Abstract
In vitro transcripts of bacteriophage RNA polymerases (RNAPs), such as T7 RNAP, often suffer from a considerable degree of 3′-end heterogeneity and, with certain promoter sequences, 5′-end heterogeneity. For some applications, this transcript heterogeneity poses a significant problem. A potential solution is to incorporate ribozymes into the transcripts at the 5′- and/or 3′-end of the target RNA sequence. This approach has been used quite widely but has required the generation of new transcription vectors or PCR-derived templates for each new RNA to be studied. To overcome this limitation, we have created two general plasmids for producing homogeneous RNA transcripts: one encodes a 3′- hepatitis delta virus (HDV) ribozyme and the other, used in combination with a two-step PCR, allows the production of double [5′-hammerhead (HH) and 3′-HDV] ribozyme constructs. A choice of cloning and run-off transcription linearisation restriction enzyme sites ensures that virtually any RNA sequence can be cloned and transcribed from these plasmids. For all the RNA sequences tested, good yields of transcript were obtained. These plasmids provide the tools for the simple, rapid creation of new RNA-coding plasmids to produce milligram quantities of homogeneous in vitro transcripts for all applications.
INTRODUCTION
The development of in vitro transcription methods using phage RNA polymerases (RNAPs), in particular T7 RNAP, to produce in vitro RNA transcripts (1–4) opened up many new avenues to RNA researchers. However, the nucleic acid transcripts produced often bear highly heterogeneous 3′-ends as both DNA and RNA polymerases have a tendency to fall off the template before they reach the end of the DNA, or to add non-coded ‘n + 1’ nucleotide(s) to the 3′-end of the nascent nucleic acid (1,2,5,6). Problems of heterogeneity at the 5′-end can also occur in a highly template-dependent manner (7,8).
One of several methods may be used to generate transcription templates: PCR, chemical synthesis or cloning DNA coding the RNA into a suitable bacterial vector. To generate templates from bacterial plasmid DNA, the sequence of interest is cloned downstream of a phage promoter and a restriction site placed at its 3′-end to enable the generation of a linearised template for run-off transcription. Transcript 3′-end heterogeneity is generally reduced relative to synthetic templates and PCR-generated templates, but is not eliminated; non-coded ‘n + 1’ nucleotides may still be incorporated. The actual amount appears to be sequence dependent but can be up to 50% (5,9 and unpublished observations). The additional potential problem of 5′-end heterogeneity also remains. The unwanted RNA species in heterogeneous mixtures can be extremely difficult to remove completely, especially for longer RNA sequences, where they do not resolve well on polyacrylamide gels. The use of self-cleaving RNA sequences (‘ribozymes’) has been widely reported as a means of avoiding these unwanted side products and producing RNA samples with homogeneous 5′- and 3′-ends (10–12). The hammerhead (HH) and hepatitis delta virus (HDV) ribozymes are the most generally amenable for this use, and their sequence and structural requirements are understood (13–15). The HDV ribozyme self-cleaves 5′ to its G + 1 nucleotide, making it ideal for placing at the 3′-end of a target RNA. The incorporation of a HH ribozyme at the 5′- or 3′-end of a target RNA is not straightforward since part of the ribozyme structure (a base-paired arm flanking the cleavage site) incorporates the target sequence requiring a unique complement in each case. This problem can only be overcome by the creation of a new HH sequence (and transcription vector) for each target RNA sequence.
We have created two general plasmids for in vitro transcription of defined RNA sequences. The first vector is tailored to the production of transcripts with homogeneous 3′-ends suitable for most uses. The second has been constructed for use in conjunction with a two-stage PCR method to allow the rapid production of double 5′-HH/3′-HDV ribozyme transcription vectors.
MATERIALS AND METHODS
Construction of 3′-HDV constructs
HDV and initial target RNA (adenovirus VA RNAI from template pT7Ad2VA) coding DNAs were generated by PCR and ligated into pUC19 plasmid. In total, six other RNA coding sequences were cloned into this transcription vector between the EcoRI and NheI restriction sites (see Fig. 1A). Double-stranded DNA inserts coding new target RNA sequences were generated by one of three different methods. Short sequences were chemically synthesised with ‘sticky ends’, phosphorylated and ligated directly into the empty vector. For longer target sequences, either standard PCR or ‘recursive’ PCR (16) methods were used. Additional unique run-off restriction enzyme sites (XhoI and EcoRV) were introduced into plasmids coding VA RNAI and a 58 nt rRNA fragment. Insert orientation and sequence were confirmed by appropriate restriction enzyme digests followed by automated DNA sequencing.
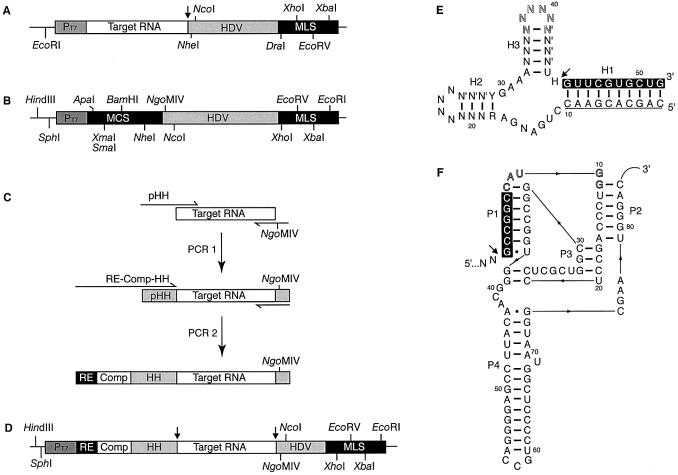
Figure 1.
Plasmids for synthesising RNA transcripts with homogeneous ends. (A) The 3′-HDV transcription plasmid construct; PT7 is the bacteriophage T7 RNAP promoter sequence and HDV is the modified HDV ribozyme RNA. Unique restriction enzyme sites for RNA gene insertion (EcoRI and NheI or NcoI) and sites for template linearisation for run-off in vitro transcription reactions (DraI, XhoI, EcoRV and XbaI) are indicated. (B) pRZ transcription vector construct with the multiple cloning sites [MCS: ApaI, XmaI (SmaI), BamHI, NcoI and NheI] and multiple linearisation sites (MLS: XhoI, EcoRV, XbaI, EcoRI) indicated; HDV and PT7 are as in (A). (C) A general two-stage PCR strategy to produce inserts coding the target RNA and hammerhead ribozyme (HH) with matched complementary sequence (Comp) and a chosen MCS restriction site (RE). A partial HH ribozyme sequence (pHH) is introduced during the first round PCR and used as a template tag in the second round of PCR. (D) The final double ribozyme pRZ-Target RNA transcription vector construct. (E) The consensus HH ribozyme sequence (15); N = any nucleotide, N′ = nucleotide complementary to N, R = purine, Y = pyrimidine, H = any nucleotide except G. The sequence is shown with the 5′-target of the hMRP sequence (black background) and complementary nucleotides (underlined) required for H1 stem formation and cleavage. The nucleotides in the hairpin of stem H3 (outline font) can bear restriction enzyme sequences that allow useful further manipulations of the DNA (described in Results). (F) The modified genomic HDV ribozyme sequence bearing an NgoMIV site in the P1 stem (black background) and an upstream NcoI site (outline lettering). The sites of ribozyme self-cleavage are marked with an arrow in the relevant parts.
Construction of the 5′-HH/3′-HDV ribozyme vector (pRZ)
The functional cassette of the pRZ vector was generated by PCR designed to reposition (by one nucleotide) a native NgoMIV site and introduce the flanking 5′-multiple cloning sites (5′-MCS) and 3′-multiple linearisation sites (3′-MLS) into the PCR product (see Fig. 1B). The DNA produced (∼150 bp) was inserted into pHST7 2BD-1 (17) via ApaI and EcoRI restriction enzyme sites. QuikChange mutagenesis (Stratagene) was used to introduce three point mutations into the 3′-side of the HDV ribozyme P1 stem to restore base-pairing ability and abolish a second native NgoMIV site, creating the plasmid ‘pHST7-RZ’. The functional cassette was then removed from pHST7-RZ, inserted into pUC18 via SphI and EcoRI sites, and the sequence of the resulting plasmid, ‘pRZ’ (Fig. 1B), confirmed by automated DNA sequencing.
Production of pRZ-NME1 and pRZ-hMRP vectors
The target RNA coding inserts were generated via a two-stage PCR and each inserted into pRZ between the ApaI and NgoMIV restriction sites to create the ‘pRZ-NME1’ and ‘pRZ-hMRP’ plasmids (Fig. 1C and D). The first stage PCR amplified the target RNA sequence, Saccharomyces cerevisiae RNase MRP RNA (NME1 RNA, 339 nt) or Homo sapiens RNase MRP RNA (hMRP RNA, 265 nt), from genomic and plasmid DNA templates, respectively. The forward primers for the first round PCR (Fig. 1C) were designed to be complementary to the target sequence (20–30 bp) with an additional flanking region (20–30 bp), encoding part of a HH ribozyme sequence (pHH), intended as a template tag for a second round of PCR. The forward primers for the second round of PCR (50–60 bp) were designed to introduce the entire HH ribozyme sequence and complementary region (designed specifically for each target RNA) along with a suitable restriction site for cloning into the MCS of the pRZ vector (Fig. 1C). The reverse primer was complementary to the desired sequence (20–30 bp) with a flanking NgoMIV restriction site for cloning into the pRZ vector, and was used in both the first and second round PCRs.
RNA transcription reactions
Plasmid templates were digested with the appropriate restriction enzyme to generate linear DNA templates for run-off transcription. After complete digestion, the restriction enzymes were heat deactivated according to the manufacturer’s recommendation, phenol–chloroform extracted, ethanol precipitated and resuspended in TE buffer at a concentration of 0.5 or 1.0 µg/µl.
The 3′-HDV RNA in vitro transcripts were synthesised using 20–50 µg of T7 RNAP (prepared ‘in-house’) in 50–100 µl reactions using optimal solution conditions previously determined for VA RNAI (18) or general RNA transcription (19) at 37°C for 3 h. Transcription and purification of RNAs from large-scale in vitro transcription reactions (>1 ml) were performed as described previously (20).
Transcription from the double ribozyme 5′-HH/3′-HDV templates (pRZ-NME1 and pRZ-hMRP linearised with XhoI) was performed in 50 µl reactions containing 80 mM HEPES buffer pH 7.5, 40 mM dithiothreitol (DTT), 1 mM spermine, 0.05 µg/µl DNA template, 5 mM rNTPs, 20–45 mM MgCl2, 0.05 U of yeast inorganic pyrophosphatase (YIP) and 20–50 µg of T7 RNAP. Following transcription, aliquots of each reaction were removed and adjusted to 40 mM MgCl2 and subjected to three rounds of thermal cycling (1 min at 72°C, 5 min at 65°C and 10 min at 37°C per round). Transcription reactions were scaled up using the same conditions; however, to avoid precipitation of the RNA at the gel surface, it was necessary to dialyse the transcriptions prior to loading onto preparative denaturing gels.
The products of each transcription reaction were resolved on 5 or 8% denaturing polyacrylamide gels. The incorporation of trace [α-32P]UTP (1 µCi/50 µl) in the transcription mixture allowed the visualisation and quantification of bands on a phosphorimager.
RESULTS
Construction of 3′-HDV plasmids and preparation of in vitro transcripts
The 3′-HDV vector was created by inserting target (VA RNAI) and HDV coding DNA sequences into pUC19 downstream of a T7 RNAP promoter sequence with various restriction enzyme sites for cloning and linearisation of the template. The genomic HDV ribozyme sequence was used but with a shortened P4 stem and the first helical region (P1 stem) altered to create a unique NheI site in the plasmid for cloning new sequences. The transcription plasmid initially contained 3′-DraI and XbaI sites for generating linear template. The latter, however, was found to give a very poor yield of transcript for several different RNAs (data not shown) and, as a result, the DraI site was the only viable site for generating linear transcription templates. To ensure that the vector would be compatible with any RNA sequence, additional run-off transcription sites, XhoI and EcoRV, were introduced to create the final 3′-HDV plasmid (Fig. 1A). Curiously, when further separated from the HDV sequence and/or DraI site, linearisation at the XbaI site produced templates that gave good yields of RNA; the final 3′-HDV vectors thus have four useful sites for generating transcription templates.
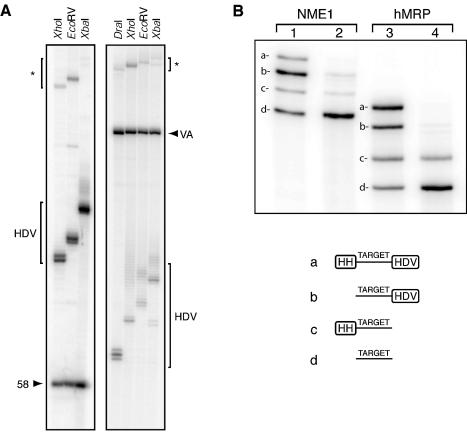
Transcription reactions were performed for a 58 nt RNA fragment and adenovirus VA RNAI using each possible restriction site to generate linear template (the 58 nt RNA sequence encodes a DraI site, so this was not used). For each RNA, a single band corresponding to the target RNA was observed regardless of restriction site used (Fig. 2A). In denaturing gels, VA RNAI, which is thought to contain a denaturation-resistant structure, migrates with an apparent size of 220 nt (21). The HDV ribozyme RNA was observed as two or more bands, presumably corresponding to differing degrees of 3′-heterogeneity for each run-off site. As expected, the gel mobility of the HDV ribozyme RNA increases regularly with the increasing transcript length from each sequential 3′-run-off site (from DraI through to XbaI). To date, five other RNA sequences have been ligated into the 3′-HDV vector using one of three different methods, depending primarily on the length of the target transcript (see Materials and Methods). Each produced target transcripts as single bands of the expected size (not shown). The self-cleavage efficiency of the HDV ribozyme used here is extremely high under the two different sets of transcription conditions, and does not require any buffer exchange, temperature cycling or extended incubations.
Figure 2.
RNA transcription reactions from the 3′-HDV and 5′-HH/3′-HDV plasmid templates. (A) Aliquots (0.5–1.0 µl) of transcription reactions from 3′-HDV templates after 3 h incubation at 37°C separated on 8% acrylamide denaturing gels. Target RNA bands are marked with an arrowhead and labelled: VA (adenovirus VA RNAI, 155 nt) and 58 (58 nt rRNA fragment). HDV ribozyme RNA (HDV) and precursor RNA (*) bands are also indicated. (B) Transcription reactions from the pRZ-NME1 and pRZ-hMRP constructs were analysed by 5% denaturing PAGE (lanes 1 and 2, and 3 and 4, respectively). Transcription reactions were carried out at 5 mM rNTPs/20 mM MgCl2 (lanes 1 and 3). Following transcription, reactions were adjusted to 40 mM MgCl2 and subjected to three rounds of thermal cycling (lanes 2 and 4). The RNA species are indicated and shown schematically under the gel: (a) HH–target–HDV, (b) target–HDV, (c) HH–hMRP and (d) target RNA.
Design of the 5′-HH/3′-HDV ribozyme vector pRZ
The pRZ vector was based upon the genomic HDV sequence but with the native NgoMIV site found in the 5′-side of the P1 stem repositioned from the G2 to the G1 position, resulting in an additional NcoI site that may also be used for cloning purposes (Fig. 1F). Base pairing of the NgoMIV sequence with the 3′-side of the P1 stem was fully restored by mutagenesis. Restriction enzyme sites were introduced to both the 5′- and 3′-sides of the HDV ribozyme to create the MCS and MLS, respectively (Fig. 1B). Sites within the MCS were chosen for their large overhangs, to aid in cloning, with the exception of SmaI, which produces blunt ends and is intended to overcome any compatibility problems with target sequences during cloning. The choice of MLS sites was based on the type of fragment ends produced (5′-overhangs or blunt ends) and enzyme cost as for the 3′-HDV vector.
Cloning of a new gene into the pRZ vector
A general two-stage PCR strategy was used to rapidly create the target pRZ-NME1 and pRZ-hMRP transcription vectors. Initial PCRs (PCR1) were shown to be highly successful using both genomic and plasmid-based templates. The second round PCRs used gel-purified PCR1 DNA as template and resulted in a single band in each case that was subsequently cloned into the pRZ vector. The HH ribozymes used in pRZ-NME1 and pRZ-hMRP are based upon the consensus HH ribozyme sequence (15) (shown in Fig. 1E with the hMRP target sequence and complementary region). The NME1 HH ribozyme and the hMRP HH ribozyme differ both in their overall sequence and in the specific target sequence and length of their H1 stems (18 and 10 bp, respectively). The two-stage PCR method enables the design of almost any HH ribozyme sequence, allowing each to be tailored to the individual target RNA. A thorough consideration of the target sequence at the design stage allows useful modifications to the HH sequence. Incorporation of a suitable restriction enzyme site into the H3 hairpin of the HH ribozyme (nucleotides in outline font in Fig. 1E) allows the ligation of a synthetic double-stranded DNA insert with the appropriate restriction site overhangs between this restriction site and the chosen MCS restriction site. This has two potential advantages: first, existing vectors can be easily modified allowing the HH ribozyme sequence and length of the helices to be altered if necessary, and, secondly, in the event of difficulties with the second round of PCR, this allows a sequential or three end ligation along with the first round PCR product. Our second round PCRs were entirely successful, making this method unnecessary. However, as PCR is often highly sequence dependent, the availability of a second cloning strategy ensures that the required transcription vector can be produced should any difficulties arise.
Transcription reactions using pRZ templates
The transcription of RNA can be highly sequence dependent, requiring the optimisation of each new construct before large-scale transcription. Each template was individually optimised for overall transcription and ribozyme cleavage by performing small-scale transcriptions, varying the rNTP and magnesium ion ratios. In both cases, high concentrations of magnesium stimulated ribozyme cleavage but reduced the efficiency of transcription, resulting in poor overall yields of RNA (not shown). Transcription at lower magnesium concentrations (20 mM MgCl2 with 5 mM rNTPs) resulted in good overall yields of RNA despite some non-cleaved products (Fig. 2B, lanes 1 and 3). The yield of target RNAs could be further increased by enhancing correct folding and ribozyme cleavage through denaturing/renaturing thermal cycles in the presence of an elevated magnesium concentration (adjusted to 40 mM; Fig. 2B, lanes 2 and 4). After employing thermal cycling, we have gel purified ∼1.5 mg/ml transcription of NME1 RNA and ∼750 µg/ml transcription of hMRP RNA (each from transcriptions containing 50 µg of DNA template per ml). Yields of the NME1 RNA were higher due to both more efficient transcription and more effective HH ribozyme cleavage (Fig. 2B). Analysis with Imagequant software (Molecular Dynamics) shows that the NME1 HH ribozyme (18 bp H1 stem) cleaves to >80%, whereas the hMRP HH ribozyme (10 bp H1 stem) was shown to cleave to >70% following thermal cycling. We attribute the difference in HH ribozyme cleavage efficiency to folding problems with the hMRP HH ribozyme due to the shorter H1 stem. Both target RNAs have secondary structures that contain a base-paired stem involving their 5′- and 3′-ends which may compete with correct HH ribozyme H1 stem formation. We highlight this as a design consideration, as different target RNAs will require the H1 stem of the HH ribozyme to be tailored not only to the target RNA sequence but also with some consideration of their structure at the 5′-end.
DISCUSSION
Several methods have been reported that reduce or remove the problem of transcript heterogeneity, particularly at the 3′-end, where heterogeneity is more common. The chemical synthesis of templates containing one or preferably two modified residues (2′-OCH3) at their 3′-end can reduce the amount of ‘n + 1’ run over transcripts (22), and the method has been extended to RNAs of moderate length (e.g. tRNAs) by using overlapping DNA oligonucleotides as a template for Klenow fragment extension (23). Similarly, PCR extension has been used to generate DNA templates bearing the HDV ribozyme at the 3′-end of a given sequence with a T7 promoter placed upstream of the target (9). Extension methods can generate transcription templates rapidly, but suffer some significant drawbacks: they are generally limited to the synthesis of small RNAs, the PCR must be repeated each time the template is required or in many reactions to generate large quantities, and errors incorporated during the PCR are reproduced in the RNA transcript. Such errors cannot be easily detected. Template-directed RNase H cleavage (24) has been used as a more direct method to remove the 5′-leader sequence of an RNA transcript and could equally be employed at the 3′-end. However, this method represents a considerable additional step and relies upon the use of chemically modified oligonucleotides to direct the RNase H cleavage.
The incorporation of specific ribozyme sequences at the 5′- and/or 3′-end of transcripts is a preferred method to quickly and easily improve RNA sample homogeneity for a specified RNA sequence (10,11). Plasmid-based templates can be easily produced in large quantity from a single bacterial culture and, unlike PCR-based methods, do not suffer the likelihood of sequence errors. However, the creation of such plasmid templates for each new RNA sequence has often been a time-consuming process. We have designed two vectors that significantly simplify the production of templates for producing run-off transcripts with homogeneous 3′- or 5′- and 3′-ends. These plasmids bear HDV ribozymes with engineered cloning sites that allow the HDV ribozyme to remain in the vector so that it can be omitted from the cloning procedure, dramatically simplifying the production of 3′-HDV transcription templates. A similar vector has been further tailored for use in conjunction with a two-stage PCR method to quickly and easily produce transcription templates bearing 5′-HH and 3′-HDV ribozymes.
The 3′-HDV plasmid encodes a modified HDV ribozyme at the 3′-end of the target RNA and is designed such that the self-cleavage reaction yields the target RNA transcript with authentic sequence. New RNA coding DNA inserts can be easily cloned into this plasmid between unique restriction enzyme sites. Two unique sites for cloning are available at the junction of the target RNA and HDV ribozyme sequences (NheI and NcoI) to avoid incompatibility with targets that contain either site. We have used this general plasmid system to produce transcription vectors for seven RNAs of varying length (20–203 nt) and sequence. With all but one target sequence tested, the HDV ribozyme cleavage was extremely efficient (even in the case where it was poorer, a good yield of target transcript was obtained). The plasmid also contains several unique restriction sites for run-off transcription, the choice of which was guided by two factors: first, that the resulting template should either be blunt ended or have a 5′-overhang to avoid complementary strand transcripts (25) and, secondly, the cost of the restriction enzyme itself. The MLSs confer two significant benefits: incompatibility with sites occurring naturally in the target RNA is avoided and gel migration of the HDV ribozyme can be varied to overcome the potential problem of resolving ribozyme and target RNA bands where they are of similar size. In principle, therefore, there is no limit to the RNA sequences that can be transcribed from this plasmid.
There is generally less potential for transcript heterogeneity at the 5′-end compared with the 3′-end, but with certain sequences or for some applications the problem is significant. Again heterogeneity can be overcome using a self-cleaving ribozyme, and incorporation of a 5′-ribozyme has two further benefits of particular use in some circumstances. First, where target sequence authenticity is important, the 5′-HH ribozyme removes the necessity for specific nucleotides (preferably one or more G) at the 5′-end. Leader sequences can be used that give good yields of transcript, which are subsequently removed by ribozyme self-cleavage. Secondly, it eliminates the need to remove the 5′-triphosphate group from an RNA transcript before use in end labelling procedures as a 5′-OH RNA is produced. For NME1 RNA, we were unable to remove the 5′-triphosphate using an alkaline phosphatase treatment, but we subsequently achieved >99% labelling efficiency using RNA derived from the pRZ-NME1 construct (unpublished observations).
The pRZ vector retains the key features of the 3′-HDV vector, including MLSs and an HDV ribozyme with engineered restriction sites (NgoMIV and NcoI) to simplify target sequence cloning. This construct also contains a T7 promoter and downstream MCSs designed to aid in the production of 5′-HH/3′-HDV transcription templates. The production of 5′-HH ribozyme transcription constructs is often complicated as the H1 stem of the HH ribozyme incorporates the 5′-end of the target sequence requiring a region of complementarity that must be placed upstream of the HH ribozyme itself. We have shown that a rapid and simple two-stage PCR method can be used to generate an insert bearing the required target with its own tailored HH ribozyme. The method allows flexibility in the choice of HH ribozyme sequence used, allowing the design of a different HH sequence appropriate to each target RNA. The reported vector pRZ has been designed to easily accommodate these inserts, resulting in a mature transcription vector bearing the required insert and both the transcription promoter (T7) and an accurately positioned 3′-HDV ribozyme.
We have used two examples of relatively large structural RNAs, of interest to us, to emphasize some of the considerations in the design and production of these 5′-HH/3′-HDV transcription vectors and highlight the optimisation of RNA transcription required from such templates. These constructs allow the production of milligram quantities of RNA transcripts with homogeneous 5′- and 3′-ends, suitable for the most demanding of applications.
Acknowledgments
ACKNOWLEDGEMENTS
Plasmids pT7Ad2VA and pGEM 3Zf(+) hMRP were the generous gifts of Professor Mike Mathews (University of Medicine and Dentistry of New Jersey) and Dr Ger Prujin (University of Nijmegen), respectively. This work was supported by a Wellcome Trust career development fellowship (G.L.C.) and The Medical Research Council career development grant G9900092 (J.M.A.). We are also grateful to the UMIST LSI for additional studentship support for S.C.W.
REFERENCES
- 1.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milligan J.F. and Uhlenbeck,O.C. (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol., 180, 51–62. [DOI] [PubMed] [Google Scholar]
- 3.Krieg P.A. and Melton,D.A. (1987) In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol., 155, 397–415. [DOI] [PubMed] [Google Scholar]
- 4.Melton D.A., Krieg,P.A., Rebagliati,M.R., Maniatis,T., Zinn,K. and Green,M.R. (1984) Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res., 12, 7035–7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kholod N., Vassilenko,K., Shlyapnikov,M., Ksenzenko,V. and Kisselev,L. (1998) Preparation of active tRNA gene transcripts devoid of 3′-extended products and dimers. Nucleic Acids Res., 26, 2500–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Draper D.E., White,S.A. and Kean,J.M. (1988) Preparation of specific ribosomal RNA fragments. Methods Enzymol., 164, 221–237. [DOI] [PubMed] [Google Scholar]
- 7.Pleiss J.A., Derrick,M.L. and Uhlenbeck,O.C. (1998) T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA, 4, 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helm M., Brule,H., Giege,R. and Florentz,C. (1999) More mistakes by T7 RNA polymerase at the 5′ ends of in vitro-transcribed RNAs. RNA, 5, 618–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schurer H., Lang,K., Schuster,J. and Morl,M. (2002) A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res., 30, e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price S.R., Ito,N., Oubridge,C., Avis,J.M. and Nagai,K. (1995) Crystallization of RNA–protein complexes. I. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J. Mol. Biol., 249, 398–408. [DOI] [PubMed] [Google Scholar]
- 11.Ferre-D’Amare A.R. and Doudna,J.A. (1996) Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res., 24, 977–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price S.R., Oubridge,C., Varani,G. and Nagai,K. (1998) Preparation of RNA: Protein Complexes for X-Ray Crystallography and NMR. Oxford University Press, New York, NY. [Google Scholar]
- 13.Lilley D.M. (1999) Structure, folding and catalysis of the small nucleolytic ribozymes. Curr. Opin. Struct. Biol., 9, 330–338. [DOI] [PubMed] [Google Scholar]
- 14.Doherty E.A. and Doudna,J.A. (2001) Ribozyme structures and mechanisms. Annu. Rev. Biophys Biomol. Struct., 30, 457–475. [DOI] [PubMed] [Google Scholar]
- 15.Eckstein F., Kore,A.R. and Nakamaye,K.L. (2001) In vitro selection of hammerhead ribozyme sequence variants. Chembiochem, 2, 629–635. [DOI] [PubMed] [Google Scholar]
- 16.Prodromou C. and Pearl,L.H. (1992) Recursive PCR: a novel technique for total gene synthesis. Protein Eng., 5, 827–829. [DOI] [PubMed] [Google Scholar]
- 17.Vos S., Berrisford,D.J. and Avis,J.M. (2002) Effect of magnesium ions on the tertiary structure of the hepatitis C virus IRES and its affinity for the cyclic peptide antibiotic viomycin. Biochemistry, 41, 5383–5396. [DOI] [PubMed] [Google Scholar]
- 18.Pe’ery T. and Mathews,M.B. (1997) Synthesis and purification of single-stranded RNA for use in experiments with PKR and in cell-free translation systems. Methods, 11, 371–381. [DOI] [PubMed] [Google Scholar]
- 19.Gurevich V.V. (1996) Use of bacteriophage RNA polymerase in RNA synthesis. Methods Enzymol., 275, 382–397. [DOI] [PubMed] [Google Scholar]
- 20.Conn G.L., Gutell,R.R. and Draper,D.E. (1998) A functional ribosomal RNA tertiary structure involves a base triple interaction. Biochemistry, 37, 11980–11988. [DOI] [PubMed] [Google Scholar]
- 21.Clarke P.A., Pe’ery,T., Ma,Y. and Mathews,M.B. (1994) Structural features of adenovirus 2-associated RNA required for binding to the protein kinase DAI. Nucleic Acids Res., 22, 4364–4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao C., Zheng,M. and Rudisser,S. (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA, 5, 1268–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherlin L.D., Bullock,T.L., Nissan,T.A., Perona,J.J., Lariviere,F.J., Uhlenbeck,O.C. and Scaringe,S.A. (2001) Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA, 7, 1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 24.Lapham J. and Crothers,D.M. (1996) RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA, 2, 289–296. [PMC free article] [PubMed] [Google Scholar]
- 25.Schenborn E.T. and Mierendorf,R.C.,Jr (1985) A novel transcription property of SP6 and T7 RNA polymerases: dependence on template structure. Nucleic Acids Res., 13, 6223–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]