 and the first index of PA2, and dBmid is the distance between PB1 and PB2. (iii) The distances dAstart ≤ µend or dBstart ≤ µend, where dAstart is the distance between the start index of PA1 and the beginning of A, and correspondingly for B. The overlap thus begins in one of the reads. (iv) The distances dAend ≤ µend or dBend ≤ µend, where dAend is the distance between the end index of PA1 and the end of A, and correspondingly for B. The overlap thus ends in one of the reads.
The construction of a raw multiple alignment begins with a starting read Rs, which is divided into non-overlapping windows, Wk·q, k = 0, 1, 2,…, |Rs|/q where Wi corresponds to the window that starts in position i in the starting read and is of length q. Since q generally is greater than 10, the division into non-overlapping windows reduces the number of computational operations in this step by at least an order of magnitude compared to the number of operations needed if a sliding window approach was used. All v-neighbors of all windows Wk·q of Rs are computed and constitute the candidate multiple alignment. All v-neighbors belonging to the same read are grouped. Each group is analyzed to determine whether it fulfils the overlap criteria stated above, i.e. if the v-neighbors can be organized into patterns and the patterns meet the distance constraints.
The margin µend is needed, since the starting read is divided into non-overlapping windows. Without the margin, the starting index of approximate matches at the start and end of the overlaps would have to be located at multiples of q in order to be found. In addition, our method for retrieving v-neighbors (described below) only considers base substitutions in the location of approximate matches. This leads to undetected overlaps where one or both of the reads contain insertions and deletions (indels) at the beginnings and ends. The use of a margin µend reduces this effect. The margin µmid is needed in order to allow indels in one or both of the reads between the first and last patterns, since the distance between the patterns in each read may not be equal if the reads have indels along the alignment.
Optimization of raw multiple alignments
It is crucial for both the DNP analysis and the subsequent error correction step that the multiple alignment is locally optimized. This is done using the ReAligner algorithm (8).
The multiple alignment is further refined by masking unique parts of reads that originate from boundaries between unique and repeated parts of the target sequence. When unique parts of reads are present in a multiple alignment together with repeats, the DNP method will correctly identify these parts as unique and assign a large number of DNPs in the unique parts. However, the probability of assigning false DNPs in the repeat reads that overlap the unique parts increases with increasing number of DNPs in unique parts. It is therefore desirable to mask these unique parts, while keeping the repeat part of the read in the multiple alignment for analysis by the DNP method.
We perform a pair-wise comparison of all reads against the starting read Rs by sliding a window of length lw along Rs. In each window, the number of mismatches between Rs and an overlapping read Ri currently being investigated is computed. The number of observed mismatches is compared to the expected number of mismatches according to the following model: the expected number of errors in each sequence window εc, εi, is the sum of Phred error rates. The expected number of mismatches is the sum of the expected number of errors, E = εc + εi. We approximate the distribution of errors in each window using the Poisson distribution. The result is that the number of mismatches is distributed under Poisson with the expectation value E in this model. To allow reads originating from different repeat copies in the same multiple alignment, we adjust the number of observed mismatches, nobs by subtracting the number of mismatches expected to arise due to single base differences between repeat copies, nadj = nobs – lw · 2 · d, where d is the maximum difference and lw is the length of the window. The probability Pr of observing nadj or more mismatches in the window is computed as
where Po(i) is the probability function for a Poisson variable with mean E. If Pr is below a threshold Prmax, the current window is masked. After the filtering step, the multiple alignment is locally re-optimized.
Error correction
The statistical methods described in (4) for separation of nearly identical repeats can be employed to correct erroneous base calls in shotgun sequence reads. Since the statistical computations are thoroughly described in (4), we will not present them here. The statistics are based on an analysis of at least a pair of columns in a multiple alignment. The number of observed coinciding deviations from consensus is compared with what to expect from a Poisson distribution.
When all DNPs in the multiple alignments have been set, the error correction is performed. For every column with a minimum coverage cmin, the consensus base is computed as the most frequent base. In all columns where consensus can be unambiguously determined, all bases that differ from consensus and are not DNPs are corrected into the consensus base.
Iteration
Since the multiple alignments are constructed with regard to the starting reads, all possible information about the regions that these reads span is obtained. All similar regions in corresponding repeat copies will be present under the starting read, and hence completely analyzed. Reads extending to the left and right of the starting read will only be analyzed in part. Parts of reads that have been present under a starting read do not have to be analyzed again. The remaining parts that are not yet analyzed are used as starting reads and new multiple alignments are constructed dynamically using the longest un-analyzed read section remaining in the database as the starting read. The process is repeated until no fragments with an un-analyzed region longer than the minimum overlap length lminol remain in the database. Note that lminol ≥ 2 · |P|, since two patterns are required in the multiple alignment construction step.
Computation of approximate q-grams
Approximate string matching is one of the most widely studied subjects in computer science and several algorithms have been developed that have numerous applications in biology. A good introduction to the subject is given by Navarro (9). An example of an algorithm for retrieving inexact word matches using suffix trees is described by Sagot (10). Our algorithm is based on the distribution of the unique indices that can be computed for different q-grams. A q-gram is denoted by x1x2…xn…xq, where xn∈Σ = {A,T,G,C} and bi is the letter index, bA = 0, bT = 1, bG = 2 and bC = 3. If |Σ| = σ is the alphabet size, there are σq possible different q-grams. For each q-gram, we can compute a unique q-gram index, Iq using
so that 0 ≤ Iq < σq. To construct a database of input sequence reads Si, all indices Iq are computed for all possible q-grams of the input sequences, Si. The starting positions Si,j of each q-gram are stored into a table, τ, that is indexed by Iq. There are
starting positions, where k is the number of sequences.
The problem is to return all the v-neighbors of a q-gram Q, i.e. the set of all q-grams present in the table τ differing at most in v positions compared to Q. The potential number of such q-grams can be computed using
With increasing v, the number of q-grams potentially matching Q grows rapidly, whereas the number of different q-grams in the database remains constant and will not contain all possible combinations when σq is fairly large compared to the amount of in data. For this reason, it is advantageous to avoid computing indices Iq of q-grams that are not present in the database. This is achieved in two ways: (i) in table τ, at every Iq, a gram pointer is set to point to the next non-empty Iq. (ii) Instead of individually computing indices of every q-gram that potentially matches Q, a set of index intervals, Ilow – Ihigh, are determined that include the potentially matching q-grams. Using the intervals together with the gram pointers in the database table, it is possible to rapidly omit empty index regions, i.e. q-grams that do not exist in the database.
The simplest way to compute the locations of the intervals is to calculate them recursively as shown in Figure 2.
Figure 2.
Pseudo code for computing v-neighbor intervals for a given q-gram.
Every position pos along a q-gram is substituted. If more than one difference is allowed, positions to the right of pos are substituted in order, through a recursive function call with the corresponding index of the substituted q-gram. The for-loop at line 2 initially traverses the whole q-gram from left to right. The recursive function call at line 12 of find_neighbors introduces an additional substitution to the right of pos if more substitutions are allowed. The get_lower_and_upper_limits function calculates the upper and lower limit within which the Iq values differing only in the positions to the right of pos are located. An integer division at line 1, and the following multiplication at line 2, give the lowest Iq value having the same k bases as the substituted q-gram, where k = pos,…,q and sigma = σ, the alphabet size. Adding the same number to the lower limit gives the upper limit to that Iq. The step size is given by sigma∧(pos-1), and represents the distance between two adjacent Iq values differing only at position pos. The if-statement at line 4 checks the gram pointer at the lower limit of the interval to determine whether there are any q-grams present within the interval. If not, no further substitutions need to be made for pos. The compute_start_index function calculates the Iq value of having an ‘a’ in position pos. This is achieved through the modulo operation at line 1 and the addition of the lower limit of the Iq values. The Iq values obtained by substituting the bases at this position are calculated by adding the step_size to the Iq value at line 14 in the code. This is repeated in the while-loop at line 7 until the upper limit is reached. pick_interval retrieves all non-empty entries in an interval of length sigma∧diff_left by following gram pointers until the upper limit of the interval is reached.
The intervals Ilow – Ihigh depend on q and v and have different lengths as follows. For all v, v = 0, 1,…, q – 1, there are
intervals of length σv–k respectively.
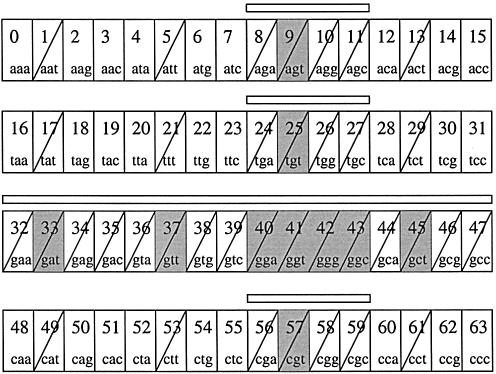
The prototype implementation uses this recursive method to find the intervals. An example of the locations of the different intervals for a q-gram ‘ggt’ with corresponding index Iq = 41, with σ = 4 and q = 3, is shown in Figure 3 when v = 1 and v = 2, respectively. For v = 1, one interval of length four and six intervals of length one are distributed over τ. For v = 2, one interval of length 16, three of length four and nine of length one are present.
Figure 3.
An example of the different intervals for ‘ggt’, Iq = 41, when q = 3 and σ = 4. The gray boxes indicate indices for v = 1. Over-struck boxes indicate indices that have v = 2. Bars indicate contiguous intervals when v = 2.
Time complexity
The time complexity of the error correction method is linear with respect to the number of sequences in the dataset. Also, the method is linear with respect to the number of repeats in the target sequence. We argue that the number of multiple alignments required does not grow with the number of repeat copies in the target sequence, and that the number of sequences in each multiple alignment grows linearly with the number of repeats. Error correction is mainly needed in the case of repeated sequences, and it is important that the running time of the method is linear under these conditions.
The following definitions are used when discussing the time complexity of the method: the length L of the target sequence; the number of sequences n; the mean length l of the sequence reads; the shotgun coverage c; the repeat copy number r ≥ 1, where r = 1 denotes entirely non-repeated sequence, r = 2 denotes a target sequence consisting of two instances of a sequence of length L/2, etc.; the repeat copy length rlen = L/r. For simplicity, only non-repeated and entirely repeated target sequences are considered when discussing time complexity. Since L = n · l/c, and L = r · rlen, it is sufficient to express the time complexity in terms of r when l, c and rlen are constant.
The time complexity Ctot for analyzing all reads in a dataset depends on the number of constructed multiple alignments nal and the time complexity Cal for constructing one multiple alignment so that
Ctot ∝ nal · Cal.
One multiple alignment can be constructed in linear time depending on the number of candidate overlaps nr of the starting read, i.e. Cal ∝ nr, and thus
Ctot ∝ nal · nr.
Using the above definitions, nal ∝ L ∝ r. Note that this is under the assumption that the number of words in the database nτ << σq. nr consists of the true and apparent overlaps of the starting read, as well as false overlaps. It is evident that the number of true and apparent overlaps in a multiple alignment increases linearly with r. The number of false overlaps increases linearly with r in the worst case, when nτ approaches σq. In this case, a greater q must be chosen. Thus, nr ∝ r, and
Ctot ∝ r2.
We can reduce this quadratic dependence to linear by marking the starting read and the sections of other reads present under the starting read as analyzed. Only un-analyzed fragments of reads are then used as seeds for new multiple alignments. This reduces the running time approximately by a factor c, the shotgun coverage. Construction of a single multiple alignment is still proportional to r, but the amount of analyzable sequence remaining after the analysis is additionally reduced by r in each subsequent analysis step. In other words, nal ∝ r/r. Thus,
Ctot ∝ r2/r = r ∝ L ∝ n3
The ReAligner algorithm, which is applied after constructing the raw multiple alignment, is linear (7). Therefore, the total time complexity is proportional to the number of reads in the data set if q is properly chosen.
RESULTS
We have developed a method to correct sequencing errors in shotgun sequence reads using DNPs, as well as a novel algorithm to rapidly and efficiently find overlaps that are used to construct the multiple alignments required for the DNP analysis. In order to test the performance of the method, a prototype software, MisEd, was implemented in C++ and run on a set of simulations, as well as on real shotgun data. The simulated shotgun projects were constructed by the gen_seq and sim_gun programs, which were developed in house for testing fragment assembly programs (4). The gen_seq program produces a DNA sequence with specified characteristics. The sequence is read by the sim_gun program that simulates the shotgun process. Sequence error probabilities can be imported from real shotgun project quality files, or a simple flat error rate can be used.
Detection of DNPs and error correction
A number of simulated shotgun sequencing projects were constructed and MisEd was run on these sequences to correct the errors. All simulations were repeated 10 times with different seeds for the gen_seq and sim_gun programs. In each simulation set, the sequencing error rates were taken from real shotgun project quality files. The resulting mean quality of the simulated shotgun sequence reads was 96.3% and the mean read length 539 bases after trimming, yielding an average coverage of 9×. The maximum error rate allowed in the trimming step was 11%. The simulated sequencing errors introduced by the sim_gun program consisted of 80% substitutions, 10% insertions and 10% deletions. The simulation sets differed in the number of repeat copies, the repeat copy length, the length of flanking unique sequence and the presence of single base differences between repeat copies. When single base differences were present in the target sequence, the difference between any two repeat copies was 1.0%. All repeat copies were positioned in tandem. Table 1 shows the characteristics of the simulated projects.
Table 1. The simulation sets used to test error correction.
| Sim. no. | Repeat copy no. | Repeat copy length | Unique seq. length | Repeat diff. (%) |
|---|---|---|---|---|
| 1 | 0 | n/a | 150 000 | n/a |
| 2 | 10 | 2000 | 0 | 0.0 |
| 3 | 10 | 2000 | 0 | 1.0 |
| 4 | 10 | 2000 | 2000 | 0.0 |
| 5 | 10 | 2000 | 2000 | 1.0 |
| 6 | 50 | 2000 | 0 | 1.0 |
| 7 | 30 | 2000 | 0 | 1.0 |
| 8 | 6 | 10 000 | 0 | 1.0 |
| 9 | 6 | 10 000 | 40 000 | 1.0 |
Each set was run 10 times with different seeds for the gen_seq and sim_gun programs.
The following set of parameters was used in all runs. Trimming: lw = 35, sw =3, nw = 4, εmax = 0.11, lminhq = 50. Construction of multiple alignments: q = 12, v = 2, ngrams = 2, µend = 2·q=24, µmid = 2·q=24, lminol = 4·q = 48. Multiple alignment filtering: lw = 24, d = 0.05, Prmax = 0.001. DNP analysis: Pmaxtot = 0.001, Dmin = 3, where Dmin is the minimum number of deviating bases on a column for it to be analyzed as a DNP candidate, and Pmaxtot is the probability threshold for DNP assignment. Error correction: cmin = 3. All test were performed on a 933 MHz Intel Pentium III PC with 1.5 GB of RAM running Linux RedHat 7.1.
The results of the error correction are shown in Table 2. In all simulated sets the read quality increased after error correction, from the initial 96.3% to at least 99.7%. The major influence on performance is whether single base differences between repeat copies are present or not. The overall performance is lower in the simulation sets containing single base differences. This is mainly due to the fact that the DNP method produces false positives as well as false negatives. For example, in simulation set 3, the percentage of detected DNPs was 84, while the remaining 16% of the number of single base differences were falsely corrected. In addition, 74 (1.1%) false DNPs remained uncorrected. This is also true for sets 5 and 6.
Table 2. The results of the error correction method.
| Sim. no. | Errors corr. (%) | Qual. after corr. (%) | Detected DNPs (%) | False DNPs (%) | Analyzed region (%) |
|---|---|---|---|---|---|
| 1 | 99.30 | 99.97 | n/a | 0 (n/a) | 89 |
| 2 | 99.35 | 99.98 | n/a | 12 (100) | 96 |
| 3 | 95.01 | 99.81 | 84 | 74 (1.1) | 96 |
| 4 | 99.13 | 99.97 | n/a | 38 (100) | 95 |
| 5 | 95.55 | 99.84 | 86 | 189 (2.5) | 95 |
| 6 | 92.56 | 99.73 | 73 | 862 (2.7) | 97 |
| 7 | 93.51 | 99.76 | 75 | 320 (1.7) | 97 |
| 8 | 95.96 | 99.85 | 87 | 181 (0.8) | 97 |
| 9 | 97.17 | 99.89 | 86 | 207 (0.95) | 97 |
For each simulation, the following is reported: percentage of errors corrected, average sequence quality after error correction, percentage of DNPs detected in reads, total number of false positives (and percentage) and the percentage of region that has been analyzed.
The difference between sets 3 and 5 is that set 5 contains flanking unique sequence as well as repeats, while set 3 only contains repeats. This corresponds to an increase in the amount of false DNPs in set 5. Further analysis of set 5 showed that the majority of the additional false DNPs compared to set 3 (93%) had been assigned to unmasked unique parts of reads that originated from boundaries between unique and repeat parts of the target sequence, that the multiple alignment filtering step had overlooked. While these ‘false’ DNPs did not represent single base differences between repeat copies, they were rightly identified as unique when present in multiple alignments together with repeats. When these ‘false’ DNPs were excluded from the analysis, the false positive rate dropped to 1.6% in simulation set 5. This effect was not observed in set 9.
Error correction was performed on columns having a coverage of three or more bases. Furthermore, a minimum overlap length is required in order for a read to be included in a multiple alignment. Since each analysis step may leave parts of reads un-analyzed, some of these remaining parts may be shorter than the minimum overlap length specified. These parts, as well as the low coverage parts, will not be analyzed. This effect is shown in Table 2, where the different percentages of the analyzed regions are shown.
Comparison of performance on real and simulated data
The quality of the simulations was investigated by performing error correction on both real and simulated data. Two manually curated, 16 711 bp (contig C1) and 30 983 bp (contig C2) contigs from a Trypanosoma cruzi genomic clone (data available at: http://cruzi.cgb.ki.se/mised.html), consisting of 252 and 469 reads, respectively, were used as the real dataset. These contigs did not contain repeats. Simulated datasets were constructed by using the consensus sequences of these two real contigs as template sequences. The quality values from the real datasets were used in the simulations, and each simulation was performed three times. The parameters used in these runs were identical to the previous ones, except that v = 3 was used. The results are shown in Table 3. As expected, the results for simulated data were not identical to the results of real data. Although sim_gun attempts to simulate shotgun sequencing as accurately as possible, some effects are not accounted for. For example, sim_gun distributes the sampled reads randomly along the target template, whereas this is not always the case in real sequencing projects. Another contributing factor is that in simulated data, the quality values correspond to actual error rates, whereas in real data, quality values are approximated from the shapes of electropherograms. For these reasons, a slightly lower performance of MisEd is expected when real data are used. For both contigs in this comparison, a decrease in performance was observed. The performance dropped 7.2% in contig C1 and 2.1% in the longer contig C2.
Table 3. Results of the comparison between real and simulated shotgun data.
| Contig | Quality before | Errors corrected | Analyzed region |
|---|---|---|---|
| C1 real | 96.6 | 90 | 89 |
| C1 sim | 96.7 | 97 | 86 |
| C2 real | 97.0 | 95 | 89 |
| C2 sim | 96.6 | 97 | 85 |
Comparison of MisEd and EULER
The EULER assembler (2) is an example of an assembly program that includes an error correction step. This correction algorithm identifies sequencing errors by locating words of varied length and correcting the words that are present in a multiplicity below a predefined threshold. The performance of MisEd was compared to that of EULER’s error correction for simulated shotgun data. EULER error correction was chosen for comparison with our method because it is, to our knowledge, the only stand-alone error correction program available.
The simulated datasets were constructed by using the consensus sequences of the manually curated contigs C1 and C2 as templates and the corresponding quality files for error simulation. Repeated datasets were also constructed using the quality values from C2. These sets consisted of repeat copies of length 5000 bp, repeated six times in tandem and differing 1% between any two repeat copies. Each simulation was performed three times. Two different stringencies in trimming were used in the simulations, allowing a maximum error of 11 and 3%. This yielded average qualities of 96.5 and 98.9%, and shotgun coverages of 9× and 6×, respectively (Table 4).
Table 4. Simulations used in comparison of MisEd and EULER.
| Sim no. | Shotgun coverage after trimming | Quality after trimming (%) |
|---|---|---|
| 10 | 9 | 96.7 |
| 11 | 9 | 96.6 |
| 12 | 6 | 99.0 |
| 13 | 6 | 98.9 |
| 14 (rep) | 9 | 96.5 |
| 15 (rep) | 6 | 98.9 |
The results of the comparison between MisEd and EULER are shown in Table 5. In all cases, MisEd performs better than EULER. The difference in performance is lower under more stringent trimming conditions. This is expected, since a prerequisite of EULER is that the input data are trimmed to 97–99% quality. The results also show that the difference in performance for repeated data is similar to that for non-repeated data.
Table 5. Results of the comparison between EULER and MisEd.
| Sim no. | False negatives/positives; Euler (%) | False negatives/positives; MisEd (%) | Errors reduced total; EULER (%) | Errors reduced total; MisEd (%) | Analyzed region; MisEd (%) | False negatives analyzed region; MisEd (%) | Errors reduced analyzed region; MisEd (%) | Errors in un-analyzed region; MisEd (%) |
|---|---|---|---|---|---|---|---|---|
| 10 | 38/1.3 | 17/0.5 | 61 | 83 | 86 | 3 | 97 | 15 |
| 11 | 37/1.4 | 16/0.5 | 62 | 83 | 85 | 2 | 97 | 14 |
| 12 | 12/1.7 | 9/0.2 | 86 | 91 | 88 | 1 | 98 | 8 |
| 13 | 12/1.5 | 10/0.1 | 87 | 90 | 88 | 2 | 98 | 8 |
| 14 (rep) | 36/5.7 | 10/4.5 | 61 | 87 | 88 | 2 | 94 | 8 |
| 15 (rep) | 11/27.9 | 7/18.4 | 69 | 79 | 89 | 3 | 82 | 5 |
Simulations 10–13 consist of non-repeated data. Simulations 14 and 15 consist of repeated data. In simulations 10, 11 and 14, a maximum error of 11% was allowed in trimming. In simulations 12, 13 and 15, a maximum error of 3% was allowed.
Simulations 14 and 15 indicate that the performance of MisEd drops with increased trimming, whereas the result in EULER is the opposite. This may be explained by the fact that the DNP method is less sensitive with more stringent trimming, i.e. lower coverage and shorter reads. The undetected DNPs will thus be erroneously corrected. This effect cannot be seen in non-repeated data, since it does not contain DNPs.
MisEd provides information concerning the parts of reads that have been analyzed for error correction. This is not the case for EULER. The results of MisEd are markedly improved if the actually analyzed parts of reads are considered, as can be seen in Table 5.
The distribution of errors in reads was investigated for simulations 14 and 15. In simulation 14, 63% of the reads contained at most one error after MisEd error correction. In simulation 15, the corresponding result was 72%. After EULER correction, 28 and 60% of the reads contained at most one error. When considering only the parts actually analyzed by MisEd, the results for simulations 14 and 15 were 73 and 80%, respectively.
Performance in overlap detection
The performance of the multiple alignment construction method was investigated. Simulated shotgun sequencing was performed five times on a 100 kb random sequence, using the quality file from contig C2 with 20% of the introduced errors as indels. A maximum error of 11% was allowed in trimming. The average sensitivity was 93.1%, while the specificity was 100.0%. These results take the minimum overlap length into account.
Running time
The pattern matching algorithm was tested separately to observe the effect on running time for different numbers of substitutions, v. This was performed by simulating shotgun sequencing with 9.8× coverage on a random target sequence of length 300 kb. For each read, all v-neighbors of every q-gram present in the read were located for different v. The running time of locating all the v-neighbors after database construction was measured. No multiple alignments were constructed and no error correction was performed.
Table 6 shows the results for different choices of v. All tests were repeated five times and the average time after subtraction of the average time needed to construct the database, 19.53 s, was reported. The results showed that the running time increased with increasing v, but also that the number of v-neighbors found grew at a significantly higher rate. This is clearly seen in the last column, where an increase in the number of v-neighbors detected per second was observed for greater values of v. This behavior is expected, since the lengths and the number of intervals increase with increasing v (equation 2). Using the gram pointers, longer stretches can be omitted when locating the v-neighbors.
Table 6. Running times for the pattern matching algorithm for q = 12 and different v.
| v | No. neighbors | No. neighbors found | Avg. running time (s) | v-neighbors/s |
|---|---|---|---|---|
| 0 | 1 | 246 000 | 0.82 | 300 000 |
| 1 | 37 | 2 347 258 | 4.2 | 560 000 |
| 2 | 631 | 28 288 787 | 43 | 660 000 |
| 3 | 6571 | 281 685 270 | 360 | 780 000 |
| 4 | 46 666 | 1 988 512 822 | 2200 | 900 000 |
For each choice of v, the following is reported: the potential number of v-neighbors for each q-gram (computed using equation 1), the number of v-neighbors present in the dataset, the average running time after subtraction of the average time needed to construct the database (19.53 s), the number of v-neighbors found per second.
The running time of the prototype error correction software, MisEd, was tested with regard to sequence length and repeat copy number. The results are shown in Figure 4. The parameters were the same as in the simulation sets above. Our implementation runs in seemingly linear time as expected from equation 3. For sequences containing repeats, the running time increases. This is expected, since the size of the multiple alignments grows linearly with increasing repeat copy number and the local optimization of multiple alignments is the most computationally expensive step in the algorithm.
Figure 4.
Running times of MisEd for increasing sequence lengths and repeat copy numbers. The contiguous line represents non-repeated sequence, the dashed line represents simulations with a constant repeat copy number of 10 and varied length, the dotted line represents simulations with varied copy number and a constant copy length of 10 000 bases.
DISCUSSION
We have designed a method for error correction in shotgun sequence fragments. The algorithm utilizes the DNP method to avoid correcting single base differences between repeat copies in the target sequence. A prototype software, MisEd, that implements this method has been developed. The program reads shotgun sequences and quality files in FASTA format, and the output is a corrected set of sequences and a list of detected single base differences between repeats.
Different simulation sets have been chosen to assess the performance of the method for various kinds of shotgun target sequences: completely non-repeated, partially repeated and entirely repeated. For the sets containing repeat elements, the performance has been tested for both identical repeats and repeats differing 1% between any two repeat copies. The difference of 1% was chosen in order to test the limit of the method. The comparison of simulated data with real data indicates that the performance of the error correction method on real data is well estimated by the use of simulated data.
After error correction, the quality is above 99.7% in all cases. Generally, the quality is somewhat lower in datasets that contain differences between repeat copies. This is explained by the fact that the method will correct all the single base differences that the DNP method has failed to detect, and will not correct erroneously detected DNPs. In other words, the false positives in the DNP method will be errors left uncorrected, and the false negative DNPs will be errors introduced by the method. This is a minor effect that will not affect the utility of this method for shotgun assembly. Furthermore, in some cases the multiple alignment filtering method will not filter out all the unique parts of reads sampling boundaries between repeated and unique regions in the target sequence. However, the assignment of ‘false’ DNPs in these regions does not pose a problem, since these positions do not constitute errors that should be corrected.
It is necessary to use inexact matching in order for this method to be sufficiently sensitive in the location of candidate overlaps for the raw multiple alignment, while maintaining high specificity. The high specificity is desirable, since as little time as possible should be spent in the most computationally expensive step, optimization of the multiple alignments. A high sensitivity is also desirable, since the DNP method requires that both true and apparent overlaps are present in the multiple alignment, and its performance increases with the amount of information present.
Since the error rates in a read increase towards the ends, and we trim the reads as little as possible, there is little probability of finding overlaps with our approach if exact matches are used. Also, single base differences between repeat copies further decrease the probability of finding exact matches. We have examined the possibility of using exact matching instead of approximate. Two approaches were considered. The first was to use our method, but with exact matches. In order to achieve a similar sensitivity as in our method with inexact matches in simulated, random sequence (100 kb, 10× coverage), the length of the exact match could not exceed 4 bases. This typically yielded a specificity below 1%. On the other hand, in order to achieve a similar specificity, a match length of at least 12 was required. This in turn yielded a sensitivity of only 40%.
An alternative method is to find candidate overlaps by locating exact matches anywhere along the potential overlap. In this case, a large number of overlap candidates need to be assessed in order to weed out false matches due to short repeats such as microsatellites. A common method, used by e.g. Phrap that we have previously used in TRAP (5), is to extend the exact match by dynamic programming. Even in simulated data without microsatellites, the trade-off between sensitivity and specificity before dynamic programming using this approach is similar to the one above. Using a word length of 15 in order to obtain a specificity over 99% resulted in 60% sensitivity. On the other hand, obtaining a sensitivity above 93% yielded a specificity below 1% at word length 7. In practice, more than 99% of all dynamic programming assessments were performed on false overlaps in the latter case. This time consuming step is not needed in our approach, since we use approximate matches both to locate candidates as well as to verify them. In the above example with matchlength 7, the overlap phase in Phrap was five to 10 times slower than our method. In this example we did not mark any of the reads analyzed, which would further decrease the running time of our method.
The linear time complexity described above relies on the assumption that the multiple alignments can be constructed in linear time. In order to achieve this, the index word size q must be chosen properly. Another way to accomplish linear time alignment construction is to pre-process the input data by computing all pair-wise overlaps between the reads in the dataset. This approach, used in Arachne and TRAP, requires a quadratic pre-processing step [O(r2)] prior to construction of multiple alignments. The time needed for construction of one multiple alignment is in this case proportional to c · r. The method that we present also performs the construction of multiple alignments in O(c · r) time, but does not require prior determination of pair-wise overlaps. The actual construction of one multiple alignment will in practice be slower, but the quadratic pre-processing step has been eliminated.
The purpose of applying an error correction program to a set of shotgun sequence fragments is to produce better results in a subsequent assembly. Sequencing errors mainly cause problems if the target sequence is repeated. If this is not the case, most assembly algorithms will readily produce accurate results despite the presence of sequencing errors. However, when the target sequence is repeated and the repeats are nearly identical, the sequencing errors will, to the common assembly algorithms, be indistinguishable from single base differences. Since most genomes contain many repeats, it is of great importance that the input sequences are as error free as possible.
How the corrected dataset should be utilized is dependent on the assembly program used to assemble the corrected sequences. Assembly programs use different strategies to handle repeats. Some applications use quality values as a primary means to separate repeats, while other programs rely on other strategies. An example of a program that relies heavily on quality values is Phrap, which computes likelihood ratios to determine whether mismatches in pair-wise overlaps between reads are due to sequencing errors or single base differences between repeats. In order to achieve better results using Phrap after error correction with MisEd, the quality values of the bases should be modified to reflect the quality of the corrected sequence. We are currently investigating how the quality values of the corrected sequences should be modified in order to fit the repeat separation model used by Phrap.
The EULER assembler does not rely on quality values, and the algorithm itself expects error free data as input. Therefore, an error correction method is already incorporated into EULER. The results of the comparison of MisEd and EULER error correction show that by employing the regions of the reads that MisEd actually analyzes, a better result can be obtained. The best result for EULER was in simulation 13, where 87% of the errors were corrected at a coverage of 6× after trimming. This can be compared to the results of MisEd in simulation 11, with less trimming and a resulting higher coverage of 9×, where 83% of the errors were corrected. If only the analyzed parts are considered (85%), the effective coverage is 7.7× with 97% of the errors corrected. The advantage of MisEd is thus that a higher accuracy can be achieved while still maintaining a higher coverage in a subsequent assembly step. As shown, MisEd produces a larger number of almost error free reads, i.e. reads that contain at most one error. We predict that MisEd is readily usable as an error correction step prior to assembly with EULER.
Arachne is a recently developed assembly program that also contains an error correction step before assembly. Like MisEd, Arachne constructs multiple alignments and locates deviations from consensus as candidates for correction. However, columns are only considered one at a time, which results in low sensitivity regarding detected single base differences if a high specificity is to be maintained, as shown in (4). Using MisEd before assembly, the performance of Arachne will probably be improved.
It will be important to test this method in practice. However, a thorough investigation of the performance of different assembly algorithms when presented with error-corrected data is a major undertaking and outside the scope of this paper.
CONCLUSIONS
The method of error correction presented here exploits two novel approaches. The first is that the DNP method can be used to correct errors in shotgun sequence data. Since sequencing errors mainly cause complications in the presence of highly similar repeats, it is essential to avoid correcting single base differences between such repeat copies. The DNP method is a way to assert that a minimal number of real differences are erroneously corrected.
Secondly, a method has been developed for rapid construction of the multiple alignments needed by the DNP method. The key for this step is a rapid algorithm for locating v-neighbors of q-grams. Recursive computation of the intervals where v-neighbors of a certain q-gram can be located, and the use of gram pointers, allows for rapid retrieval of inexact matches present in the database. Multiple alignments can subsequently be constructed dynamically using q-grams, without previous pair-wise matching of sequence reads. The algorithm is made faster by only allowing un-analyzed sections of reads in consecutive multiple alignments.
The MisEd program is available from the authors upon request. We are currently incorporating the multiple alignment construction method into a new version of our in-house assembly program.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Daniel Nilsson for valuable comments. This study was supported by The Swedish Technology Research Council, Ulf Petterson and the Beijer Foundation, NIAID (5 U01 AI 45061) and the Knut and Alice Wallenberg Foundation.
REFERENCES
- 1.Eichler E. (1999) Repetitive conundrums of centromere structure and function. Hum. Mol. Genet., 8, 151–155. [DOI] [PubMed] [Google Scholar]
- 2.Pevzner P.A., Tang,H. and Waterman,M.S. (2001) An eulerian path approach to dna fragment assembly. Proc. Natl Acad. Sci. USA, 98, 9748–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batzoglou S., Jaffe,D., Stanley,K., Butler,J., Gnerre,S., Mauceli,E., Berger,B., Mesirov,J. and Lander,E. (2002) ARACHNE: a whole-genome shotgun assembler. Genome Res., 12, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tammi M.T., Arner,E., Britton,T. and Andersson,B. (2002) Separation of nearly identical repeats in shotgun assemblies using defined nucleotide positions, DNPs. Bioinformatics, 18, 379–388. [DOI] [PubMed] [Google Scholar]
- 5.Tammi M.T., Arner,E. and Andersson,B. (2003) TRAP: Tandem repeat assembly program, produces improved shotgun assemblies of repetitive sequences. Comput. Methods Programs Biomed., 70, 47–59. [DOI] [PubMed] [Google Scholar]
- 6.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 7.Hamming R.W. (1950) Error-detecting and error-correcting codes. Bell Syst. Tech. J., 29, 147–160. [Google Scholar]
- 8.Anson E.L. and Myers,E.W. (1997) Realigner: a program for refining DNA sequence multi-alignments. J. Comp. Biol., 4, 369–383. [DOI] [PubMed] [Google Scholar]
- 9.Navarro G. (2001) A guided tour to approximate string matching. ACM Comput. Surveys, 33, 31–88. [Google Scholar]
- 10.Sagot M.-F. (1998) Spelling approximate repeated or common motifs using a suffix tree. In Lucchesi,C.L. and Moura,A.V. (eds), Proceedings of the 3rd Latin American Symposium, Campinas, Brazil. Springer-Verlag, Berlin, Number 1380, 374–390. [Google Scholar]