Abstract
Here we report the construction of a yeast genetic screen designed to identify essential residues in tRNAArg. The system consists of a tRNAArg knockout strain and a set of vectors designed to rescue and select for variants of tRNAArg. By plasmid shuffling we selected inactive tRNA mutants that were further analyzed by northern blotting. The mutational analysis focused on the tRNA D and anticodon loops that contact the aminoacyl-tRNA synthetase. The anticodon triplet was excluded from the analysis because of its role in decoding the Arg codons. Most of the inactivating mutations are residues involved in tertiary interactions. These mutations had dramatic effects on tRNAArg abundance. Other inactivating mutations were located in the anticodon loop, where they did not affect transcription and aminoacylation but probably altered interaction with the translation machinery. No lethal effects were observed when residues 16, 20 and 38 were individually mutated, despite the fact that they are involved in sequence-specific interactions with the aminoacyl-tRNA synthetase. However, the steady-state levels of the aminoacylated forms of U20A and U20G were decreased by a factor of 3.5-fold in vivo. This suggests that, unlike in the Escherichia coli tRNAArg:ArgRS system where residue 20 (A) is a major identity element, in yeast this position is of limited consequence.
INTRODUCTION
Mutant tRNAs have been widely used to study the mechanism of recognition between tRNAs and the corresponding aminoacyl-tRNA synthetases (aaRS). The 1990s were the years of in vitro studies that involved T7 RNA polymerase transcription of mutant tRNA genes. This powerful approach allows the synthesis of all the desired mutations at any position. However, due to the large number of possible mutations, this approach is time consuming and costly. Moreover, the absence of base modifications in the T7 RNA polymerase products can act negatively or positively on aaRS recognition (1–3) or modify the physico-chemical properties of these molecules and thus the assay conditions (4). In vivo approaches have been less frequently used. Amber, ochre and opal tRNA suppressors are efficient tools to monitor the real in vivo amino acid acceptance, but they suffer from sequence constraints at the anticodon level (5–7). Efficient tools were developed to monitor and analyze the in vivo amino acid acceptance of these suppressors, such as incorporation in a reporter protein followed by N-terminal sequencing (5) or northern blot analysis under acidic conditions (8). This last method is particularly efficient to measure the in vivo aminoacylation level and verify the tRNA identity. More recently, tRNA knockout strains have been used as a selection tool to study the structure–function relationships of tRNAs in vivo (9–11).
Actually, a powerful approach to study the critical nucleotides of tRNA would require the rapid preparation of large libraries of mutated RNAs and an in vivo screen of the libraries in order to select tRNA genes for the desired phenotype. A typical example of such a study was described recently in which a combinatorial tRNA library was built and was screened in a tRNAAla knockout strain (12).
Here we describe the search for critical residues of yeast tRNAArg4CCG, one of the two minor isoacceptors of the arginine system. Previous structural investigations have shown that yeast ArgRS interacts with the major form tRNAArg2ICG in a canonical class I manner. The enzyme contacts the tRNA on the D loop side and interacts with the major groove of the acceptor stem and anticodon loop. The direct contacts between the two partners involve the terminal adenosine with the catalytic site, several residues of the D loop and some residues of the anticodon loop (13). In terms of amino acid identity, residues that confer arginine acceptance are C35 and G36 or U36, as shown by kinetic analyses with in vitro synthesized transcripts (14).
In the present work we used an in vivo approach in order to identify the critical residues of tRNAArg that interact with ArgRS. First, we created a knockout strain of the minor tRNAArg4CCG isoacceptor by disrupting its single gene in Saccharomyces cerevisiae. Then, a randomly mutated library of the same tRNA was generated and tested by plasmid shuffling in the knockout strain. In this way, 17 lethal mutants were isolated and were further analyzed for their tRNA content by northern blotting under acidic conditions. To understand the function of several non-essential interactions that were observed in the crystal structure of ArgRS:tRNAArg, we also analyzed the RNA contents of five viable mutants. Among them, the U20A and U20G mutants exhibited a 3.5-fold decrease in their steady-state levels of aminoacylation, demonstrating that the yeast cells can survive and grow with reduced amounts of aminoacylated tRNA.
MATERIALS AND METHODS
Enzymes, chemicals and oligonucleotides
Restriction enzymes, T7 DNA polymerase, bacterial alkaline phosphatase and polynucleotide kinase were from New England Biolabs (USA). [α-35S]dATP, [γ-32P]dATP, [14C]l-arginine and Hybond-XL nylon membrane were from Amersham Pharmacia (UK). 5-Fluoroorotic acid (5-FOA) was from Toronto Research Chemicals (Canada). Oligo nucleotides were synthesized by NAPS (Germany) and Genset (France).
Vectors, bacterial and yeast strains
Escherichia coli TB1 [F– ara Δ(lac-proAB) hsdR (rk– mk+) rpsL(Strr) [φ80, dlac Δ (lacZ)M15] was used as a recipient for cloning procedures. Saccharomyces cerevisiae YBAM2 2n (ura3-52 lys2-801am trp1-Δ63 his3-Δ200 leu2-Δ1 ade2-Δ450 ade3-Δ1483) was used for the disruption of the tRNAArg4 gene. The resulting strain, S.cerevisiae YAL5 [YBAM2, tr4::HIS3, pAL5 (tR4+, Ura+, Ade3+)] was used to assay the mutant phenotypes and to extract RNA for northern blot analysis. This strain carries a knockout of the tRNAArg4CCG gene (tR4) and is rescued by a plasmid (pAL5) carrying the native gene. The bacterial and yeast strains were grown and transformed according to standard procedures.
A 0.65 kb PCR fragment containing the tRNAArg4 gene was cloned into several plasmids: pRS315-tR4 (tR4+, Leu+), pRS314-tR4 (tR4+, Trp+), pAL5 (tR4+, Ura+, Ade3+) a derivative of pALR10 (15). pRS315-tR3 was constructed by cloning a 0.4 kb HindIII fragment encoding the native tRNAArg3mcm5UCU gene (16). This vector was used to generate the pRS315-tR3CCG vector by site-directed mutagenesis; this vector is Leu+ and codes for a chimeric tRNAArg3 with a tRNAArg4 anticodon (tRNAArg3CCG).
In situ disruption of the gene encoding tRNAArg4
The 1.8 kb fragment encoding the His3 gene was inserted in the NsiI site of the tRNAArg4CCG gene located on the pRS315-tR4 vector. This new vector served as a source for the 2.45 kb DNA fragment containing the disrupted tR4 gene. By double homologous recombination the tR4 gene of the diploid yeast strain YBAM2 was disrupted (17). His+ colonies were selected and were induced to form spores on sporulation plates. Then, tetrads were dissected with a Singer apparatus and haploids were plated on YPD plates. Only two His– viable spores were expected in the case of an essential gene. Then, to generate the haploid strain YAL5, disrupted for tR4 and rescued by a plasmid copy of tR4, the diploid colonies showing 2:2 segregation were transformed by plasmid pAL5 (tR4+, Ura+, Ade3+) and sporulation was again induced. Among the four viable spores, two gave the expected His+, red and 5-FOAs phenotype corresponding to the haploid strain designated YAL5. The integration event was further confirmed by PCR analysis of the genomic DNA. The sect+ character, resulting from the introduction of another plasmid tR4 gene, was tested after introduction in trans of pRS314-tR4.
Random mutagenesis and in vivo selection
A 32 nt long deoxyoligonucleotide was used to generate a library containing randomly mutated tR4 genes (18). This primer was designed in order to generate statistically one mutation per tR4 gene. It had the sequence GTGCA ATGGT TAGCA TGCAT TCTTC CGGTG GC. The underlined positions contained a mixture of 90% of the wild-type nucleotide and 10% of a mixture of the three other nucleotides. This primer was complementary to positions 10–41 of tR4.
An aliquot of 10 ng of the pRS314-tR4 library was used to transform the YAL5 strain on minimal medium supplemented with adenine (limiting concentration 2 µg ml–1), uracil (20 µg ml–1), lysine (20 µg ml–1) and leucine (60 µg ml–1). The concentration of 10 ng was determined in order to obtain 50–100 colonies per 88 mm diameter Petri dish. After 72 h incubation at 30°C, the Trp+ red (Sec–) colonies were isolated and screened for 5-FOA resistance. The Trp+ 5-FOA sensitive (5-FOAs) colonies were selected and were designated as containing an inactive tR4 gene on the pRS314 vector.
Site-directed mutagenesis
To complete the first round of in vivo selection, site-directed mutagenesis was performed with 23 different primers by the Kunkel procedure (18). The primer sequences were as follows (mutated positions in bold).
U16C, CTAGT GCAAC GGTTA GCATG; U16A, CTAGT GCAAA GGTTA GCATG; U16G, CTAGT GCAAG GGTTA GCATG; U20A, GTGCA ATGGA TAGCA TGC; U20C, GTGCA ATGGC TAGCA TGC; U20G, GTGCA ATGGG TAGCA TGC; U20Aa, GCAAT GGTAA GCATGC; U20Ca, GCAAT GGTCA GCATGC; U20Ga, GCAAT GGTGA GCATGC; A21C, GCAAT GGTTC GCATG CATTC; A21G, GCAAT GGTTG GCATG CATTC; A21T, GCAAT GGTTT GCATG CATTC; U32A, GCATG CATTC ATCCG GTGG; U32C, GCATG CATTC CTCCG GTGG; U32G, GCATG CATTC GTCCG GTGG; U33A, CATGC ATTCT ACCGG TGGC; U33C, CATGC ATTCT CCCGG TGGC; U33G, CATGC ATTCT GCCGG TGGC; G37A, GCATT CTTCC GATGG CTGTG ATCC; G37C, GCATT CTTCC GCTGG CTGTG ATCC; U38A, CATTC TTCCG GAGGC TGTGA TCC; U38C, CATTC TTCCG GCGGC TGTGA TCC; U38G, CATTC TTCCG GGGGC TGTGA TCC.
Analysis of in vivo aminoacylation
Yeast clones carrying mutated tR4 genes were cured of the rescuing pAL5 plasmid (tR4+, Ura+, Ade3+). For this, the mutated clones were transformed by pRS315-tR3CCG and plated on minimal medium supplemented with adenine (limiting concentration 2 µg ml–1), uracil (20 µg ml–1) and lysine (20 µg ml–1). After incubation, red/white sectored colonies were observed. White sectors were isolated since they correspond to cells cured of pAL5: YAL5 containing pRS314-tR4 and pRS315-tR3CCG. An aliquot of 100 ml of the same liquid medium was inoculated and grown to an A700 of 2, then cells were harvested by centrifugation. All following steps were performed on ice. Cell pellets were resuspended in 5 ml of extraction buffer (0.3 M sodium acetate, pH 4.5, 10 mM EDTA) and subjected to two phenol extractions (pH 4.5). After ethanol precipitation the RNA samples were dissolved in 200 µl of extraction buffer and quantified by absorbance measurement at 260 nm.
A sample of 30 µg of total RNA was subjected to northern blot analysis. Aliquots of 15 µl were supplemented with 5 µl of loading buffer (0.1 M sodium acetate, pH 4.5, 8 M urea, 0.2% bromophenol blue and 0.2% xylene cyanol) and separated on a 1 mm thick 6.5% polyacrylamide gel (19:1 acrylamide:bisacrylamide) containing 8 M urea in 0.1 M sodium acetate buffer pH 5. Electrophoresis was performed at 18 W (∼400 V) at 4°C for 24 h. The portion of the gel between the two dyes was transferred and baked at 80°C to a nylon membrane (Hybond-XL) using a gel dryer. Prehybridization was performed for 4 h at 60°C in a 40 ml solution (1× Denhardt’s solution, 5× SSPE, 0.5% SDS). Hybridization was performed for 12 h at 60°C in 15 ml of the same solution, in the presence of a tRNAArg probe, a 5S probe (as an internal control) and a tRNALeuUAA probe (to control the aminoacylation level of the samples). Each probe was 5′ end 32P-labeled. The probes were: tRNAArg4CCG, 5′-CTCGA ACCCG GATCA CAGCC, annealing to positions 39–59; 5S, 5′-ACCCA CTACA CTACT CGGTC AGGCT CTTAC; tRNALeuUAA, 5′-GGATG CGAGG TTCGA ACTCG CGCGG. Washing was performed in 2× SSPE, 0.5% SDS solution for 20 min at 50°C. Signals were quantified using a Fuji Bioimager Bas2000.
Transcription with T7 RNA polymerase and aminoacylation
The gene for tRNAArg4CCG and the different mutants were constructed with four DNA oligonucleotides that were ligated between the HindIII and BamHI restriction sites of plasmid pUC18. Transcription using T7 RNA polymerase was performed according to standard procedures (19). After phenol extraction and ethanol precipitation, transcripts were purified using denaturing PAGE. Full-length tRNAs were eluted from gel slices using a Schleicher & Schüll (Dassel, Germany) electroelution apparatus.
Aminoacylation assays were carried out at 37°C in 50 mM HEPES (pH 7.5), 30 mM KCl, 25 µM [14C]l-arginine (25– 300 Ci mol–1), 10 mM ATP, 10 mM MgCl2, 0.1 mg ml–1 bovine serum albumin, 5 µM tRNA transcript and 30 nM ArgRS, as described previously (15).
RESULTS
Gene cloning and in situ disruption
The yeast genome contains 19 tRNAArg genes distributed as six tRNAArg2ACG genes, eleven tRNAArg3UCU genes and two unique genes for tRNAArg1CCU and tRNAArg4CCG each. We chose to disrupt the tRNAArg4CCG gene, whose decoding capacity is not redundant with the other tRNAArg. The tRNAArg4CCG gene is located on chromosome XII and is flanked by ORFs YLR343w and YLR344w. By PCR amplification we isolated a 647 bp HindIII fragment which was cloned in the same site of pRS315 (15). The tRNA gene was disrupted by inserting a 1.8 kb DNA fragment encoding the HIS3 gene. Finally, this DNA fragment encoding the tRNA gene disrupted by HIS3 (tr4::HIS3) was excised from the pRS315 vector and used to replace the wild-type copy of the tRNA gene in the yeast strain YAMB4. The tetrads resulting from meiosis and sporulation were dissected: two spores were unable to grow whereas the two growing spores were His–. This was the first evidence showing that the tRNAArg4CCG gene is an essential gene. When transformed with a rescuing plasmid encoding the tRNAArg4CCG gene (pAL5: Ura+, Ade3+, tRNAArg4CCG gene) the disrupted diploid gave four viable spores after segregation. Two of them were His– and were able to grow on 5-FOA medium, whereas the other two were His+ and 5-FOAs, demonstrating that, when disrupted for the tRNAArg4 gene, yeast cells cannot lose the rescuing plasmid encoding the tRNA gene. We conclude that the tRNAArg4CCG gene is an essential gene. The resulting disrupted strain was called YAL5.
Construction of a genetic screen based on the knockout strain
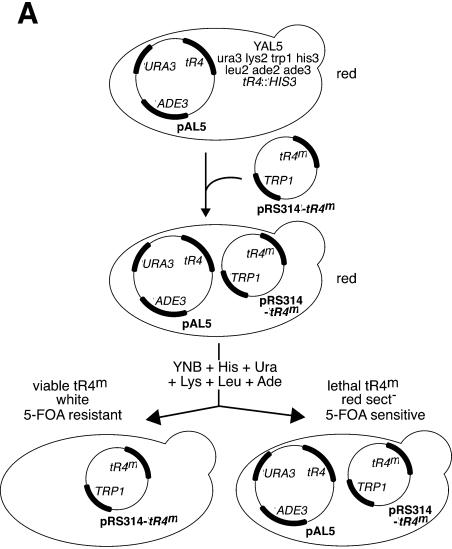
tR4 is an essential gene, as shown by the inability of spores that are disrupted for tR4 to grow. By introducing a copy of tR4 [on pAL5 (tR4+, Ura+, Ade3+)] before meiosis we can rescue the mutant spores and thus generate haploid strains disrupted for tR4 and harboring a plasmid copy of tR4. We used this property to design a genetic approach for selection of tRNAArg4CCG mutants affected in their functions (Fig. 1). For the recipient yeast cell, a genetic background (Ade2–, Ade3–) was chosen in order to give a red pigmentation when rescued by the Ade3+ vector (pAL5) and grown on medium containing limiting amounts of adenine (see Materials and Methods). Moreover, the presence of URA3 in this vector confers sensitivity to 5-FOA on the transformed cells. The screening procedure is based on the exchange of tR4 carried by pAL5 for another tR4 carried by a pRS314 vector (tR4+, Trp+). This exchange can occur when the second tR4 encodes a native tRNA and when the cells are spread on suitable growth medium. The exchange of pAL5 by pRS314-tR4 is followed by a loss of red pigmentation (colonies appear white/red sectored, sect+), due to ADE3 loss, and by 5-FOA resistance, due to URA3 loss. Using this approach, it becomes possible to select from a library of mutated tR4 genes those coding for an inactive tRNAArg4CCG. These clones will appear red and non-sectored (red, sect– and 5-FOAs) since the rescuing pAL5 is essential for viability and cannot leave the transformed cell. Thus, this convenient selection for lethal tR4 is based on visual discrimination of the colonies.
Figure 1.
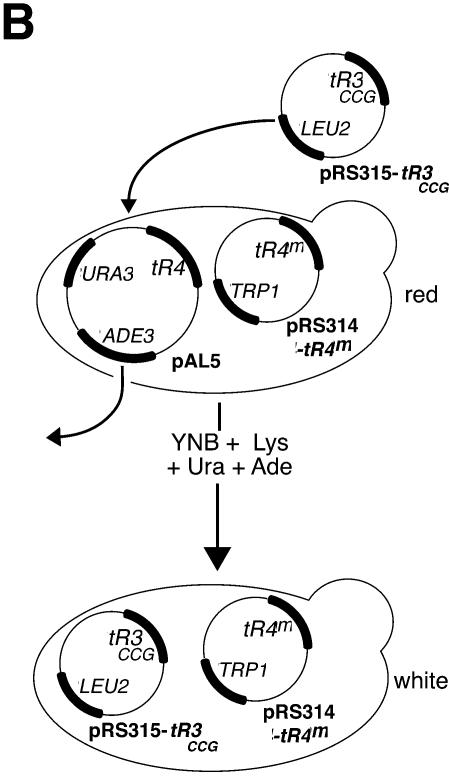
Schematic representation of the plasmid shuffle procedures used for the selection and analysis of tRNAArg4 mutated genes. (A) Selection of lethal tRNAArg4 genes. The tR4-disrupted strain YAL5 (tr4::HIS3), rescued by pAL5 (tR4, ADE3, URA3), was transformed by the mutated tR4 library (pRS314-tR4m). The resulting yeast strain was red according to the Ade3+ Ade2– phenotype. In the presence of a functional tRNAArg4 encoded by pRS314-tR4m, pAL5 was lost and the resulting strain became Ade3– Ade2– (white or white/red sectored colonies). In the presence of an inactive tR4m (lethal tR4), pAL5 could not be lost and the colonies remained non-sectored red (red sect– colonies). (B) To analyze the aminoacylation levels of tRNAArg4CCG by northern blot the rescuing native tR4 gene from pAL5 (tR4, ADE3, URA3) was shuffled from the red strains by transformation with pRS315-tR3CCG (tR3CCG, LEU2). In this way, the northern blot signal measured with the tRNAArg4CCG probe was specific to the mutated tR4 alleles.
Analysis of the selected random mutations
By the shuffle screen described above, we isolated 5000 colonies co-transformed with pAL5 and with the mutated library pRS314-tR4. We found 33 red, sect– colonies that were unable to lose the rescuing pAL5 vector. From these clones we extracted the pRS314-tR4 plasmids and confirmed the red sect– phenotype by re-introducing them into the YAL5 strain. Then the tR4 alleles and flanking regions were sequenced. Twenty-nine single substitutions, one single nucleotide deletion and three double mutants were identified. The mutants appeared one or more times: A14C, one occurrence; A14G, one occurrence; A14U, four occurrences; A15C, two occurrences; A15G, two occurrences; A15U, six occurrences; G18A, one occurrence; G18C, two occurrences; G18U, four occurrences; G19A, two occurrences; G19C, one occurrence; G19U, two occurrences; G37U, one occurrence; ΔA15, one occurrence; A14U/A15G, two occurrences; A15C/G19U, one occurrence. In fact, only six different positions in tRNAArg were selected as lethal mutants, whereas 12 different positions statistically were degenerate in the oligonucleotide used for the mutagenesis. Lethal mutations affected mainly nucleotides of the D loop, particularly those involved in tertiary and triple interactions. Only one mutation of position 37 of the anticodon loop was found to have a lethal effect. Despite repetitive screens, no different lethal mutants could be isolated from the randomly mutated library. To ensure that the library is complete and exhaustively screened we sequenced 48 clones of the library that confirmed the random distribution of the mutations. Nevertheless, in order to strictly screen the 12 selected positions we constructed the missing substitutions by site-directed mutagenesis and assayed them in vivo.
Site-directed mutagenesis to complete the first set of identified essential nucleotides
Twenty-three new mutants were built by site-directed mutagenesis. In the shuffle assay 19 mutants were viable (sect+, 5-FOAr), but only four were lethal (red, sect–, 5-FOAs); these were A21G and A21U in the D loop and U33G and U33A in the anticodon loop. Thus, the in vivo selection was very efficient, since only four mutants escaped the first selection procedure.
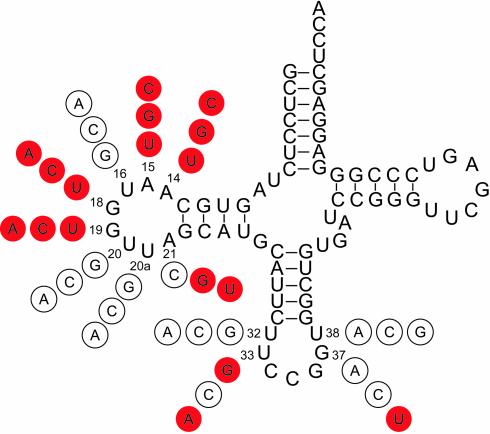
In summary, 17 lethal mutations were selected among the 36 mutations generated at 12 positions. Positions 14, 15, 18 and 19 did not accept any other nucleotide than the native one. In contrast, positions 21, 33 and 37 can tolerate some mutations (Fig. 2).
Figure 2.
Secondary structure of S.cerevisiae tRNAArg4CCG. Numbering is according to Sprinzl et al. (32). The encircled nucleotides are the mutations found at corresponding positions. Red filled circles indicate the lethal mutations that gave inactive tRNAs in vivo. They exhibited a non-sectored red phenotype. Empty circles contained nucleotides that gave active tRNAs in vivo (red-white sectored phenotype).
Northern blot analysis under acidic conditions
Lethal mutations can inactivate the tRNA function at different levels. Transcription, processing, stability, modification, aminoacylation and interaction with the translation factors and the ribosome are steps that can be crucially affected by single point mutations. Northern blot analysis is a powerful method to investigate the global effect of mutations in vivo. Extraction of RNA under acidic conditions and acid gel separation allows the measurement of the aminoacylation levels. Acid conditions preserve the ester bond between the tRNA and the amino acid and thus aminoacylated tRNA can be separated from uncharged tRNA and visualized by hybridization with a specific probe (8). In the case of lethal mutations, the yeast strain contains, in addition to the mutated tRNAArg4CCG (from pRS314-tR4), the rescuing native tRNAArg4CCG (from pAL5), which might also hybridize with the labeled probe. To distinguish between the two species an appropriate choice of oligonucleotide and appropriate washing temperatures can be used. However, this makes the comparison between the different mutants difficult. To circumvent this problem we substituted the rescuing native tRNAArg4CCG by a tRNAArg3CCG which is able to read the codon CGG usually read by tRNAArg4. In this way, only the lethal tRNAArg4CCG from pRS314-tR4 will be detected by northern blot assay, with a single probe and annealing protocol for all mutants. tRNAArg3CCG was engineered by site-directed mutagenesis and cloned into pRS315 (Leu+). Lethal clones were transformed by pRS315-tR3CCG (tR3CCG+, Leu+) and shuffled for the rescuing plasmid pAL5 (tR4+, Ura+, Ade3+) on Ura-containing medium (Fig. 1).
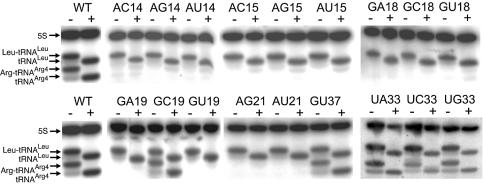
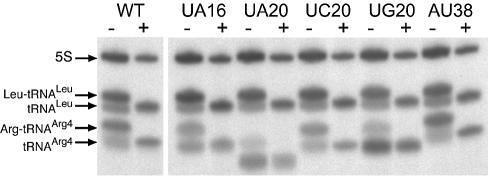
Northern blot analysis revealed the absence of mutated tRNAArg4 in most of the clones exhibiting a lethal phenotype (Fig. 3). Only four mutants exhibited a normal level of mutated tRNA (G19C, U33A, U33G and G37U), with a surprising normal level of aminoacylation, despite their lethal phenotype. The genes for these mutants were transcribed by T7 RNA polymerase and were assayed for their aminoacylation properties (data not shown). They were aminoacylated with parameters comparable to the native one except that the Km value of mutant G19C was increased 14-fold (Km = 1.4 µM). We also analyzed the content of aminoacyl-tRNA in several viable mutants for which an effect on the aminoacylation efficiency was suspected. These were mutants at positions 16, 20 and 38. The complete set of mutants was assayed for position 20 (U20A, U20C and U20G) and a unique transversion mutation was tested for each of the other positions (U16A and A38U). The expression level of the four mutants was comparable to the wild-type level, but a significant decrease in the charging level was observed for mutants U20A and U20G (22 and 20% aminoacylation, respectively, compared to the 69% of wild-type tRNAArg4) (Fig. 4). Moreover, mutant U20A migrates faster than the other tRNA, suggesting an incomplete denaturation of its tertiary structure in the low temperature conditions used to run the acid urea gels. Another reason for the fast mobility could be a change in the modified base content of this tRNA.
Figure 3.
Northern blot analysis of tRNA samples electrophoresed on a 6.5% polyacrylamide gel containing 8 M urea at pH 5.0 and 4°C. Total RNA from lethal mutants was extracted under acidic conditions, migrated and transferred to a Hybond membrane. The blot was probed with a mixture of 32P-labeled oligonucleotides complementary to 5S RNA (control for RNA content), to tRNALeu (control for the acidic extraction procedure) and to tRNAArg4CCG. + and – marks refer to alkaline treatment used to deacylate the RNA. Most of the extracts from mutants of the D loop are deprived of tRNAArg4CCG, in contrast to those of the anticodon loop that are expressed and aminoacylated. Except for U33C, all the RNA samples are from lethal mutants.
Figure 4.
Northern blot analysis of tRNA samples separated on a 6.5% polyacrylamide gel containing 8 M urea at pH 5.0 and 4°C. + and – refer to alkaline treatment used to deacylate the RNA. The three U20 mutants and one representative U16 and A38 mutant were analyzed. U20A and U20G were only charged up to 22 and 20%, respectively, compared to 69% for the wild-type tRNA.
DISCUSSION
In vivo approaches are useful to screen for genes carrying a desired phenotype among large libraries of degenerate genes. tRNA suppressors have been widely used for studies of aaRSs recognition of their cognate tRNA. But they suffer from the presence of non-cognate anticodon nucleotides that sometimes strongly determine another identity. Therefore, many tRNA suppressors are glutamine or lysine inserting suppressors (5), a phenomenon that singularly reduces the interest of these studies.
As tRNAs are essential for the cell, it was tempting to use their genes as genetic tools to analyze tRNA function. However, tRNA genes are small and are often present as several copies, which are two major difficulties to create knockout strains or to isolate conditional strains. For these reasons, only a few strains containing mutated chromosomal tRNA genes have been described. A thermosensitive E.coli tRNATrp mutant was described, because of its unique copy in the bacterial genome (20). This strain was later used to study the identity elements of E.coli tRNATrp (21). More recently, two E.coli knockout strains were engineered for tRNAAla and tRNAAsp (9,11). They required inactivation of three tRNA genes in each case and the resulting bacterial strains were rescued by plasmid copies of the corresponding tRNAs.
In this work we chose the arginine system of S.cerevisiae to build a new tRNA knockout strain. In the genome of S.cerevisiae, 19 tRNAArg gene copies are present (22). Four distinct isoacceptors are found that decode the six different Arg codons. Two tRNAs, tRNAArg2ICG and tRNAArg3mcm5UCU, are preponderantly found in yeast cells where they read CG(C/U/A) and AG(G/A) codons, respectively. They are encoded by 6 and 11 gene copies, respectively. A third isoacceptor is tRNAArg1CCU, which reads the AGG codon. It is encoded by a single gene copy and is dispensable, as shown by the viable phenotype of the knockout strain (23). Indeed, AGG codons can be read by tRNAArg2ICG. The last tRNA is the minor tRNAArg4CCG, which reads the rarest Arg codon CGG (3.9% of the Arg codons) and which is encoded by a single gene copy. By disrupting the gene encoding tRNAArg4CCG we thought we could selectively impair reading of the 9827 CGG codons present in the yeast genome and thus have a lethal phenotype that could be rescued by a plasmid copy of the tRNA gene. The resulting tr4::HIS3 disruptant was lethal, indicating that tRNAArg4CCG is the sole tRNA species decoding the CGG codon. The disrupted strain was rescued by a plasmid-encoded tRNAArg4CCG and a shuffle method was used to screen a library of mutated copies of tR4. Thanks to colored yeast genetic markers, it was possible to visually analyze the plasmid shuffling and thus discriminate between the viable and the lethal mutated tR4 genes. We focused the mutational analysis on the tRNA parts that are single stranded and in close vicinity to the synthetase in the complex, namely the D loop and the anticodon loop. We deliberately excluded the double-stranded regions of the stems since mutations here easily generate lethality by destabilizing effects on the tRNA structure. Also, the anticodon triplet was excluded from our mutational analysis, since it cannot be modified without changing the decoding properties of the tRNA.
Specific contacts between ArgRS and tRNA residues 16, 20 and 38 are not essential
Next to the anticodon triplet, three other residues of tRNAArg2ICG interact specifically with ArgRS (13). These are residues 16 and 20 of the D loop and residue 38 next to the anticodon bases. Oxygen atoms of the U16 ring interact with Lys543 (Nζ), Trp60 (N) and Arg36 (NH2). U20 (O2, N3, O4) atoms interact with Asn106 (Oδ1) and Gln111 (Nε2), whereas A38 (N6) interacts with the terminal carboxyl group of the protein (Met607-OT2) (13).
Among the three residues, residue 20 deserves most attention. In most organisms residue 20 is a well-conserved adenine that crucially contributes to the arginine identity, as shown by studies on E.coli (24–27). However, fungi like S.cerevisiae or Schizosaccharomyces pombe and Neurospora crassa and some mitochondrial tRNAArg contain C or D instead of A at position 20. In tRNAArg3UCU from S.cerevisiae it was shown that this residue does not play a role in the arginine identity but acts as an antideterminant against E.coli ArgRS (28). The in vivo study on tRNAArg4CCG performed here confirms the dispensable character of this interaction with the synthetase. The three mutants at position 20 were able to complement the tRNAArg4 knockout strain, suggesting that this interaction is a simple remnant of evolution. Nevertheless, the analysis of the in vivo aminoacylation levels of these tRNA species showed a 3.5-fold drop for the two transversion mutations U20A and U20G (Fig. 4). This shows that the interactions that occur with U20 are not simple relics but still play an active role in the aminoacylation of tRNAArg4CCG. Moreover, the results show that yeast cells can grow with a reduced amount of acylated tRNA, whereas it is usually believed that in vivo 100% of the tRNA molecules are in the acylated form and are in complex with eEF1A. In fact, it seems that in the arginine system the in vivo aminoacylation level never reaches 100% but only 69% for tRNAArg4CCG, 68% for tRNAArg1CCU, 71% for tRNAArg2ICG and 74% for tRNAArg3mcm5UCU in different genetic backgrounds (data not shown). For now, we can only speculate on the reason for this partial in vivo aminoacylation. A low stability of the arginyl–ester bond might be the simplest explanation for a decreased level of aminoacylated tRNAArg. Another reason might be that ArgRS activity is too low in vivo, suggesting that aminoacylation of tRNAArg might easily become a limiting step for the translation machinery.
Mutations of positions 16 and 38 exhibited no lethal phenotype whatever the mutation was. No difference was visible on the RNA blot profiles when compared with native tRNAArg4. Here again, the results show that the contacts observed in the structure of the ArgRS:tRNAArg2 complex do not play a preponderant role, at least for the aminoacylation of tRNAArg4. Additionally, the tRNAArg4CCG isoacceptor is the only arginine isoacceptor with U38 instead of A38. Thus, one can imagine that tRNAArg4CCG uses a different binding mode of its anticodon loop, with a different distribution of the synthetase:tRNA interactions. It could therefore be interesting to crystallize the ArgRS:tRNAArg4 complex, to see whether the binding mode is conserved.
Lethal mutations in the D loop lead to drastically reduced tRNA levels
Fourteen lethal mutants were identified in the D loop, in contrast to only three in the anticodon loop. Positions 14, 15, 18, 19 and 21 did not tolerate any change, except a unique A21C substitution. Due to their location, a first explanation might be that these mutations affect the tertiary interactions that stabilize the folded structure. It is well known that a complex network of interactions contribute to the stability of the tertiary structure of tRNA. The residues involved in these interactions are very conserved, like U8:A14 (reverse Hoogsteen pairing), G18:ψ55 and G19:C56 (distorted Watson–Crick base pair), or partially conserved, like the Levitt trans base pair G15:C48 (A15:U48 in tRNAArg4). In tRNAArg4, most of these interactions gave lethal phenotypes when they were disrupted by any change. In addition, they were not detected by northern blot, suggesting that these tRNAs are not produced or are rapidly degraded due to an unstable structure. A second cause of lethality might be reduced transcription. Transcription of eukaryotic tRNA genes is established by the RNA polymerase III transcription machinery that binds to the internal conserved sequences A and B mapped to nucleotides 13–20 and 51–64 (29). These sequences closely coincide with two conserved sequence blocks present in all eukaryotic tRNA genes, responsible for tRNA folding. The A and B blocks are bound simultaneously by the various subunits of the TFIIIC factor that recruit RNA polymerase III (30). Thus, lethal mutations in positions 14, 15, 18 and 19 might affect transcription, but also later steps, like the processing or the stability of the transcripts. Such effects, in addition to transcription rate, have been shown with mutations that disrupt the tertiary interactions (31).
Among the selected mutants, the G19C lethal mutant is remarkable for two reasons. It is expressed and aminoacylated (Fig. 3), suggesting that the mutation may affect the following steps, like interactions with the translation machinery for instance (elongation factor or the ribosome). The in vitro aminoacylation parameters of this mutant were only affected at the Km level (increase of 14-fold), an effect that could be explained by some structure change induced by the mutation. However, this affinity decrease measured in vitro had no effect on the steady-state level of aminoacylation in vivo.
The universally conserved U33 can be substituted
U33 is a quasi-universally conserved residue found in the anticodon loop of tRNAs (32). The U-turn motif associated with this residue was the first structural motif characterized in tRNA anticodon loops (33). It is defined by the formation of a hydrogen bond between U33 N3-H and O-P of residue 36 and by a stacking interaction of the aromatic cycle of U33 and the OR atom of residue 35, leading to a turn in the phosphodiester backbone following U33. Despite this conserved structural role, a U33C viable mutant was isolated in tRNAArg4, whereas U33A and U33G were lethal. All were aminoacylated as efficiently as native tRNA (Fig. 3), suggesting that altered interaction with the ribosome coding site might explain the lethal phenotype. Interestingly, the same mutants were analyzed in amber suppressor su7 for their ability to interact with the translation apparatus (34). Substitution U33C caused only a 2-fold decrease in suppression efficiency, a relatively modest effect that is in agreement with the viable phenotype that was observed in tRNAArg4. This corresponds to a 20-fold increase in the Kd value for ribosomes (35). On the other hand, when purines were introduced at position 33 a more pronounced increase of one order of magnitude was observed (34). In agreement with these effects, we observed a lethal phenotype when both A33 and G33 mutants were tested in the tR4 complementation assay.
Recent studies based on phylogenetic, structural and molecular dynamics data established a correlation between the presence of the conserved 33:35 interaction and a non-Watson–Crick isosteric base pair at position 32:38 (36). This arrangement would allow the formation of a canonical hairpin structure required for recognition by the ribosome and for the cognate interaction with the mRNA codon. However, this structural motif would also favor unfolding during the interaction with aaRSs when the tRNA anticodon bases are specifically recognized by the enzyme. U33 is found associated with a conserved pyrimidine at position 32 (98% of the occurrences), with a conserved purine at position 37 (71% of the occurrences) and with a strong preference for a purine at position 38. Earlier it was noticed that the first (position 32) and last residues (position 38) of the seven-membered anticodon loop are involved in a bifurcated hydrogen bond contact and form a ‘pseudo’ base pair (33). Most of the base pairs that are found at these positions cannot form classical Watson–Crick pairs, with the exception of the U·A base pair (32). Even in this case a classical pairing is not used, as shown by the crystal structure of tRNACys complexed with elongation factor EF-Tu, where the pair Ψ32·A38 does not form a Watson–Crick arrangement, but is isosteric to Ψ·C and C·A observed in yeast tRNAAsp and yeast tRNAPhe. Among the six mutants that we constructed at positions 32 and 38 (U32·A38, U32·C38, U32·G38, A32·U38, C32·U38 and G32·U38) we found two Watson–Crick pairs and four non-Watson–Crick pairs. Some of them are rarely found, like A32·U38 (0.4%), C32·U38 (1.3%) and G32·U38 (0.6%) (36). However, all these mutants were functional and able to complement the tRNAArg4 knockout strain, suggesting that they can pair with isosteric properties compatible with the formation of a canonical hairpin structure. In fact U·A, U·C and G·U pairs should be isosteric to C·A, as observed in the crystal structure of yeast tRNAPhe, whereas U·G, C·U and A·U pairs should be isosteric to a U·U pair, as observed in the crystal structure of the tRNAGln complexed with GlnRS (36,37).
Another crucial residue for the anticodon loop of tRNAArg4CCG is residue 37, which is methylated at position 1 (38). A G37U mutant is lethal in the knockout strain despite expression and aminoacylation levels comparable to native tRNAArg4. This strongly suggests a defect at the translation level. Residue 37 is almost always modified and it has been proposed that this modification is essential for reading frame maintenance (39,40) and binding efficiency with the ribosome (41). Only very few tRNAs have a U at position 37, and among them most are mitochondrial tRNAs (32), thus it was not completely unexpected to find this inactive phenotype.
Concluding remarks
We have used a genetic screen to select from among mutated tRNA genes those that were transcribed into molecules inactive in translation of CGG arginine codons. A first class of inactive tRNAs exhibited transcription or stability deficiencies as shown by their absence from RNA samples. The second class of inactive tRNAs probably resulted from a decrease in their interaction capacity with the translation machinery. A third class of mutations that removed sequence-specific interactions between tRNA residues 16, 20 and 38 and ArgRS did not lead to lethal effects. Only moderate effects on the steady-state level of aminoacylation of U20 mutants were observed. This suggests that interactions with the anticodon triplet play the major role in arginine identity, as demonstrated by in vitro studies (14). Further in vivo investigations will focus on the anticodon triplet of tRNAArg4 in order to explore the effects of the mutations on the arginine identity. The main advantage of the cellular assay is that it can take into account many cellular parameters that might contribute to amino acid identity. Among them, competition with other aaRS, the enzyme/substrate ratio and interaction with other cellular compounds or structures are parameters that differ with the in vitro assay conditions and that could significantly influence the aminoacylation of mutants (42,43).
Acknowledgments
ACKNOWLEDGEMENTS
We want to express our gratitude to Dr Jean Gangloff for constant advice and precious encouragement. We want to thank S. Barends for careful reading of the manuscript. We are grateful to Prof. J. Cavarelli for stimulating discussions. This work was supported by the Centre National de la Recherche Scientifique and grants from the European Union (4ème Programme des Biotechnologies ‘Design of RNA Domains, Substrates or Inhibitors of tRNA Recognising Proteins’).
REFERENCES
- 1.Muramatsu T., Nishikawa,K., Nemoto,F., Kuchino,Y., Nishimura,S., Miyazawa,T. and Yokoyama,S. (1988) Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature, 336, 179–181. [DOI] [PubMed] [Google Scholar]
- 2.Sylvers L.A., Rogers,K.C., Shimizu,M., Ohtsuka,E. and Söll,D. (1993) A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry, 32, 3836–3841. [DOI] [PubMed] [Google Scholar]
- 3.Pütz J., Florentz,C., Benseler,F. and Giegé,R. (1994) A single methyl group prevents the mischarging of a tRNA. Nature Struct. Biol., 1, 580–582. [DOI] [PubMed] [Google Scholar]
- 4.Kholod N.S., Pan’kova,N.V., Mayorov,S.G., Krutilina,A.I., Shlyapnikov,M.G., Kisselev,L.L. and Ksenzenko,V.N. (1997) Transfer RNAPhe isoacceptors possess non-identical set of identity elements at high and low Mg2+ concentration. FEBS Lett., 411, 123–127. [DOI] [PubMed] [Google Scholar]
- 5.Normanly J., Kleina,L.G., Masson,J.M., Abelson,J. and Miller,J.H. (1990) Construction of Escherichia coli amber suppressor tRNA genes. III. Determination of tRNA specificity. J. Mol. Biol., 213, 719–726. [DOI] [PubMed] [Google Scholar]
- 6.Varshney U. and RajBhandary,U.L. (1990) Initiation of protein synthesis using a termination codon. Proc. Natl Acad. Sci. USA, 87, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McClain W.H., Foss,K., Jenkins,R.A. and Schneider,J. (1990) Nucleotides that determine Escherichia coli tRNAArg and tRNALys acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc. Natl Acad. Sci. USA, 87, 9260–9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshney U., Lee,C.P. and RajBhandary,U.L. (1991) Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of E. coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem., 266, 24712–24718. [PubMed] [Google Scholar]
- 9.Gabriel K., Schneider,J. and McClain,W.H. (1996) Functional evidence for indirect recognition of G.U in tRNAAla by alanyl-tRNA synthetase. Science, 271, 195–197. [DOI] [PubMed] [Google Scholar]
- 10.Moulinier L., Eiler,S., Eriani,G., Gangloff,J., Thierry,J.C., Gabriel,K., McClain,W.H. and Moras,D. (2001) The structure of an AspRS-tRNAAsp complex reveals a tRNA-dependent control mechanism. EMBO J., 20, 5290–5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClain W.H. and Gabriel,K. (2001) Construction of an Escherichia coli knockout strain for functional analysis of tRNAAsp. J. Mol. Biol., 310, 537–542. [DOI] [PubMed] [Google Scholar]
- 12.Choi H., Otten,S. and McClain,W.H. (2002) Isolation of novel tRNAAla mutants by library selection in a tRNAAla knockout strain. Biochimie, 84, 705–711. [DOI] [PubMed] [Google Scholar]
- 13.Delagoutte B., Moras,D. and Cavarelli,J. (2000) tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J., 19, 5599–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sissler M., Giegé,R. and Florentz,C. (1996) Arginine aminoacylation identity is context-dependent and ensured by alternate recognition sets in the anticodon loop of accepting tRNA transcripts. EMBO J., 15, 5069–5076. [PMC free article] [PubMed] [Google Scholar]
- 15.Geslain R., Martin,F., Delagoutte,B., Cavarelli,J., Gangloff,J. and Eriani,G. (2000) In vivo selection of lethal mutations reveals two functional domains in arginyl-tRNA synthetase. RNA, 6, 434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt O., Mao,J., Ogden,R., Beckmann,J., Sakano,H., Abelson,J. and Söll,D. (1980) Dimeric tRNA precursors in yeast. Nature, 287, 750–752. [DOI] [PubMed] [Google Scholar]
- 17.Rothstein R. (1983) One-step gene disruption in yeast. Methods Enzymol., 101, 202–211. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel T.A. (1985) Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl Acad. Sci. USA, 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampson J.R. and Uhlenbeck,O.C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA, 85, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanofsky C. and Soll,L. (1977) Mutations affecting tRNATrp and its charging and their effect on regulation of transcription termination at the attenuator of the tryptophan operon. J. Mol. Biol., 113, 663–677. [DOI] [PubMed] [Google Scholar]
- 21.Pak M., Willis,I.M. and Schulman,L.H. (1994) Analysis of acceptor stem base pairing on tRNATrp aminoacylation and function in vivo. J. Biol. Chem., 269, 2277–2282. [PubMed] [Google Scholar]
- 22.Mewes H.W., Albermann,K., Bähr,M., Frishman,D., Gleissner,A., Hani,J., Heumann,K., Kleine,K., Maierl,A., Oliver, S.G., Pfeiffer,F. and Zollner,A. (1997) Overview of the yeast genome. Nature, 387, 7–65. [DOI] [PubMed] [Google Scholar]
- 23.Reijo R., Cho,D. and Huffaker,T. (1993) Deletion of a single-copy tRNA affects microtubule function in Saccharomyces cerevisiae. Genetics, 135, 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClain W.H. and Foss,K. (1988) Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science, 241, 1804–1807. [DOI] [PubMed] [Google Scholar]
- 25.Schulman L.H. and Pelka,H. (1989) The anticodon contains a major element of the identity of arginine transfer RNAs. Science, 246, 1595–1597. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K., Himeno,H., Asahara,H., Hasegawa,T. and Shimizu,M. (1992) In vitro study of E.coli tRNAArg and tRNALys identity elements. Nucleic Acids Res., 20, 2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimada A., Nureki,O., Goto,M., Takahashi,S. and Yokoyama,S. (2001) Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase. Proc. Natl Acad. Sci. USA, 98, 13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu W., Huang,Y., Eriani,G., Gangloff,J., Wang,E. and Wang,Y. (1999) A single base substitution in the variable pocket of yeast tRNAArg eliminates species-specific aminoacylation. Biochim. Biophys. Acta, 1473, 356–362. [DOI] [PubMed] [Google Scholar]
- 29.Galli G., Hofstetter,H. and Birnstiel,M. (1981) Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature, 294, 626–631. [DOI] [PubMed] [Google Scholar]
- 30.Paule M.R. and White,R.J. (2000) Transcription by RNA polymerase I and III. Nucleic Acids Res., 28, 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traboni C., Ciliberto,G. and Cortese,R. (1984) Mutations in Box B of the promoter of a eucaryotic tRNAPro gene affect rate of transcription, processing and stability of the transcripts. Cell, 36, 179–187. [DOI] [PubMed] [Google Scholar]
- 32.Sprinzl M., Horn,C., Brown,M., Loudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quigley G. and Rich,A. (1976) Structural domains of transfer RNA molecules. Science, 194, 796–806. [DOI] [PubMed] [Google Scholar]
- 34.Curran J. and Yarus,M. (1986) Base substitutions in the tRNA anticodon arm do not degrade the accuracy of reading frame maintenance. Proc. Natl Acad. Sci. USA, 83, 6538–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashraf S., Sochacka,E., Cain,R., Guenther,R., Malkiewicz,A. and Agris,P. (1999) Single atom modification (O→S) of tRNA confers ribosome binding. RNA, 5, 188–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auffinger P. and Westhof,E. (1999) Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol., 292, 467–483. [DOI] [PubMed] [Google Scholar]
- 37.Rould M.A., Perona,J.J. and Steitz,T.A. (1991) Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature, 352, 213–218. [DOI] [PubMed] [Google Scholar]
- 38.Björk G.R., Jacobsson,K., Nilsson,K., Johansson,M.J., Bystrom,A.S. and Persson,O.P. (2001) A primordial tRNA modification required for the evolution of life? EMBO J., 20, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Björk G., Wikstrom,P. and Bystrom,A. (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science, 244, 986–989. [DOI] [PubMed] [Google Scholar]
- 40.Hagervall T.G., Tuohy,T.M.F., Atkins,J.F. and Björk,G.R. (1993) Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol., 232, 756–765. [DOI] [PubMed] [Google Scholar]
- 41.Yarian C., Townsend,H., Czestkowski,W., Sochacka,E., Malkiewicz,A., Guenther,R., Miskiewicz,A. and Agris,P. (2002) Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem., 277, 16391–16395. [DOI] [PubMed] [Google Scholar]
- 42.Swanson R., Hoben,P., Sumner-Smith,M., Uemura,H., Watson,L. and Söll,D. (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science, 242, 1548–1551. [DOI] [PubMed] [Google Scholar]
- 43.McClain W., Jou,Y., Bhattacharya,S., Gabriel,K. and Schneider,J. (1999) The reliability of in vivo structure-function analysis of tRNA aminoacylation. J. Mol. Biol., 290, 391–409. [DOI] [PubMed] [Google Scholar]