Abstract
The assembly of plasmid-encoded proteins at a unique site (oriT) on the plasmid R1162, to form a complex called the relaxosome, is required for conjugative transfer of the plasmid and for negative regulation of neighboring promoters. Two-dimensional chloroquine gel electrophoresis was used to show that oriTs are physically coupled at the relaxosome. This interaction requires all the relaxosome proteins, which are assembled into a structure resulting in a decrease in the average linking number of the plasmid DNA in the cell. Molecules with higher superhelical densities are preferentially selected for assembly of the relaxosome. Genetic data obtained earlier indicate that the molecular coupling reported here is a ‘handcuffing’ reaction that contributes to the regulation of adjacent plasmid promoters. However, although these promoters affect the expression of the genes for replication, plasmid copy-control is regulated independently. This is the first time ‘handcuffing’ has been observed at an oriT, and its possible significance for transfer is discussed.
INTRODUCTION
Conjugative mobilization of the broad host-range plasmid R1162 requires the assembly of three, plasmid-encoded proteins at the origin of transfer (oriT), to form a structure called the relaxosome. One of these proteins, the transesterase MobA, reversibly cleaves one of the DNA strands. This reaction results in covalent joining of the protein to the 5′ end of the DNA, by means of a tyrosyl phosphodiester linkage (1,2). The protein–DNA complex is an essential intermediate in transfer. In addition, there are two small proteins which stimulate the activity of MobA. MobB increases the stability of the relaxosome (3), by a mechanism not yet understood, and MobC promotes separation of the DNA strands at oriT (4). This localized disruption of the helical DNA is required for binding of MobA to the DNA and for the subsequent transesterification (2,5).
The importance of the relaxosome goes beyond its role in transfer. Adjacent to oriT are three promoters (6), which together are responsible for transcription of the three mob genes and at least one of the genes for replication (Fig. 1A). These promoters are negatively regulated at oriT by the Mob proteins (7). In the absence of these proteins, the relaxosome is either not assembled or is unstable, and the promoters are derepressed (7,8). The increased rate of transcription has a toxic effect on the cell, presumably due to overexpression of one or more of the plasmid gene products (8). Some in-frame deletions in mobB result in partially defective proteins that are still active in transfer, but which exhibit poor regulation at oriT (8). Interestingly, the toxic effect of these mutations, as well as the increased rate of transcription, are reduced by additional copies of oriT, cloned in trans (8). This indicates that repression at oriT is enhanced by a second copy of this locus on another molecule. The interaction of plasmid molecules at oriT is further suggested by the observation that MobB enhances intermolecular recombination at oriT (8). This recombination does not require nicking within the relaxosome; possibly, the relaxosomes increase the frequency of recombination by bringing the oriTs together.
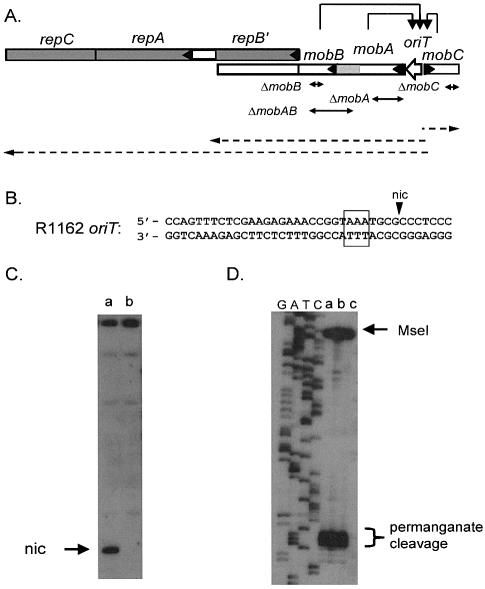
Figure 1.
(A) Region of R1162 containing the mob and rep genes (indicated by the open and filled rectangles, respectively, with an arrow head at the beginning of each gene). The product of repB′ is made as a fusion product with MobA, and is also translated separately (6). Thus, mobA is transcribed through mobB, but the two gene products are coded in separate reading frames. Transcripts are indicated by the dashed arrows. These transcripts originate from promoters located between oriT and mobC (6), and are negatively regulated by the proteins of the relaxosome (7), as indicated by the arrows. The approximate locations of the deletions in the mob genes are also shown. The patterned region of mobA encodes the region of the protein that interacts with MobB. (B) Base sequence of R1162 oriT, showing cleavage site (nic); bases within the box are sensitive to oxidation by permanganate. (C) Primer extension through oriT for (a) R1162, (b) pUT1579. The top strand in B was the template. (D) Primer extension on permanganate-treated template DNA, with the bottom strand of (B) as template. Plasmid DNAs were (a) R1162, (b) pUT1292, which contains a mutated oriT that forms relaxosome but is not cleaved (29 and S.Zhang, unpublished data) and (c) pUT1579. DNA was digested with MseI, which cleaves distal to the permanganate-sensitive region, prior to synthesis. On the left of the figure is a DNA dideoxy sequencing ladder generated from the oligonucleotide used in the primer extension.
A coupling between plasmid molecules, mediated by site-specific DNA binding proteins, is characteristic of a mode of regulation referred to as ‘handcuffing’ (9). This mechanism has been invoked most often to explain the control of plasmid copy-number. The replicative origins of many plasmids contain directly-repeated DNA segments, or iterons, which bind an initiator protein that is generally plasmid-specific (10). These iterons are required to load the initiator at the origin, and in some plasmids there are additional copies, which are not required for initiation of replication, but which are part of the mechanism for copy-control. How binding of initiator protein at essential and accessory iterons contributes to copy-control has not been obvious (11). Genetic and physical evidence has indicated, for some plasmids at least, that initiator proteins bound on different molecules interact to form a ‘handcuffing’ complex that inhibits initiation (9,11–13).
We show here that the ‘handcuffing’ mode of regulation occurs at oriT, and that the structure of the relaxosome involved in this handcuffing involves DNA that is topologically constrained at oriT.
MATERIALS AND METHODS
Bacterial strains, growth conditions and plasmids
For routine cloning and preparation of plasmid DNA, we used Escherichia coli K-12 strain MV10 (C600 ΔtrpE5) (14). Plasmid DNA for two-dimensional agarose gel electrophoresis was prepared from E.coli B strain AS19, which is highly permeable to novobiocin, used in these studies (15). Bacteria were grown in broth medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl), supplemented as required with ampicillin (100 µg/ml), chloramphenicol (25 µg/ml) or streptomycin sulfate (25 µg/ml). The plasmid pAO-SLO, which consists of a 42 bp oligonucleotide containing the lac repressor binding site cloned into the pBR322 derivative pAO (17), was kindly provided by D. Chattoraj. The ampicillin-resistance gene is the only major transcriptional unit on the plasmid: both the tetracycline-resistance and rop genes of pBR322 are deleted (16). For the construction of pAO-oriT, a 93 bp, oriT-containing segment of R1162 DNA was amplified by PCR with the primers 5′-GCGCGCGACGTCGCCGTTAGGCCAGTTTC and 5′-GCGCGCGACGTCCTGCAGCTATCATGGAGGCACAGC, digested with AatII, and used to replace the cloned lacO AatII fragment of pAO-SLO. The derivatives of R1162 used in this work are listed in Table 1; the deletions in the mob genes are indicated in Figure 1A.
Table 1. Derivatives of R1162 used in this work.
| Plasmid | Properties | Source |
|---|---|---|
| pUT1579 | R1162 mobA(Y25F) | Oligonucleotide-directed mutagenesis (E.Becker, unpublished data) |
| pUT1578 | R1162 mobA(Y25F) ΔmobC | PflMI fragment exchange between pUT1579 and pUT1568 (4) |
| pUT1210 | R1162 ΔmobA | (18) |
| pUT1292 | R1162 oriT (nic–) | By oligonucleotide-directed mutagenesis |
| pUT1309 | R1162 ΔoriT | (3) |
| pUT2000 | R1162 mobA(Y25F) ΔmobB | PflmI fragment exchange between pUT1579 and pUT1371 (8) |
| pUT2001 | R1162 mobA(Y25F) ΔmobAB | PflmI fragment exchange between pUT1579 and pUT1562 (8) |
| pUT221 | mobB | R1162 DNA, containing mobB, cloned in pACYC184 (3) |
Oxidation of DNA with permanganate
Cleared lysates (17) of plasmid-containing strains were treated with permanganate as described previously (18). Primer extension by limited thermocycling was carried out as described (3) with 32P-end-labeled primers.
Chloroquine agarose gel electrophoresis
One-dimensional gel electrophoresis was essentially according to the method of Shure et al. (19). Gels (1% agarose) were 13.5 × 20 × 0.5 cm. Gel buffer was TPE (50 mM Tris–H3PO4, pH 7.2, 2.8 mM EDTA) containing 14 µg/ml chloroquine. Electrophoresis was at 1.25 V/cm for about 16 h. DNA was visualized by staining or by hybridization and autoradiography. Hybridization probes were prepared with the Rediprime II kit (Amersham) according to the instructions of the manufacturer. Template was PstI-digested pAO DNA; probes were uniformly labeled with α-[32P]dCTP.
For two-dimensional gels, AS19 cells containing the desired plasmids were grown overnight at 37°C in broth medium supplemented with antibiotics, as required. The cells were then diluted 1:1000 into 20 ml fresh medium, and incubated at 37°C for about 3 h to OD600 = 0.2. Novobiocin was then added to 100 µg/ml and incubation continued for 30 min. Plasmid DNA was isolated by the Qiagen miniprep procedure.
Electrophoresis was carried out basically according to the procedure of Wu et al. (20), as also described by Park et al. (13), with small modifications. The 1% agarose gel was 13.5 × 12 × 0.7 cm, and the buffer was TBE (0.089 M Tris, 0.089 M boric acid, 0.0024 M Na2EDTA). Electrophoresis in the first dimension was at 90 V for 7 h. The gel was then rotated, placed in TBE containing 14 µg/ml chloroquine and electrophoresis continued at 45 V for 14 h in the dark.
RESULTS
Coupling of oriT loci on plasmid dimers
Proteins linking the duplicated sites on plasmid dimers will result in DNA looping. Wu and Liu (16) have pointed out that if a promoter is isolated in the loop, diffusional resolution of the positive and negative supercoils generated by transcription will be prevented. If in addition the cells are treated with an inhibitor of DNA topoisomerase II, there will be no enzymatic mechanism available to eliminate the positive supercoils, which will therefore accumulate. The positively supercoiled topoisomers can be identified by two-dimensional chloroquine gel electrophoresis. This was first demonstrated by Wu and Liu for the tetrameric lac repressor, which can bind simultaneously to the two lacO sites created by dimerization of the plasmid pAO-SLO (16). In addition, the method has been used to show pairing between partitioning sites of plasmid P1 (21), and also ‘handcuffing’ at the replicative origin of this plasmid (13). We used this method to test for molecular coupling at oriT by the relaxase proteins.
We replaced the 21 base-pair (bp) lacO segment in the plasmid pAO-SLO (16) with a 104 bp fragment containing the minimal 38 bp oriT of R1162 (22) (Fig. 1B). As a source of the Mob proteins, we constructed pUT1579, a derivative of R1162 containing a mobA mutation that replaced tyrosine, the active nucleophile in the protein–DNA transesterification reaction (2), with phenylalanine. The substitution prevented strand nicking at oriT (Fig. 1C), so that the covalently-closed topoisomers were not depleted due to the cleavage reaction. However, the conservative Y25F substitution did not affect assembly of the relaxosome, since there was still localized disruption of the DNA helix within the complex (Fig. 1D), as shown by the sensitivity of the unpaired bases to permanganate oxidation (18). Moreover, the mutation changed neither the degree nor the pattern of this sensitivity (Fig. 1D, lanes a and c).
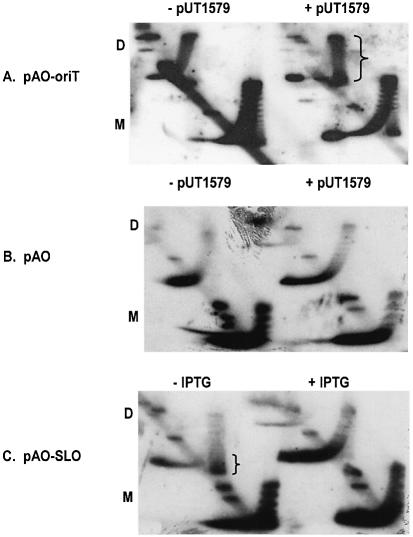
Both pAO-oriT and pUT1579 were introduced into E.coli strain AS19 (15), and the cells were then treated during exponential growth with novobiocin, an inhibitor of topoisomerase II. Plasmid DNA was isolated after treatment and the profile of topoisomers displayed by two-dimensional agarose gel electrophoresis. As a positive control, we also examined the topoisomers of pAO-SLO in the presence of active (–IPTG) or inactive (+IPTG) lac repressor. The results (Fig. 2A) show that although pUT1579 had little obvious effect on the topoisomer profile of pAO-oriT monomers, there was a substantial increase in the population of positively supercoiled dimers (indicated by the bracket in Fig. 2A). This change requires a cloned copy of oriT, since pUT1579 had no effect on dimers of the plasmid pAO, which lacks oriT DNA (Fig. 2B). In addition, the population of positively supercoiled molecules of pAO-SLO reflected the presence of active lac repressor in the cell and was unaffected by pUT1579 (Fig. 2C). We conclude that there is an interaction between paired oriT loci, and that this interaction is sufficiently strong to prevent the cancellation, by diffusion along the molecule, of local changes in supercoiling due to transcription.
Figure 2.
Two-dimensional chloroquine gel electrophoresis of pAO-based plasmids. ‘D’ and ‘M’ refer to the locations of the dimer and monomer forms. The populations of positively supercoiled topoisomers for pAO-oriT in the presence of pUT1579, and for pAO-SLO in uninduced cells (–IPTG), are indicated by the brackets.
Molecular coupling at oriT requires all the relaxase proteins
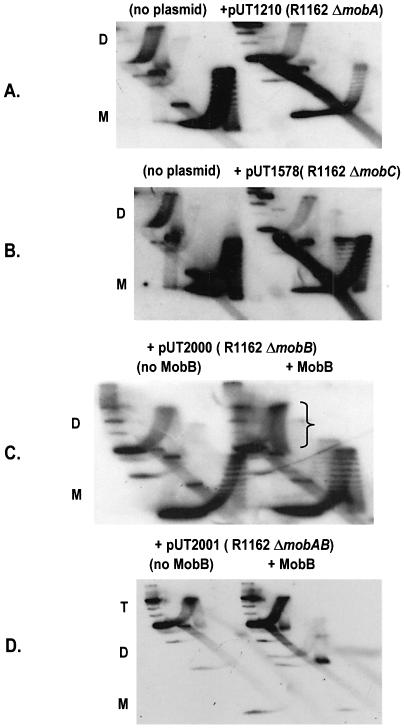
We next determined which of the mob proteins are required for pairing of the oriT sites in dimers. Inactivating deletions in either mobA (pUT1210) or mobC (pUT1578) prevented the accumulation of positively supercoiled topoisomers (Fig. 3A and B). We have shown previously that MobB stabilizes the relaxosome by a mechanism requiring a region of MobA distinct from that involved in strand cleavage and rejoining (8). This region is encoded by the segment of mobA adjacent to mobB, just upstream in the direction of transcription (Fig. 1A). Thus, although a 162 bp, in-frame deletion within mobB (ΔmobB in Fig. 1A) results in relaxosomes defective for transfer and regulation, the deletion is complemented in trans by a plasmid (pUT221) encoding MobB (8). In a parallel way, this mobB deletion, in the plasmid pUT2000, abolished coupling, but this was reversed by pUT221 in the cell (Fig. 3C).
Figure 3.
Effect of deletions in R1162 mob genes on accumulation of positively supercoiled topoisomers of pAO-oriT. Cells contained pUT221 (Table 1) for complementation with MobB (C and D). Abbreviations are as in Figure 2; ‘T’ refers to the location of trimer molecules prevalent for pUT2001 and the bracket indicates the enriched population of positively supercoiled topoisomers.
We also tested for coupling the plasmid pUT2001, which contains a larger (294 bp) in-frame deletion (ΔmobAB in Fig. 1A), extending into the adjacent region of mobA. This deletion is not complemented by pUT221 for transcriptional regulation (8). The pAO-oriT DNA isolated from AS19 cells containing pUT2001 was largely in the form of higher multimers, with monomers being nearly undetectable. This was occasionally observed for all the strains, and probably reflects the lack of a system for resolving multimers of pAO. No multimers were formed in a recA strain (X.Zhang, unpublished). Coupling on higher multimers should still cause the formation of positively supercoiled topoisomers, but this was not observed, even in the presence of pUT221 (pUT2001, Fig. 3D). We conclude that an intact relaxosome is required for linking the two oriTs. In addition, the interaction between MobA and MobB that is required for regulation of the promoters adjacent to oriT is also required for coupling.
The relaxosome proteins binding at oriT change the topoisomer profile of plasmid DNA extracted from healthy cells
In general, initiation proteins assemble at iteron-containing origins to form initiation complexes. In each case, the proteins may include those that are replicon-specific, as well as those recruited from the host. It appears that one function of the complex is to topologically constrain the DNA, causing a weakening of the helical structure and facilitating strand separation and entry by components of the replication machinery (23). Since there is also localized strand separation in the R1162 relaxosome, we asked whether here, too, a complex was assembled that constrained the duplex DNA.
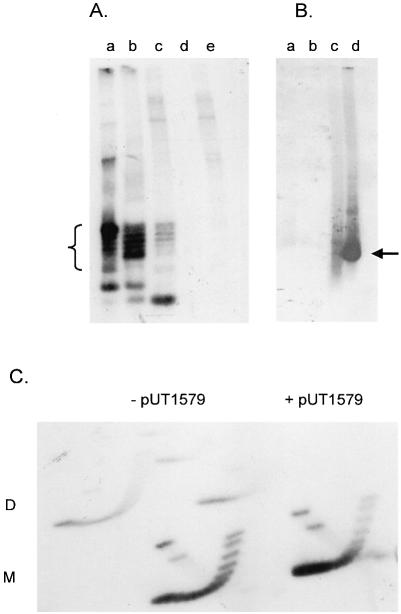
Plasmid pAO-oriT DNA was isolated from exponentially-growing MV10 cells and the topoisomers were separated by one-dimensional chloroquine gel electrophoresis (Fig. 4A, lane a). The same sample applied to a two-dimensional gel showed that these topoisomers were negatively supercoiled, as expected for plasmids replicating under normal conditions (Fig. 4C, left). When pUT1579 was also present, the molecules of pAO-oriT migrated more rapidly in the gel, indicating that on average they were more negatively supercoiled (Fig. 4A, lane b). This change in the topoisomer profile was also seen, although less clearly, in a two-dimensional gel (Fig. 4C, right). Molecules with greater superhelical density migrated more rapidly in the first dimension (from top to bottom in the figure). These molecules were relatively more abundant in the presence of pUT1579.
Figure 4.
(A) One-dimensional chloroquine gel electrophoresis of pAO-oriT DNA extracted from MV10 cells lacking other plasmids (lane a), or containing pUT1579 (lane b) or pUT1309 (lane c). Lanes d and e contain plasmid DNA from cells lacking pAO-oriT but containing pUT1579 or pUT1379, respectively. DNA was visualized by staining with ethidium bromide; topoisomers of pAO-oriT are indicated by the bracket. (B) One-dimensional chloroquine gel electrophoresis of pAO-oriT DNA isolated from E.coli strain MV10. After electrophoresis, the gel was stained to locate the bands, and the DNA then transferred to a membrane for hybridization without prior denaturation and depurination. Cells contained pUT1579 alone (lane a), pUT1309 alone (lane b), pAO-oriT and pUT1579 (lane c) or pAO-oriT and pUT1309 (lane d). The location of DNA transferring efficiently to the membrane is indicated by the arrow. (C) Two-dimensional chloroquine gel electrophoresis of pAO-oriT extracted from cells without (left) or with (right) pUT1579.
The different plasmid DNA topoisomers did not have the same activity for strand cleavage and rejoining within the relaxosome. We examined the topoisomer profile for pAO-oriT isolated from cells also containing pUT1309 (Table 1). This plasmid is deleted for oriT, but is otherwise identical to R1162 and encodes all the relaxase proteins, including a MobA protein active in nicking. The profile is shown in Figure 4A, lane c. A greater proportion of the more rapidly migrating species was depleted, suggesting that they had been preferentially nicked. In addition, there was an intense, rapidly-migrating band in the gel. This consisted mostly of single-stranded plasmid DNA derived from the nicked molecules and denatured by the alkaline lysis step during purification. The plasmid DNA samples were applied to a gel as in Figure 4A, but after electrophoresis and staining, the gel was blotted and probed without denaturation or depurination of the DNA. Only this band strongly hybridized with the probe, indicating that it was single-stranded and thus able to transfer efficiently to the blotting membrane (Fig. 4B, lane d).
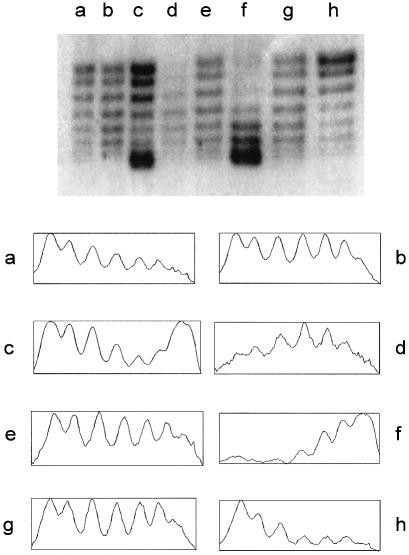
Inactivation of any of the mob genes decreased the average superhelical density, compared to when all the Mob proteins were present. Plasmids isolated from strains lacking either MobA, MobB or MobC had a lower average superhelical density (Fig. 5, compare lanes a, b and g with lane c). The deletion in mobB could be complemented in trans (Fig. 5, lanes f and g); the larger deletion, extending into mobA, was not complemented (lanes d and e). The topoisomer profiles of pAO-oriT DNA from cells lacking R1162, or containing pUT1578 (R1162 ΔmobC), were similar (Fig. 5, compare lanes a and h). When MobC was present in the cell, there was a relative increase in the more highly supercoiled topoisomers, when either MobA (Fig. 5, lane b) or MobB (Fig. 5, lanes e and g) were absent. Thus, MobC by itself exerts an effect on the distribution of topoisomers. Results to be reported elsewhere (S.Zhang and R.Meyer, in preparation) confirm that MobC alone can bind to plasmid DNA, resulting in a change in supercoiling of this DNA in the cell.
Figure 5.
(Top) One-dimensional chloroquine gel electrophoresis of pAO-oriT from MV10 strains. Cells also contained pUT1578 (lane a), pUT1210 (lane b), pUT1579 (lane c), pUT2001 and pUT221 (lane d), pUT2001 (lane e), pUT2000 and pUT221 (lane f), pUT2000 (lane g), or no other plasmids (lane h). (Bottom) Densitometric tracings of the topoisomer populations shown in the gel. The horizontal scale is approximately 400 pixels in each case; the vertical scale extends from the minimum to maximum value for the pixels in each case (vertical scale range is between 80 and 180).
When the relaxosome is assembled, the increase in superhelical density does not occur by equal recruitment of all the pre-existing topoisomers. It is those that are already most highly supercoiled that are preferentially selected. This is shown by the relative depletion of these species in cells containing active relaxosome (Fig. 5, lanes b and c). This is consistent with the results in Fig. 4A, lane c, which indicate that the more highly supercoiled plasmids are selected for cleavage. Additionally, however, when MobB is encoded by a separate plasmid, and is no longer under control of the regulated promoters at oriT, there was a shift of all the topoisomers to the more highly supercoiled forms (Fig. 5, lane f). In this case, MobB is no longer co-regulated with MobA and MobC. This deregulation allows the formation of relaxosomes on molecules having less supercoiling.
Regulation of plasmid copy-number is independent of coupling at oriT
Transcription initiated from the promoters adjacent to oriT are responsible for expression not only of the mob genes, but also for a gene essential for replication, repB. In addition, some of these transcripts extend through repA and repB (24) (Fig. 1A). Thus, coupling at oriT could regulate plasmid copy-number by controlling the overall expression of the rep genes. Regulation of copy-number would then be indirect, rather than by the sequestration and inhibition of an initiator protein, as proposed for the plasmids P1 and R6K (9,13). Consistent with this idea, deletions in the mob genes of the closely related plasmid RSF1010 were reported to raise the plasmid copy number approximately 3-fold (7).
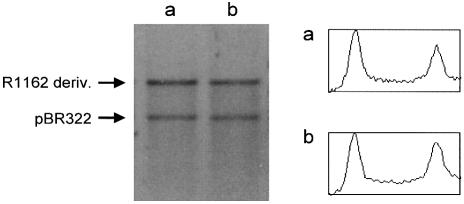
We compared the copy-number of R1162 (mobAY25F) with a derivative of this plasmid deleted for oriT, but with the adjacent promoters intact. In each case, plasmid DNA was extracted from log-phase cells, linearized and the amounts then compared with co-extracted pBR322 DNA (Fig. 6). We found no significant difference between the copy-numbers of the two R1162 derivatives. Therefore, regulation at oriT is not a major determinant of plasmid copy-number.
Figure 6.
Copy-number of R1162 in the absence (a) and presence (b) of regulation at oriT. Plasmid DNA was isolated from log-phase cells and linearized with AflIII. Cells contained pUT1309 and pBR322 (lane a) or pUT1579 and pBR322 (lane b). DNA was visualized by staining with ethidium bromide after electrophoresis through 1% agarose. Densitometric tracings are shown as described in Figure 5.
DISCUSSION
Our results show that the positive and negative supercoils generated by transcription are not resolved by diffusion along the DNA of oriT-containing dimer molecules. Thus, the molecules are coupled at oriT, and we have shown this requires assembly of the relaxosome proteins at this site. We attribute this coupling to the association of two oriTs by means of the relaxosome proteins. However, in principle, any constraint at oriT would have the same effect: for example, the plasmids could be anchored at oriT to a component of the cell membrane, perhaps as part of an initial step in transfer. However, there are two arguments against this idea. First, the anchoring of monomers, containing a single copy of oriT, should prevent resolution of the supercoils induced by transcription, but in our experiments an enhanced population of positively supercoiled topoisomers was observed only for multimers (Fig. 2). Second, earlier results showed that partial deregulation at oriT due to mutations in mobB could be reversed by extra copies of oriT in trans (8). In view of the coupling of oriTs at plasmid dimers, we interpret this to mean that ‘handcuffing’ at oriT is involved in repression of gene expression from the adjacent promoters.
Since molecular ‘handcuffing’ can be a mechanism for measuring and responding to plasmid copy-number, an attractive idea is that initiation of R1162 replication is controlled in this way. However, there was little increase in copy-number in the absence of regulation at oriT. Our results are different than those reported by Frey et al. (7), who detected up to a 3-fold increase in copy-number when mob genes were inactivated by deletion. Part of the difference observed by these authors could be due to the reduction in nicking at oriT for their mutated plasmids. This would result in a greater yield during purification of plasmid DNA by methods favoring the recovery of covalently-closed DNA.
Does handcuffing at oriT have any significance for conjugative transfer? For a single mating pair, more than one copy of R1162 is transferred (25). One way for this to occur is to bring more than one plasmid molecule to the vicinity of the conjugative pore by means of the handcuffing reaction. Alternatively, a single molecule in the donor might remain at the conjugative pore, and thus be available to feed into the transfer apparatus strands generated by replacement synthesis. Recent experiments (26) argue against the idea of a single molecule remaining associated with the pore. When there is a single plasmid molecule in the donor, multiple rounds of transfer still occur. However, there is a lag between successive rounds of transfer, which is more consistent with the selection of individual molecules for transfer, rather than with a long-lived transfer ‘machine’ (26). Thus, handcuffing could play a role in promoting multiple rounds of transfer when there is more than one plasmid copy in the cell. In this connection, it is interesting that molecular pairing at the par locus of plasmid P1 has also been detected by two-dimensional chloroquine gel electrophoresis (21). It is reasonable to think that handcuffing is involved not only in control of gene expression, but also in other functions where the gathering of plasmid molecules would be desirable.
The DNA of plasmid pAO-oriT has a higher negative average superhelical density when isolated from cells containing the Mob proteins (Fig. 4A and C). This is most likely due to a decrease in negative supercoiling of the plasmid DNA brought about by the assembly of the relaxosome. Such a decrease would be sensed by topoisomerase II, which would then introduce additional negative supercoils in the DNA. The relaxosome can cause a change in supercoiling in two ways, by inducing the localized opening of the helix at oriT or by constraining the path of the DNA at the relaxosome. Although there is localized strand separation at oriT (18), this is unlikely to account completely for the change in superhelical density when plasmid DNA is complexed with Mob proteins. As shown in Fig. 1D, permanganate sensitivity is limited to the AT-rich region of oriT, and encompasses no more than 10 base-pairs. Thus, we would not expect a change of more than one linking number to be attributable to this strand separation (27). In addition, we have shown previously that the extent of localized melting is unchanged when MobB is provided from another plasmid, to complement ΔmobB (8), yet this complementation results in a large overall shift toward more negatively supercoiled molecules (Fig. 5, lane f). Thus, we believe that the plasmid DNA undergoes constrained winding, stabilized by MobB, at the relaxosome.
Plasmid topoisomers with higher superhelical density are preferentially recruited to form relaxosome and thus are more likely to be cleaved at oriT (Figs 4A and 5, lane c). Wrapping of the DNA during assembly of the relaxosome could relieve the torsional strain reflected by supercoiling, and assembly on those molecules that are more highly supercoiled would be energetically favored. At the same time, these molecules would have the greatest amount of helical distortion; spontaneous strand separation at the AT-rich region of oriT (Fig. 1B) could nucleate formation of the relaxosome by providing a MobA binding site. Supercoiling might also affect the strand cleavage-rejoining equilibrium within the relaxosome (1). The rejoining of cleaved DNA in the relaxosome requires noncovalent binding of MobA to the DNA on one side of the cleavage site, and the increased tension of highly supercoiled molecules might affect this reaction. We have isolated mutations resulting in unstable relaxosomes but still allowing a high transfer frequency (8). A large proportion of the molecules isolated from the cell are nicked. Presumably, the instability of the relaxosome is suppressed by a shift in the cleavage equilibrium toward the nicked species. This suggests that those molecules which spend more time in the nicked configuration are preferentially utilized for transfer.
Finally, we note that the reporter plasmid in these experiments was derived from the high copy-number (28) plasmid pAO. The expression of the mob genes is regulated, and, as the effect of complementing supply of MobB indicates (Fig. 5, lane f), the amounts of the proteins can be limiting. Thus, the presence of active complexes suggests that their assembly is strongly cooperative.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dhruba Chattoraj for advice and strains. This work was supported by NIH Grant GM37462 to R.M.
REFERENCES
- 1.Scherzinger E., Lurz,R., Otto,S. and Dobrinski,B. (1992) In vitro cleavage of double- and single-stranded DNA by plasmid RSF1010-encoded mobilization proteins. Nucleic Acids Res., 20, 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherzinger E., Kruft,V. and Otto,S. (1993) Purification of the large mobilization protein of plasmid RSF1010 and characterization of its site-specific DNA-cleaving/DNA-joining activity. Eur. J. Biochem., 217, 929–938. [DOI] [PubMed] [Google Scholar]
- 3.Perwez T. and Meyer,R. (1996) MobB protein stimulates nicking at the R1162 origin of transfer by increasing the proportion of complexed plasmid DNA. J. Bacteriol., 178, 5762–5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang S. and Meyer,R. (1997) The relaxosome protein MobC promotes conjugal plasmid mobilization by extending DNA strand separation to the nick site at the origin of transfer. Mol. Microbiol., 25, 509–516. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee M., Rao,X.M. and Meyer,R.J. (1992) Role of the origin of transfer in termination of strand transfer during bacterial conjugation. J. Bacteriol., 174, 6659–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scholz P., Haring,V., Wittmann-Liebold,B., Ashman,K., Bagdasarian,M. and Scherzinger,E. (1989) Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene, 75, 271–288. [DOI] [PubMed] [Google Scholar]
- 7.Frey J., Bagdasarian,M.M. and Bagdasarian,M. (1992) Replication and copy number control of the broad host-range plasmid RSF1010. Gene, 113, 101–106. [DOI] [PubMed] [Google Scholar]
- 8.Perwez T. and Meyer,R. (1999) Stabilization of the relaxosome and stimulation of conjugal transfer are genetically distinct functions of the R1162 protein MobB. J. Bacteriol., 181, 2124–2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McEachern M.J., Bott,M.A., Tooker,P.A. and Helinski,D.R. (1989) Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc. Natl Acad. Sci. USA, 86, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kues U. and Stahl,U. (1989) Replication of plasmids in Gram-negative bacteria. Microbiol. Rev., 53, 491–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattoraj D. (2000) Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol., 37, 467–476. [DOI] [PubMed] [Google Scholar]
- 12.Kittel B. and Helinski,D. (1991) Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc. Natl Acad. Sci. USA, 88, 1389–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park K., Han,E., Paulsson,J. and Chattoraj,D. (2001) Origin pairing (‘handcuffing’) as a mode of negative control of P1 plasmid copy number. EMBO J., 20, 7323–7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershfield V., Boyer,H.W., Yanofsky,C., Lovett,M.A. and Helinski,D.R. (1974) Plasmid ColE1 as a molecular vehicle for cloning and amplification of DNA. Proc. Natl Acad. Sci. USA, 71, 3455–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockshon D. and Morris,D. (1983) Positively supercoiled DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res., 11, 2999–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H.-Y. and Liu,L. (1991) DNA looping alters local DNA conformation during transcription. J. Mol. Biol., 219, 615–622. [DOI] [PubMed] [Google Scholar]
- 17.Clewell D.B. and Helinski,D.R. (1969) Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl Acad. Sci. USA, 62, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S. and Meyer,R.J. (1995) Localized denaturation of oriT DNA within relaxosomes of the broad host-range plasmid R1162. Mol. Microbiol., 17, 727–735. [DOI] [PubMed] [Google Scholar]
- 19.Shure M., Pulleyblank,D. and Vinograd,J. (1977) The problems of eukaryotic and prokaryotic DNA packaging and in vivo conformation posed by superhelix density heterogeneity. Nucleic Acids Res., 4, 1183–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H.Y., Shyy,S., Wang,J.C. and Liu,L.F. (1988) Transcription generates positively and negatively supercoiled domains in the template. Cell, 53, 433–440. [DOI] [PubMed] [Google Scholar]
- 21.Edgar R., Chattoraj,D. and Yarmolinsky,M. (2001) Pairing of P1 plasmid partition sites by ParB. Mol. Microbiol., 42, 1363–1370. [DOI] [PubMed] [Google Scholar]
- 22.Brasch M.A. and Meyer,R.J. (1987) A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J. Mol. Biol., 198, 361–369. [DOI] [PubMed] [Google Scholar]
- 23.Bramhill D. and Kornberg,A. (1988) A model for initiation at origins of DNA replication. Cell, 54, 915–918. [DOI] [PubMed] [Google Scholar]
- 24.Bagdasarian M., Scholz,P., Frey,J. and Bagdasarina,M. (1986) Regulation of the rep operon expression of the broad-host-range plasmid RSF1010. In Novick,R. and Levy,S. (eds), Evolution and Environmental Spread of Antibiotic Resistance Genes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 209–223. [Google Scholar]
- 25.Rao X.M. and Meyer,R.J. (1994) Conjugal mobilization of plasmid DNA: Termination frequency at the origin of transfer of plasmid R1162. J. Bacteriol., 176, 5958–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parker C. and Meyer,R. (2002) Selection of plasmid molecules for conjugative transfer and replacement strand synthesis in the donor. Mol. Microbiol., 46, 761–768. [DOI] [PubMed] [Google Scholar]
- 27.Walter J. and Newport,J. (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA and DNA polymerase α. Mol. Cell, 5, 617–627. [DOI] [PubMed] [Google Scholar]
- 28.Twigg A.J. and Sherratt,D. (1980) Trans-complementable copy-number mutants of ColE1. Nature, 283, 216–218. [DOI] [PubMed] [Google Scholar]
- 29.Kim K. and Meyer,R.J. (1989) Unidirectional transfer of broad host-range plasmid R1162 during conjugative mobilization. Evidence for genetically distinct events at oriT. J. Mol. Biol., 208, 501–505. [DOI] [PubMed] [Google Scholar]