Abstract
Small interference RNA (siRNA) is an emerging methodology in reverse genetics. Here we report the development of a new tetracycline-inducible vector-based siRNA system, which uses a tetracycline-responsive derivative of the U6 promoter and the tetracycline repressor for conditional in vivo transcription of short hairpin RNA. This method prevents potential lethality immediately after transfection of a vector when the targeted gene is indispensable, or the phenotype of the knockdown is lethal or results in a growth abnormality. We show that the controlled knockdown of DNA methyltransferase 1 (DNMT1) in human cancer resulted in growth arrest. Removal of the inducer, doxycycline, from treated cells led to re-expression of the targeted gene. Thus the method allows for a highly controlled approach to gene knockdown.
INTRODUCTION
Chemically synthesized, 21–23 nucleotide double-stranded (ds) RNAs have enabled the application of reverse genetic methodologies to mammalian cells which were previously only available in nematode and fruit fly systems. The precise mechanism of small interference RNA (siRNA) function is not known; however, targeted genes are suppressed and this approach has been widely used. Until recently, three methodologies for siRNA have been available; chemically synthesized dsRNAs (1), ds/hairpin-structure RNAs transcribed in vitro (2) and ds/hairpin-structure RNAs transcribed in vivo from plasmid vectors (3–7). Whether synthesized in vitro or in vivo, hairpin siRNA especially with a single mismatch is thought to be processed to small temporal RNA and inhibit translation, whereas double-stranded siRNA is thought to induce targeted RNA degradation (8–10). Hairpin siRNAs transcribed from a vector are thought to suppress expression of targeted genes more efficiently than double-stranded siRNA (2).
The plasmid vector system for siRNA has advantages over the other methods. First, less expensive DNA oligomers can be utilized for vector construction; secondly, more efficient transfection is expected with a selection marker; and thirdly, the inducible transcription of siRNA is theoretically possible using a modified promoter. An RNA polymerase III (pol III) promoter, such as the U6 promoter or H1 promoter, is commonly used for in vivo transcription of siRNA, since pol III transcription is terminated by a stretch of four or five thymidines and this accurate termination is important in avoiding a non-specific reaction by transcription of another region of vector sequence. Ohkawa and Taira (11) described the successful regulation of expression by the integration of the bacterial tetracycline operon sequence (tetO) into the U6 promoter. The tetO sequence in the modified U6 promoter is bound by a tetracycline repressor (tetR) in the absence of tetracycline or a more stable derivative such as doxycycline, and addition of doxycycline inhibits tetR binding to tetO sequences (12), thus allowing transcription. We have applied this tetracycline-inducible U6 promoter for the in vivo transcription of siRNA which enables stable transfection followed by the conditional expression of siRNA.
MATERIALS AND METHODS
Construction of DNA methyltransferase 1 knockdown vector
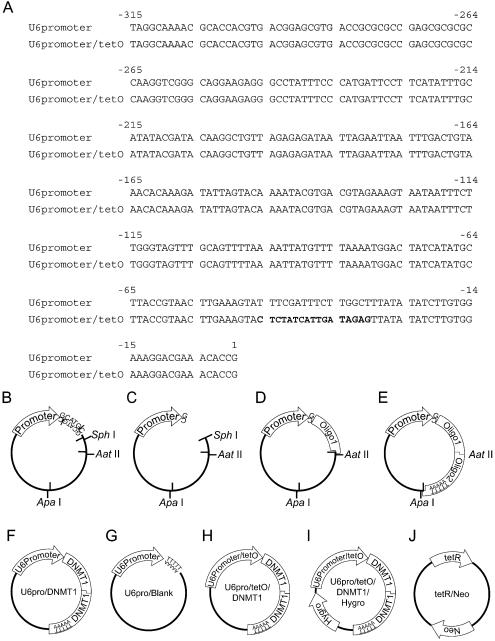
The human U6 promoter was amplified by PCR from the genomic DNA of T24 cells with the primers U6proTop (5′-AGGCAAAACGCACCACGTGACGGAG-3′) and U6proBottomSphI (5′-GCATGCGGTGTTTCGTCCTTTCCACAAGAT-3′), then ligated into a TA cloning vector, pGEM-T Easy (Promega, Madison, WI) (Fig. 1B). The plasmid was then digested with SphI (New England Biolabs, Beverly, MA) and blunted with Klenow fragment (New England Biolabs) (Fig. 1C). Oligo1 [a mixture of two DNA oligomers, DNMTO1F (5′-GGAATGGCAGATGCCAACAGGACGT-3′) and DNMTO1R (5′-CCTGTTGGCATCTGCCATTCC-3′)] was ligated following restriction digestion with AatII (New England Biolabs) (Fig. 1D). Oligo1 consists of a targeted DNA methyltransferase 1 (DNMT1) sequence (3) (nucleotides 1627–1647 of GenBank accession no. XM_ 086566) with an AatII recognition sequence. Oligo2 [a mixture of DNMTO2F (5′-CCTGTTGGCATCTGCCATTCCCTTTTTGGGCC-3′) and DNMTO2R (5′-CAAAAAGGGAATGGCAGATGCCAACAGGACGT-3′)] was ligated after sequence analysis and AatII–ApaI (New England Biolabs) digestion (Fig. 1E). The resulting plasmid, which contains the U6 promoter and codes for hairpin siRNA against DNMT1, was designated as U6pro/DNMT1 (Fig. 1F). As a control, we ligated blank oligos, BlankF (5′-TTTTTGAATTCGACGT-3′) and BlankR (5′-CGAATTCAAAAA-3′), instead of Oligo1. The resulting plasmid was designated U6pro/Blank (Fig. 1G). For the tetracycline-inducible promoter (Fig. 1A), the U6 promoter was modified by PCR with primers U6proTop and U6proBottomTetO (5′-GCATGCGGTGTTTCGTCCTTTCCACAAGATATATAACTCTATCAATGATAGAGTACTTTCAAGTTACGGTAAGC-3′) from the U6 promoter which was obtained from the above procedure. After confirmation of the correct sequence, we applied the same procedures described above to construct an inducible knockdown vector. The resulting plasmid was designated U6pro/tetO/DNMT1 (Fig. 1H). U6pro/tetO/DNMT1 was digested by ApaI, and a DNA fragment that encoded the hygromycin resistance gene was ligated into the plasmid (U6pro/tetO/DNMT1/Hygro, Fig. 1I). The tetR coding region was amplified from the Inv110 (Invitrogen, Carlsbad, CA) bacterial genome, which contains Tn10, by PCR using primers sense (5′-ATGTCTAGATTAGATAAAAGTAAAGTGATTAAC-3′) and antisense (5′-TTAAGACCCACTTTCACATTTAAGTTGT-3′). The PCR product was ligated into pGEM-T Easy and the sequence was confirmed by sequence analysis. The tetR gene was spliced out with NotI and ligated into the NotI site of an expression vector, pcDNA3.1 (Invitrogen). The resulting plasmid was designated tetR/Neo (Fig. 1J). Vector requests should be addressed to D. Takai (dtakai-ind@umin.u-tokyo.ac.jp).
Figure 1.
(A) Sequence comparison of the human U6 promoter and the modified U6 promoter with the bacterial tetracycline operon (tetO). The modified portion is indicated in bold. (B–E) Schematic illustrations of the vector construction process. (B) The U6 promoter sequence with an SphI recognition sequence (GCATG|C) was ligated into a TA cloning vector. (C) To preserve the first transcribed nucleotide as guanosine, the plasmid was digested with SphI and blunted with Klenow fragment. (D) To avoid hairpin formation of integrated sequences (3), DNA oligomers were ligated in two separate ligation reactions. Oligo1 was ligated following AatII digestion. (E) Oligo2 including the transcriptional termination signal T5 was ligated following AatII and ApaI digestion. (F–J) Schematic maps of vectors used in this study. (F) U6pro/DNMT1. DNMT1-targeting hairpin siRNA is driven by the human U6 promoter. (G) U6pro/Blank. As a control, the transcriptional termination signal T5 was ligated immediately downstream of the human U6 promoter. (H) U6pro/tetO/DNMT1. DNMT1-targeting hairpin siRNA is driven by the modified U6 promoter regulated by tetracycline. (I) U6pro/tetO/DNMT1/Hygro. U6pro/tetO/DNMT1 with the hygromycin resistance gene. (J) tetR/Neo. The tetR coding region was ligated in an expression vector with the neomycin resistance gene.
Transfection of plasmid vectors
The plasmids described above were transfected into the HCT116 human colorectal cancer cell line (ATCC, Manassas, VA) with Lipofectamine 2000 (Invitrogen). For establishment of double transfectants of U6pro/DNMT1/Hygro and tetR/Neo, once tetR/Neo was transfected into HCT116 cells and selected with neomycin, the U6pro/DNMT1/Hygro was then transfected and selected with hygromycin. Expression of the tetR gene was confirmed with RT–PCR (data not shown).
Western blot analysis of DNMT1 protein levels
Approximately 20 µg of total protein extracted with radioimmunoprecipitation assay buffer (1 × phosphate-buffered saline, 0.1% SDS, 1% NP-40 and 0.5% deoxycholic acid) was loaded onto 4–15% gradient Tris–HCl gels (Bio-Rad Laboratories, Hercules, CA), electrophoresed in Tris-glycine- SDS running buffer and transferred to a polyvinylidene difluoride membrane in Tris-glycine buffer. The membranes were hybridized with antibodies against DNMT1 (1:1000 dilution; New England Biolabs, Beverly, MA), β-actin (1:10 000 dilution; Sigma, St Louis, MO) and proliferating cell nuclear antigen (PCNA; 1:10 000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA) in Tris-buffered saline-T buffer with 5% non-fat dry milk. We used first antibodies for DNMT3A (1:500 dilution; Santa Cruz Biotechnology) and DNMT3B (1:500 dilution; Santa Cruz Biotechnology). The membranes were then washed with Tris-buffered saline-T buffer. The membranes were incubated with secondary antibodies as follows: anti-rabbit-IgG–horseradish peroxidase (HRP) (1:1000 dilution for DNMT1; Santa Cruz Biotechnology), anti-goat-IgG–HRP (1:10 000 dilution for DNMT3A and DNMT3B; EMD Bioscience, Darmstadt, Germany) and anti-mouse-IgG–HRP (1:10 000 dilution for β-actin and PCNA; Santa Cruz Biotechnology). The proteins were detected with the an enhanced chemiluminescence detection kit (Amersham-Pharmacia, Uppsala, Sweden) and by exposure to Kodak X-OMAT AR film. DNMT1 protein levels were quantitated by a Calibrated Imaging Densitometer™ GS-710 (Bio-Rad Laboratories, Hercules, CA). We performed triplicate measurements, then calculated an average percentage protein level and standard deviation (HCT116 transfected with U6pro/DNMT1 versus HCT116, and HCT116 transfected with tetR/Neo and U6pro/DNMT1 versus HCT116).
Measurements of cell growth
Cell numbers were obtained following trypsinization using the Beckman Coulter Counter™ Z Series (Beckman Coulter Inc. Particle Characterization Group, FL). Cells containing the indicated plasmids were plated into dishes. Cell numbers were counted in triplicate measurements and calculated as an average number and standard deviation.
RESULTS AND DISCUSSION
Construction of DNMT1 knockdown vector
We chose the human U6 promoter (Fig. 1A) for transcription of hairpin siRNA since it is well characterized and a tetracycline-responsive derivative is available (11). The U6 promoter containing an SphI recognition sequence was isolated by PCR and ligated into a TA cloning vector (Fig. 1B). The plasmid was digested with SphI and blunted with Klenow fragment to maintain the first transcribed nucleotide as guanosine (Fig. 1C), which is the first nucleotide of the U6-like promoter (13). Oligo1, a sequence targeted to DNMT1 with an AatII recognition sequence, was ligated following AatII digestion of the plasmid (Fig. 1D). Oligo2, a sequence complementary to Oligo1 with AatII and ApaI recognition sites, was ligated following AatII and ApaI digestion (Fig. 1E). The resulting plasmid, which contained the U6 promoter and codes for hairpin siRNA against DNMT1, was designated U6pro/DNMT1 (Fig. 1F). As a control, a T5 transcriptional termination signal was ligated immediately downstream of the human U6 promoter (Fig. 1G). The tetracycline-responsive promoter was modified from the U6 promoter by PCR (Fig. 1A) to construct the inducible knockdown vector (Fig. 1H). The inducible knockdown vector was digested by ApaI, and a DNA fragment that encoded the hygromycin-resistance gene was ligated into the plasmid (Fig. 1I). The tetR coding gene was isolated by PCR and ligated into an expression vector (Fig. 1J).
DNMT1 knockdown with plasmid vector-based siRNA method
The expression levels of DNMT1 were analyzed by western blot analysis after transient transfection of the various vectors described above under non-selective conditions to verify the efficacy of the knockdowns. Figure 2A shows the suppression of DNMT1 by the in vivo transcribed hairpin siRNA from U6pro/DNMT1. Three days after transfection, DNMT1 expression in U6pro/DNMT1-transfected HCT116 cells was reduced by 54% (54 ± 18% versus untreated HCT116 cells). As the target sequence worked as well as in a previous description (3), the same target sequence was ligated downstream of the tetracycline-responsive promoter. DNMT1 expression was reduced by 23% (23 ± 15% versus untreated HCT116 cells) in U6pro/tetO/DNMT1-transfected cells 3 days after treatment with 10 µg/ml of doxycycline (Fig. 2B). The incomplete suppression seen in this experiment may have been due to a <100% transfection efficiency.
Figure 2.
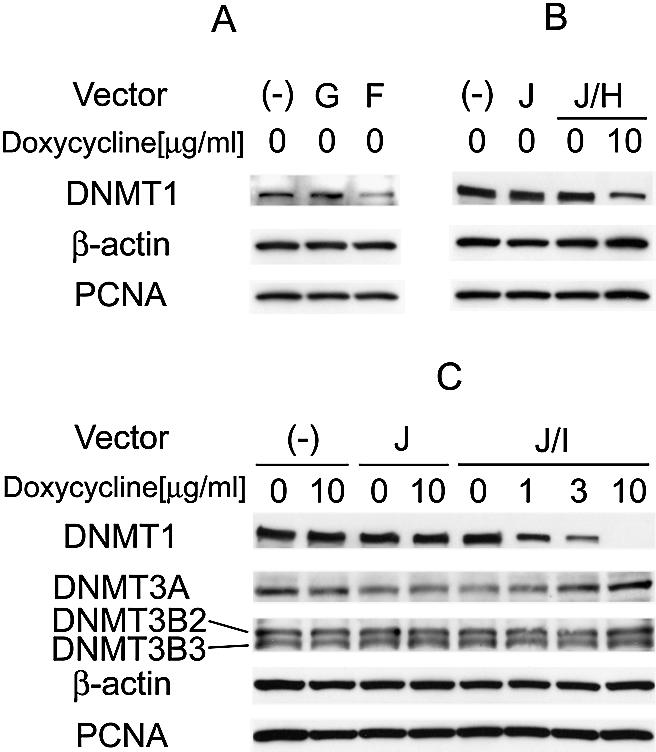
Western blot analysis of DNMT1 expression in cells transfected with various vectors. Vectors transfected into HCT116 cells are indicated with the symbols used in Figure 1. As a control, expression of β-actin and PCNA is shown. (A) Suppression of DNMT1 by siRNA targeting DNMT1 transcribed in vivo under non-selective conditions. (B) Suppression of DNMT1 by hairpin siRNA with inducible vectors. DNMT1 expression in HCT116 cells transfected with the plasmids indicated with or without treatment with doxycycline is shown. (C) DNMT1 knockdown with the inducible vector and selection marker. Expression of DNMT1, DNMT3A and DNMT3B was determined by western blot analysis. HCT116 cells transfected with the various plasmids indicated with or without doxycycline treatment are shown. Western blot analysis showed that DNMT1 levels were decreased by addition of doxycycline in a dose-dependent fashion, with complete suppression at 10 µg/ml. Expression of DNMT3A and DNMT3B was not affected by 10 µg/ml of doxycycline, showing the specificity of the knockdown.
We then inserted the hygromycin resistance gene into U6pro/tetO/DNMT1 (U6pro/tetO/DNMT1/Hygro) to allow for the isolation of stable transfectants. Western blot analysis showed that DNMT1 levels were decreased in such transfectants by addition of doxycycline in a dose-dependent fashion, with complete suppression at 10 µg/ml (Fig. 2C). The transfectants of tetR/Neo and U6pro/tetO/DNMT1/Hygro were found to express DNMT1 mRNA (data not shown), supporting the idea that hairpin siRNA inhibits translation as previously has been described (8–10). Expression of DNMT3A and DNMT3B was not affected by 10 µg/ml of doxycycline, showing the specificity of the knockdown. Ohkawa and Taira (11) described that 1 µg/ml of tetracycline was sufficient to induce the expression from this modified U6 promoter. The fact that we required 10 µg/ml of doxycycline for complete suppression may be due to over-expression of tetR driven by the cytomegalovirus (CMV) promoter requiring higher concentrations of the drug. A weaker promoter might enable the suppression of the targeted gene with a lower concentration of doxycycline.
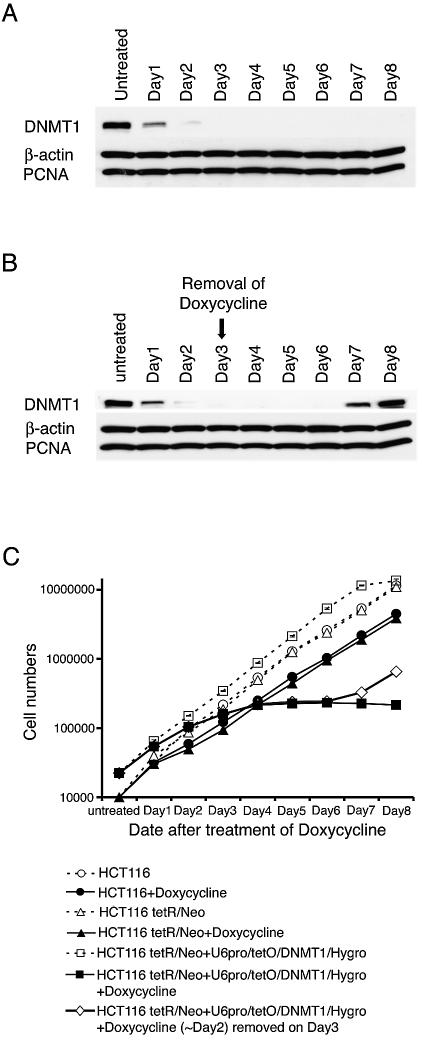
We next analyzed the time course for DNMT1 suppression following doxycycline addition (Fig. 3A). DNMT1 was reduced within 24 h after treatment and completely suppressed on the third day. Ohkawa and Taira (11) showed that transcription driven by the tetracycline-inducible U6 promoter was highest by 24 h, and McManus et al. (14) observed the maximum effect of chemically synthesized siRNA by 48 h. Our findings are compatible with both reports, in that transcription of the hairpin siRNA occurred within 24 h and a further 48 h were needed for maximum siRNA function, although this duration might differ when different molecules are targeted. Importantly, DNMT1 levels were restored when doxycycline was removed on the third day after treatment. DNMT1 began to be re-expressed on day 7, with the expression levels fully recovered by day 8. This experiment emphasized the value of the inducible vector in that it allows for transient knockdowns of target genes.
Figure 3.
Reversibility of DNMT1 knockdowns by controlled expression of siRNA. (A) Western blot analysis of cell extracts from HCT116 cells containing the inducible vector. DNMT1 protein is not evident 3 days after addition of 10 µg/ml of doxycycline. β-Actin and PCNA are not affected. (B) DNMT1 protein reappears on day 7 following removal of doxycycline on day 3. (C) Growth rates of the indicated cell types show growth arrest in cells with the knockdown vector in the presence but not the absence of doxycycline. Note that these effects are reversible following removal of the doxycycline on day 3.
The tetR/Neo and U6pro/tetO/DNMT1/Hygro transfectants after doxycycline treatment clearly showed growth arrest (Fig. 3C). Removal of doxycycline from the growth medium resulted in recovery of growth on day 7 (Fig. 3C) in concordance with the recovery of expression of DNMT1 (Fig. 3B). After treatment with doxycycline, the HCT116 cells and tetR/Neo transfectants showed similar cell growth rates (Fig. 3C). Although this observation does not exclude the possibility that the growth arrest of tetR/Neo and U6pro/tetO/DNMT1/Hygro transfectants after doxycycline treatment was due to a high concentration of doxycycline, the growth arrest could be attributed to the abolishment of DNMT1, since a recent report described that DNMT1 knockdown with antisense oligos resulted in intra-S phase arrest (15).
These results therefore establish the feasibility of using an inducible siRNA vector for transiently or permanently decreasing the expression of a protein in selected cells. The system also has the advantage that the level of protein can be titrated by varying the concentration of inducer, thus allowing the effects of intermediate levels of key proteins to be studied.
Acknowledgments
ACKNOWLEDGEMENTS
D.T. and P.A.J. are supported by National Cancer Institute grant R01 CA 82422.
REFERENCES
- 1.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 2.Yu J.Y., DeRuiter,S.L. and Turner,D.L. (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 6047–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sui G., Soohoo,C., Affar el,B., Gay,F., Shi,Y. and Forrester,W.C. (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul C.P., Good,P.D., Winer,I. and Engelke,D.R. (2002) Effective expression of small interfering RNA in human cells. Nat. Biotechnol., 20, 505–508. [DOI] [PubMed] [Google Scholar]
- 5.Miyagishi M. and Taira,K. (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol., 20, 497–500. [DOI] [PubMed] [Google Scholar]
- 6.Lee N.S., Dohjima,T., Bauer,G., Li,H., Li,M.J., Ehsani,A., Salvaterra,P. and Rossi,J. (2002) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat. Biotechnol., 20, 500–505. [DOI] [PubMed] [Google Scholar]
- 7.Brummelkamp T.R., Bernards,R. and Agami,R. (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science, 296, 550–553. [DOI] [PubMed] [Google Scholar]
- 8.Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- 9.McManus M.T. and Sharp,P.A. (2002) Gene silencing in mammals by small interfering RNAs. Nature Rev. Genet., 3, 737–747. [DOI] [PubMed] [Google Scholar]
- 10.Shi Y. (2003) Mammalian RNAi for the masses. Trends Genet., 19, 9–12. [DOI] [PubMed] [Google Scholar]
- 11.Ohkawa J. and Taira,K. (2000) Control of the functional activity of an antisense RNA by a tetracycline-responsive derivative of the human U6 snRNA promoter. Hum. Gene Ther., 11, 577–585. [DOI] [PubMed] [Google Scholar]
- 12.Hillen W. and Berens,C. (1994) Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol., 48, 345–369. [DOI] [PubMed] [Google Scholar]
- 13.Tuschl T. (2002) Expanding small RNA interference. Nat. Biotechnol., 20, 446–448. [DOI] [PubMed] [Google Scholar]
- 14.McManus M.T., Haines,B.B., Dillon,C.P., Whitehurst,C.E., van Parijs,L., Chen,J. and Sharp,P.A. (2002) Small interfering RNA-mediated gene silencing in T lymphocytes. J. Immunol., 169, 5754–5760. [DOI] [PubMed] [Google Scholar]
- 15.Milutinovic S., Zhuang,Q., Niveleau,A. and Szyf,M. (2003) Epigenomic stress response: knock-down of DNA methyltransferase 1 triggers an intra-S-phase arrest of DNA replication and induction of stress response genes. J. Biol. Chem., 278, 14985–14995. [DOI] [PubMed] [Google Scholar]