Abstract
We describe a strategy to analyze the impact of single nucleotide mutations on protein function. Our method utilizes a combination of yeast functional complementation, growth competition of mutant pools and polyacrylamide gel immobilized PCR. A system was constructed in which the yeast PGK1 gene was expressed from a plasmid-borne copy of the gene in a PGK1 deletion strain of Saccharomyces cerevisiae. Using this system, we demonstrated that the enrichment or depletion of PGK1 point mutants from a mixed culture was consistent with the expected results based on the isolated growth rates of the mutants. Enrichment or depletion of individual point mutants was shown to result from increases or decreases, respectively, in the specific activities of the encoded proteins. Further, we demonstrate the ability to analyze the functional effect of many individual point mutations in parallel. By functional complementation of yeast deletions with human homologs, our technique could be readily applied to the functional analysis of single nucleotide polymorphisms in human genes of medical interest.
INTRODUCTION
How does the information encoded in the genome and genetic variability affect fitness? This question, which seeks a relation between the genotype and the phenotype, is a fundamental question facing the biological sciences. The recently published human genome sequence (1,2) and major ongoing efforts devoted to describing the genetic variation within the human population (3) are early steps in addressing this question. One of the next steps is understanding the physiological effect of genetic diversity in the human population. Specifically, it is critical to identify and characterize the subset of genetic variations and mutations which impact on biochemical function. In order to achieve this goal several additional questions must be addressed. How do we identify functionally important mutations? How do we identify functionally important residues within a protein? How do we analyze the relation between single point mutations and the complex integrated functions in a cellular system?
The objective of this work was the development of a quantifiable technique which can be used to measure the relative impact of single nucleotide mutations on protein function. Ultimately, a technique that can be used to evaluate the effect of many individual mutations in a highly parallel manner is desired. To address these goals, we have used a functional assay in Saccharomyces cerevisiae in conjunction with a competitive growth strategy in which mutants were enriched or depleted from a culture according to the fitness of the mutant. The concept of this method was similar to those used by Steinmetz et al. (4) and Winzeler et al. (5) in which the fitness of whole gene deletion S.cerevisiae mutants was compared in various growth media. Badarinarayana et al. (6) also demonstrated the differential enrichment of insertion mutant Escherichia coli strains in selective and non-selective media, thus providing a highly parallel method to identify functionally important proteins. In each case, specially designed microarrays were used to quantify mutant enrichment.
Herein we describe a screening strategy, based on functional complementation, in which expression of S.cerevisiae phosphoglycerate kinase was restored to a PGK1 deletion strain by transforming the strain with a plasmid bearing a copy of this gene. Growth of the organism in glucose minimal medium was dependent upon the functionality of the plasmid-borne gene. Eight plasmids were constructed, each containing a different single base mutation in the PGK1 gene. These plasmids were transformed into S.cerevisiae. Pools containing various combinations of these strains were grown competitively in two different experiments. The fraction of the cultures represented by each strain in the pool was determined at various times using immobilized PCR colony (polony) technology (7). Briefly, polonies arise from individual DNA molecules polymerized into an acrylamide gel atop a glass microscope slide and thermal cycled with other PCR components. The polyacrylamide gel retards diffusion such that the resulting PCR product is localized in a spherical region surrounding the original template molecule. Individual polonies can then be identified using single base extensions (SBE) with fluorescently labeled nucleotides. This technology was selected for its ability to discriminate, quantitatively and in parallel, large numbers of DNA molecules differing by only a single base pair.
A growth model was applied to the competitive growth data and used to determine the specific growth rate for each individual strain. The calculated growth rates were qualitatively consistent with relative in vitro activities of the mutant proteins reported previously and were consistent with the growth rates of the strains when determined directly from individual mutant growth experiments.
MATERIALS AND METHODS
Strain and media
The starting strain used in all experiments was the CEN.PK381-2B haploid strain of S.cerevisiae (8). This strain carries the ura3-52 mutation and the PGK1 gene has been deleted using the short flanking sequence method. The strain is unable to use glucose as its sole carbon source.
The following media were used. YPD (9): 10 g/l yeast extract (Fisher Scientific), 20 g/l peptone (Fisher Scientific), 20 g/l dextrose (Fisher Scientific). YPGE: 10 g/l yeast extract, 20 g/l peptone, 5 g/l ethyl alcohol (Fisher Scientific), 5 g/l glycerol (Fisher Scientific). SD: 6.7 g/l yeast nitrogen base without amino acids (Difco), 20 g/l dextrose. SGE: 6.7 g/l yeast nitrogen base without amino acids, 5 g/l ethyl alcohol, 5 g/l glycerol. LB: 10 g/l Tryptone (Difco), 5 g/l yeast extract, 10 g/l NaCl. SD and SGE media were supplemented with 20 mg/l uracil (Sigma Aldrich) as required. SD and SGE media were supplemented with 150 mg/l (liquid formulations) or 300 mg/l (solid agar formulations) geneticin (G-418; Gibco BRL). LB medium was supplemented with 50 mg/l ampicillin. Solid agar media were made using 15 g/l agar (Fisher Scientific).
Nucleic acid manipulation and plasmid construction
The PGK1 gene insertion cassette was constructed with flanking 5′ BamHI and 3′ HindIII restriction sites by amplifying the wild-type gene from 200 ng of purified yeast genomic DNA (Research Genetics) using PCR (Pfu Turbo; Stratagene) (94°C for 5 min, 30 cycles of 94°C for 45 s, 60°C for 45 s, 72°C for 2.5 min and a final 72°C extension for 5 min) (MJ Research PTC200 thermal cycler). All oligonucleotide primers used in this and subsequent methods are listed in Supplementary Material. The PCR product was purified (Qiaquick PCR Purification Kit; Qiagen) to remove excess primer and template DNA.
The PGK1 gene was cloned into the p416ADH and p416CYC1 low copy number (CEN6/ARS4 ori) plasmids (10) as described (8) using the HindIII and BamHI sites and then used to transform the CEN.PK381-2B strain (11). Plasmid DNA was isolated (12) and the PGK1 coding region was sequenced. The constructs were named p416ADH/PGK1-WT and p416CYC1/PGK1-WT. The PGK1 gene was constitutively expressed from these plasmids and the plasmids differ only in the promoter used to drive protein expression; p416ADH carries the mid strength ADH promoter while p416CYC1 carries the relatively weak CYC1 promoter.
Mutant construction
Eight single base mutations (within the PGK1 coding region) of the p416CYC1/PGK1-WT plasmid were generated (QuickChange Site-Directed Mutagenesis Kit; Stratagene) according to the manufacturer’s protocol. Individual E.coli colonies arising from each mutagenesis reaction were selected and cultured. Plasmid DNA was isolated from each culture and sequenced to confirm the presence of the desired mutation and no other mutations. Each mutant plasmid was named according to the resulting mutation in the coded protein: p416CYC1/PGK1-Arg65Ser, p416CYC1/PGK1-Lys131Glu, etc. (Table 1). Each plasmid was used to transform the CEN.PK381-2B strain and, with the exception of Gly370Ala, all plasmids restored the ability of the PGK– strain to grow using glucose as the sole carbon source. Selected mutants were chosen based upon one or more of the following factors: available in vitro activity data (Arg65Ser, Glu190Gln and His388Glu), proximity to or location within a catalytic site (Lys131Glu and Gly370Ala) and proximity to other mutations for multiplexing experiments (Arg65Ser, Leu76Trp, Val114Asp and Ile124Phe).
Table 1. Individual PGK1 mutant growth experiments.
| Mutant (promoter) | µ (per h)a | Percent wild-type (CYC1) | In vitro protein activity (% native)a |
|---|---|---|---|
| Wild-type (ADH) | 0.391 ± 0.003 | 147% | 100% |
| Wild-type (CYC1) | 0.266 ± 0.024 | 100% | 100% |
| Arg65Ser (CYC1) | 0.252 ± 0.010 | 95% | 57% (50 mM SO42–) |
| 163% (0 mM SO42–) (14) | |||
| Leu76Trp (CYC1) | 0.310 ± 0.011 | 117% | N/A |
| Val114Asp (CYC1) | 0.298 ± 0.004 | 112% | N/A |
| Ile124Phe (CYC1) | 0.273 ± 0.004 | 102% | N/A |
| Lys131Glu (CYC1) | 0.264 ± 0.004 | 99% | N/A (15) |
| Glu190Gln (CYC1) | 0.177 ± 0.001 | 67% | 6% (30 mM SO42–) |
| 26% (0 mM SO42–) (16) | |||
| His388Glu (CYC1) | 0.249 ± 0.020 | 94% | 20% (50 mM SO42–) (17) |
| Gly370Ala (CYC1) | 0b | 0%b | N/A |
N/A, not available.
aGrowth medium contained 40 mM extracellular SO42–.
bAbility to grow in glucose minimal medium was not restored by transformation with a plasmid bearing the Gly370Ala mutation.
Culture growth
All liquid cultures were grown using either Innova 4080 and Series 25 incubator shakers (New Brunswick Scientific) or a 2 l BioFlow 110 (New Brunswick Scientific) bench-top fermentor. Escherichia coli shake flask cultures were conducted at 37°C and 300 r.p.m. Saccharomyces cerevisiae shake flask cultures were conducted at 28.5°C and 300 r.p.m. The first competitive growth experiment was conducted using the BioFlow 110 as follows. Precultures containing SD medium were inoculated from frozen stocks of each individual mutant. These cultures were grown overnight and used to inoculate fresh medium as necessary to maintain exponential growth. When each preculture shake flask had reached a similar cell density (OD660 ≈ 0.1–0.5), precultures were pooled such that approximately equal quantities of each mutant were present in the pool and inoculated into 1.5 l of SD medium in the BioFlow 110. Maximum growth rate was maintained in the fermentor for 100 h (∼40 population doublings) by operating in a repeated batch mode; prior to exhausting nutrients in the medium, spent medium was drained from the fermentor and fresh medium was added. The fermentor controlled the temperature at 28.5°C, pH at 5.0 and dissolved oxygen concentration at >50%.
The second competitive growth experiment was conducted in shake flasks. Preculture growth was identical to that described above. An appropriate amount of the preculture pool was used to inoculate 250 ml of SD medium such that the culture would grow exponentially for 24–36 h. Upon reaching an OD660 of ∼0.5–1.0, this culture was used to inoculate 250 ml of fresh SD medium and the procedure was repeated until the elapsed total culture time was 100 h (∼40 population doublings).
Polony slides
Plasmid DNA from the competitive growth experiment was used to quantify the relative concentrations of each mutant in culture as a function of time. Plasmid DNA was isolated from culture samples (12). The DNA was further purified using a Qiagen Miniprep Kit. Purified plasmid DNA (200–300 ng) was used as the template in a PCR reaction (HotStart Taq; Qiagen) (94°C for 15 min, 35 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 1.5 min and a final 72°C extension for 5 min) to isolate and amplify only the PGK1 coding region of the plasmid. The PCR product was used as the PCR template in the polony gels.
Polony reactions were conducted as described (7). A master mix was made containing 1.1× Jumpstart buffer (Sigma Aldrich), 0.22 µM dNTP (Stratagene), 0.22% (w/v) BSA (Sigma Aldrich), 0.22% (v/v) Tween 20 (Fisher Scientific) and 11% (w/v) 19:1 acrylamide:bisacrylamide (Fisher Scientific). To 67.2 µl of the master mix was added 3.32 µl of template DNA (100–5000 molecules), 0.75 µl of each forward and reverse primer (50 µM), 27.65 U of JumpStart Taq (Sigma Aldrich), 1.41 µl of 5% (v/v) TEMED (Pharmacia), 1.41 µl of 5% (w/v) ammonium persulfate (Pharmacia) and 5 µl of distilled water. An aliquot of 20 µl of the mixture was transferred to the well of a teflon-coated bind-silane (Pharmacia)-treated (bind-silane increases adherence of the polyacrylamide gel to the glass surface) glass slide (Erie Scientific). The well was covered with a coverslip and the gel was allowed to polymerize for 10 min. The coverslip was then covered with a hybridization well (Grace Bio-Labs), mineral oil was injected into the hybridization well and it was sealed. The slides were thermal cycled (24°C for 10 min, 94°C for 2 min, 40 cycles of 94°C for 15 s, 55°C for 15 s, 72°C for 30 s and a final 72°C extension for 2 min) using a PTC200 thermal cycler adapted for use with glass slides (16/16 twin tower block; MJ Research). Primers used in this step are named according to the region of the PGK1 gene that they amplify. The hybridization wells were then removed, the slides were washed in hexane to remove the mineral oil and the glass slide covers were removed.
Polony slides were then either SYBR Green stained (2× SYBR Green I in TBE, pH 8.0 for 10 min) and imaged or the single base extension protocol was initiated without completing this intermediate step. Slides were imaged on a ScanArray Express (GSI Lumonics).
Single base extension sequencing
Single base extension sequencing was used to identify and quantify single base mutations in the polonies. Polony slides were initially incubated in 70% (v/v) formamide in 1× SSC solution for 15 min at 70°C to denature the double-stranded polony DNA. Following the denaturing step, electrophoresis (42% w/v urea in 0.5× TBE) was conducted to remove the DNA strand not covalently bound to the gel with the 5′ acrydite. The electrophoresis was run for 2.5 h at 140 V in a standard DNA gel electrophoresis box. Slides were then washed in Wash 1E (100 mM Tris pH 7.5, 20 mM EDTA, 500 mM KCl). An aliquot of 100 µl of 1 µM sequencing primer in 6× SSPE and 0.1% (v/v) Triton X-100 (Acros Organics) was placed on the surface of the gel, covered with a hybridization well and allowed to anneal to the immobilized polony DNA (94°C for 2 min, then 50°C for 20 min). Slides were washed in Wash 1E and equilibrated for 5 min in Klenow Extension Buffer (50 mM Tris pH 7.5, 5 mM MgCl2, 0.01% v/v Triton X-100). Slides were then covered with 55 µl of an extension solution consisting of 50.67 µl Klenow Extension Buffer, 1 µl Klenow fragment (50 000 U/ml; New England Biolabs), 0.83 µl single-stranded DNA binding protein (1–5 µg/µl; USB) and 2.5 µl of either cyanine-5-dATP, cyanine-5-dUTP, cyanine-5-dCTP or cyanine-5-dGTP (10 µM; PerkinElmer Life Sciences) depending upon the desired extension. The extension reaction was allowed to proceed for 3 min at room temperature. Slides were then washed in Wash 1E and imaged.
Subsequent extension reactions were conducted in essentially the same manner. However, following the formamide treatment the electrophoresis was omitted. Slides were washed in Wash 1E and the procedure above was resumed at the sequencing primer annealing step.
In special cases where the nucleotide following the nucleotide at the position of interest was different from either the wild-type or mutant nucleotide at the target position, it was possible to extend both the mutant and the wild-type concurrently. The procedure was identical to that described above with the following exception. Both cyanine-5- and cyanine-3-labeled nucleotides were included in the extension mixture. Depending upon the extension, the cyanine-5-labeled nucleotide was used to identify either the wild-type or the mutant at the position of interest and the cyanine-3-labeled nucleotide was used to identify the other.
Polony expression profiling
Total RNA was isolated from four strains of S.cerevisiae: wild-type PGK1 (ADH promoter), wild-type PGK1 (CYC1 promoter), Arg65Ser (CYC1 promoter) and Lys131Glu (CYC1 promoter). Strains were grown in SD at 28.5°C to mid-log phase (OD660 0.5–1.0). Culture samples (50 ml) were rapidly cooled in liquid nitrogen to 4°C and pelleted. The pellets were resuspended in RNAwiz buffer (Ambion). Cells were lysed using a Mini-Bead Beater (Biospec) and RNA was isolated using the RNAwiz protocol. The RNA pellet was suspended in 20 µl of RNase-free water. Contaminating DNA was digested using DNase I according to the manufacturer’s protocol (Gibco). Total RNA levels were quantified using A260 measurements. cDNA was synthesized from 40 µg of this RNA using reverse transcriptase (Gibco) according to the method outlined by the supplier. cDNA was purified by RNase H (Gibco) digestion and treatment with 5 µl of Clontech enzyme removal resin. cDNA was ethanol precipitated and then resuspended in 25 µl of RNase-free water.
Polony slides were made using the cDNA as template. The method was identical to that described above except that two sets of polony primers were included in the polony gels. The first primer set was designed to amplify nucleotides 159–466 of the PGK1 gene. The second primer set amplified nucleotides 257–564 of the yeast AKY2 gene. After polony synthesis, the single base extension protocol described above was implemented. Two sequencing primers designed to hybridize to regions in either the PGK1 or the AKY2 polonies were included in the annealing mixture. Polonies were then extended with Klenow fragment as described above in the presence of both cyanine-3-dUTP and cyanine-5-dATP. Design of the sequencing primers allowed concurrent extension, such that cyanine-3 fluorescence identified a PGK1 polony and cyanine-5 fluorescence identified an AKY2 polony.
RESULTS
Simulations of PGK1 mutants with a flux balance model
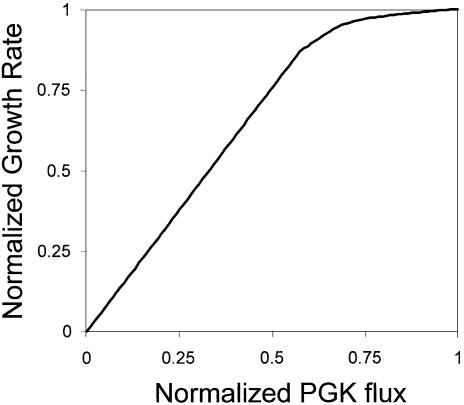
A flux balance model of the core metabolic pathways in S.cerevisiae was used to determine the optimal relationship between the flux in the phosphoglycerate reaction and the optimal growth rate [as has been previously performed with E.coli (13)]. It was determined that the growth rate and the flux were linearly related at flux levels below ∼60% of the optimal flux (Fig. 1). Therefore, we can expect to see a relationship between the effect of mutations in the PGK1 gene and growth rate in the competition experiments (the measured variable).
Figure 1.
Saccharomyces cerevisiae growth as a function of the flux in the phosphoglyerate kinase reaction. The growth rate was calculated using a FBA model of the core metabolic pathways in S.cerevisiase as described (13). The growth rate was normalized to the optimal growth rate without flux limitations. The flux in the phosphoglycerate reaction was normalized to the optimal value.
Growth of individual mutants
The specific growth rate (µ) of each individual mutant was determined by growing each separately (in triplicate) in shake flask cultures. A least squares curve fitting routine was used to determine the growth rate of each mutant strain. In all cases the quality of the curve fit was determined (R2 > 0.99). The averages and standard deviations of the three shake flasks grown for each mutant are reported in Table 1 with the in vitro mutant protein activity reported previously (14–17). All plasmids tested with the exception of p416CYC1/PGK1-Gly370Ala restored the ability of the PGK1 deletion strain to grow using glucose as the sole carbon source. It appears that the Gly370Ala mutation, which lies between the Mg-ATP and the 3-phosphoglycerate binding sites of the phosphoglycerate kinase protein (18), hinders the interaction of the two substrates such that the PGK Gly370Ala mutation is unable to complement growth in this S.cerevisiae strain.
The specific growth rates of strains grown in isola tion ranged from 0.177 (p416CYC1/PGK1-Glu190Gln) to 0.391 per h (p416ADH/PGK1-WT) or 67 to 147% that of the growth of the strain transformed with p416CYC1/PGK1-WT.
Expression profiling of individual mutants
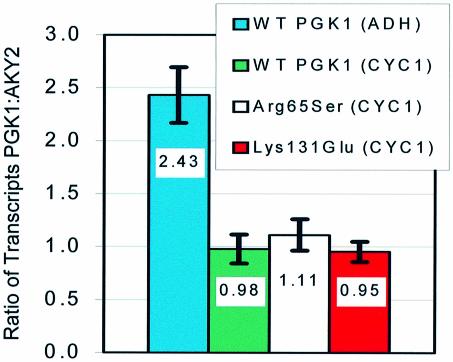
In order to demonstrate that the observed differences in growth rate of individual PGK1 mutants were not the result of different transcription rates, transcript expression levels of several of the strains used in this work were quantified using polony gels as described in Materials and Methods. The results of these experiments are shown in Figure 2 and are presented as the ratio of PGK1 to AKY2 transcripts. The AKY2 gene was used to normalize PGK1 expression data because its cellular mRNA concentrations reportedly change little under various growth conditions (e.g. elapse of the growth cycle, growth on various carbon sources, etc.) (19). Initially, strains bearing plasmids expressing wild-type PGK1 under the control of either a moderate strength promoter (ADH) or a weak promoter (mutant CYC1), but otherwise identical, were tested for PGK1 mRNA level and growth rate. The relative expression level of the two promoters/plasmids expressing wild-type PGK1 was 2.5:1 (ADH:CYC1) and agreed approximately with the value reported previously (10). Further, the growth rate of the strain expressing PGK1 under the control of the weak promoter was lower than that of the strain carrying the moderate strength promoter (Table 1). PGK1 expression levels of each mutant tested (all utilizing the CYC1 promoter) were approximately equal to that of the strain expressing wild-type PGK1 (CYC1 promoter).
Figure 2.
PGK1 mRNA expression levels of several strains used in this work. Expression from plasmids utilizing the CYC1 promoter was approximately equal in the three representative strains tested. The ratio of expression from the ADH and CYC1 promoters was 2.5:1 and agreed approximately with previously reported results. AKY2 mRNA constitutively expressed from the genomic copy of the gene was used to normalize PGK1 expression.
Taken together, these results indicate that the growth rates of strains expressing the PGK1 gene under the control of the CYC1 promoter are limited by the rates of PGK1 expression. Furthermore, observed differences in the growth rates of PGK1 mutants expressing the gene under the control of the CYC1 promoter result from changes in the specific activities of the expressed proteins rather than the level of gene expression at the mRNA level.
Competitive growth experiment
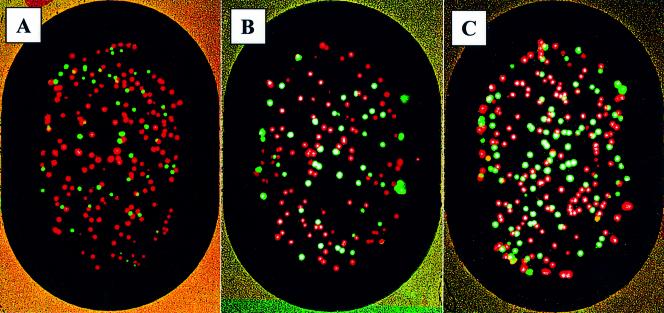
We next tested whether or not the polony assay could be used to measure many mutant growth rates in parallel. As an alternative to measuring individual growth rates of each mutant, a competitive growth experiment was conducted in which strains carrying the plasmids p416CYC1/PGK1-WT, p416CYC1/PGK1-Arg65Ser, p416CYC1/PGK1-Lys131Glu, p416CYC1/PGK1-Glu190Gln and p416CYC1/PGK1-His388Gln were pooled and grown in a single culture. Plasmid DNA was isolated from the culture at 24 h intervals and used as the PCR template in the polony gels. Polony primers were designed to amplify ∼300 bp segments of the PGK1 gene, each containing one or two of the sites at which mutations were placed. Four slides were made with each primer set. Two of these slides were extended to identify polonies with the wild-type nucleotide at the given position and the other two were similarly extended with the mutant targeted fluorescent nucleotide. The slides were then imaged, the extended primer was removed and the procedure was repeated with the mutant extension switched to the first set of slides and the wild-type extension to the second set. The images were then overlaid with the Spotfinder software (TIGR v. 1.0.0) and the number of polonies which were mutant or wild-type at the specific position was determined. Representative polony slides are shown in Figure 3.
Figure 3.
Representative polony images. Lys131Glu mutant time series from left to right: (A) 0 h, (B) 51.75 h and (C) 97 h. Polonies were initially extended with cyanine-5-dCTP (A and C) or cyanine-5-dUTP (B). The slides were then imaged and the extended primers were removed. The procedure was then repeated with cyanine-5-dCTP used to extend (B) and cyanine-5-dUTP used to extend (A) and (C). A positive extension with cyanine-5-dUTP indicates that the wild-type nucleotide is present at position 391 of PGK1 (red), while positive extension with cyanine-5-dCTP indicates the mutant nucleotide (green) at this position.
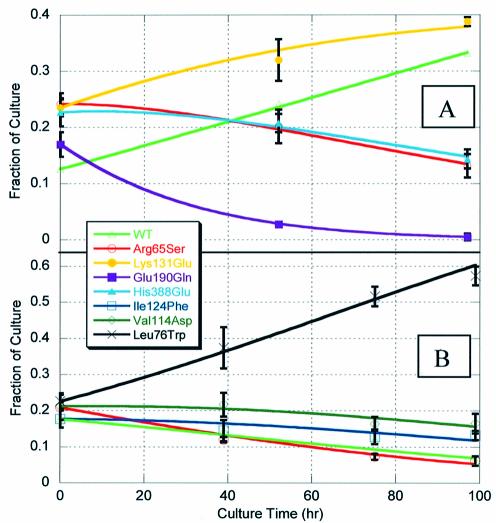
Results of this competitive growth experiment are shown in Figure 4A. Additionally, curve fits to the SBE data were performed using the exponential growth equations of the form:
Figure 4.
Fraction of each mutant strain in culture as a function of time during competitive growth experiments. Fractions were measured using polony gels and single base primer extension sequencing. The fraction of the wild-type strain was determined by subtracting the sum of the four mutant strains from 100%. Measured data are presented as the average of three to six polony slides (symbols); error bars are ±1 SD in the data at a given point. Curve fits (lines) were determined using a least squares error minimization routine as described in the text. (A) Initial growth competition run in fermentor; mutations located in three distinct regions of the PGK1 gene. (B) Second growth competition run in shake flasks; all mutations lie within a single 300 bp region of the PGK1 gene.
Xc(n,m) = [Xe(1,m)·eµn·tm]/[∑5n = 1Xe(1,m)·eµn·tm]
where Xe is a 5 × 3 matrix containing the experimentally measured percent concentrations of each mutant at three different times, µ is a 5 × 1 matrix containing the specific growth rate of each strain in the competition and t is a 1 × 3 matrix containing the times at which samples were taken and mutant concentrations measured. The wild-type growth rate was assumed to be known (as measured in the shake flask experiments) and fixed and the elements of µ were allowed to vary in order to minimize the sum of the square of the error [∑(error)2] between the calculated and measured matrix according to the equation:
∑ (error)2 = ∑3m = 1∑5n = 1[Xc(n,m) – Xe(n,m)]2
Thus, specific growth rate values for each of the PGK1 mutants were calculated from the polony and SBE data from the pooled mutant growth competition experiment.
These specific growth rates were compared to those measured in individual strain shake flask experiments in Table 2. The calculated growth rates were consistent with the measured growth rates and ranged from 0.220 (Glu190Gln mutant) to 0.261 per h (Lys131Glu mutant). Additionally, calculated mutant growth rates, relative to wild-type PGK1 (CYC1 promoter), followed the same trend as protein activity measured in vitro (Table 1).
Table 2. Comparison of specific growth rates of PGK1 mutants measured in individual shake flask experiments and calculated from the initial competitive growth experiment.
| Mutant (CYC1 promoter) | µmeasured (per h) | µcalculated (per h) |
|---|---|---|
| Wild-type PGK1 | 0.266 ± 0.024 | 0.266a |
| Arg65Ser PGK1 | 0.252 ± 0.010 | 0.250 |
| Lys131Glu PGK1 | 0.264 ± 0.004 | 0.261 |
| Glu190Gln PGK1 | 0.177 ± 0.001 | 0.220 |
| His388Glu PGK1 | 0.249 ± 0.020 | 0.251 |
aSupplied to the curve fitting routine as a fixed value.
Polony multiplexing
To demonstrate the ability to concurrently quantify strains with mutations in a DNA region amplified by a single set of polony primers, a second competitive growth experiment was conducted using strains transformed with the follow ing plasmids: p416CYC1/PGK1-WT, p416CYC1/PGK1-Arg65Ser, p416CYC1/PGK1-Leu76Trp, p416CYC1/PGK1-Val114Asp and p416CYC1/PGK1-Ile124Phe. Growth rates of the individual strains were determined as previously described. The growth competition for this experiment was conducted in shake flasks rather than in a fermentor.
Growth rates measured in individual mutant shake flask cultures are shown in Table 3. Specific growth rates ranged from ∼95 (Arg65Ser mutant) to 117% (Leu76Trp mutant) of the wild-type PGK1 carrying strain. The growth rates of the Arg65Ser mutant and the wild-type PGK1 carrying strain were re-measured as controls. The values of these growth rates were determined to be nearly identical to previous measurements.
Table 3. Comparison of specific growth rates of PGK1 mutants measured in individual shake flask experiments and calculated from the second competitive growth experiment.
| Mutant (CYC1 promoter) | µmeasured (per h) | µcalculated (per h) |
|---|---|---|
| Wild-type PGK1 | 0.266 ± 0.007a | 0.266b |
| Arg65Ser PGK1 | 0.255 ± 0.002a | 0.262 |
| Leu76Trp PGK1 | 0.310 ± 0.011 | 0.285 |
| Val114Asp PGK1 | 0.298 ± 0.004 | 0.272 |
| Ile124Phe PGK1 | 0.273 ± 0.004 | 0.271 |
aRetested in a separate experiment (from Table 1).
bSupplied to the curve fitting routine as a fixed value.
Data collection was similar to that described for the initial competitive growth experiment with the exception that a single set of polonies was made for each time point. These polonies were then repeatedly extended, imaged and denatured to determine the fraction of each mutant in culture. Results of this experiment are shown in Figure 4B. An identical error minimization routine was employed to calculate specific growth rates from the polony single base extension data. These calculated growth rates are compared to measured individual mutant growth rates in Table 3. Measured and calculated values were similar with a maximum error of ∼8% (Val114Asp mutant).
DISCUSSION
In this report we have described a simple method to analyze, in a parallel manner, the functional impact of single nucleotide mutations on a model protein, yeast phosphoglycerate kinase. The method utilizes three core technologies: (i) the available S.cerevisiae deletion strains (5); (ii) functional complementation; (iii) polony technology (7). The method as described could readily be applied to the analysis of single nucleotide mutations within numerous proteins whose deletion results in a growth defect or total dysfunction (see for example 4,5) using a plasmid shuffling strategy (20) if needed. Additionally, our technique could be adapted to functional analysis of mutations within the protein coding region or the target DNA of DNA-binding proteins (21,22), mutations within the coding regions of interacting proteins (23) and, potentially, several other systems (24). Furthermore, the ability of many yeast mutants to be complemented by human homologs (25) leads to the possibility that the techniques described herein can be used to analyze human single nucleotide polymorphisms (SNPs) of medical interest.
One of the primary strengths of the competition strategy employed in this work is its ability to discriminate between very small differences in growth rate and thus protein activity, particularly if it is used in conjunction with a weak promoter of protein expression. This point is demonstrated by comparing the Arg65Ser and His388Glu mutants. The growth rates of these mutants differ by ∼1%; however, there is a clear separation between the two mutants at 97 h (Fig. 4A). This ability to discriminate between mutants with very small differences in growth rate could be increasingly important as the number of different mutants initially put into the screen increases.
The specific growth rates derived from the competitive growth data were consistent with the specific growth rates determined in individual growth experiments (differences were less than ±10% of the measured values) with the exception of the Glu190Gln mutant (∼24% error). In the first growth competition experiment, the Glu190Gln mutant was the slowest growing and was nearly eliminated from the culture by 51.75 h. Therefore, the curve fit for this mutant was heavily weighted by points at which noise was comparable to signal. However, because absolute rather than relative error minimization was employed in the curve fitting routine, error in the Glu190Gln growth rate had little impact on the calculated growth rate of the other mutant strains (confirmed by sensitivity analysis). More frequent early culture sampling would reduce this error.
Experiments comparing the growth of strains bearing the p416ADH/PGK1-WT and p416CYC1/PGK1-WT plasmids indicate that the rate of expression of the PGK1 gene under the control of the CYC1 promoter is growth limiting, as is necessary to determine the relative fitness of each mutant from the competitive growth data. However, it appears that, even under the control of the CYC1 promoter, the expression level of PGK1 resulted in a flux above the linear portion of Figure 1. While this result does not diminish the ability of the technique to assign relative activities to mutant proteins, it precludes assigning accurate absolute activity values. It is expected that, in general, the rate of expression that will result in growth limitation is gene specific. Therefore, our approach would be more generally applicable if a tunable expression promoter such as CUP1 (26) were used in place of the fixed strength promoter used in this work. Presumably, a larger range of mutations could be compared if expression of the gene of interest were tunable. For example, small differences between highly active mutants would be more readily identified in a low protein expression regime. However, at very low expression levels protein mutants with low specific activity might not support growth. Therefore, analysis of these mutants would require higher levels of protein expression.
The polony multiplexing experiment demonstrates a second strength of our approach, the ability to measure the functional effect of many single nucleotide mutations in parallel. For example, a mutant library could be generated in which each strain carries a mutation in the PGK1 gene (all separated by 10 bp). This library would consist of approximately 120 different PGK1 mutants (1251 bp coding region). After competitive growth, the library could be analyzed by amplifying individual 200 bp segments of the PGK1 coding region on separate polony slides. Therefore, each slide would result in polonies containing one of 20 different mutations. Mutants could then be identified using single base extensions.
The method described in this report is the first step in developing a general high throughput technique to analyze the functional effect of single nucleotide mutations in a genome. The majority of genetic diversity in human DNA results from a category of single nucleotide mutations called SNPs (27). Because of the vast number of SNPs estimated to be contained within the human population (1/1000 bp), high throughput methods will be required to identify and characterize functionally important mutations. Such characterization will lead to a better understanding of the predisposition and/or susceptibility to genetically linked diseases and the variation in therapeutic drug response (28,29). Furthermore, it appears likely that in the near future it will be possible to use the type of data generated using the methods reported here, in conjunction with developing systems biology tools such as in silico metabolic models (30), to both define the underlying mechanisms of SNP-related human pathophysiology and predict disease states based upon genome sequence.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Venugopal Mikkilineni for helpful discussion. We would also like to acknowledge an NIH COBRE grant (RR15588-03) to J.S.E., a US Department of Energy Office of Biological and Environmental Research grant (DE-FG02-01ER63200) to J.S.E. and a DARPA grant to G.M.C.
REFERENCES
- 1.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J., Sutton,G.G., Smith,H.O., Yandell,M., Evans,C.A., Holt,R.A. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 2.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 3.Masood E. (1999) US firm’s bid to sequence rice genome causes stir in Japan … as consortium plans free SNP map of human genome. Nature, 398, 545–546. [DOI] [PubMed] [Google Scholar]
- 4.Steinmetz L.M., Scharfe,C., Deutschbauer,A.M., Mokranjac,D., Herman,Z.S., Jones,T., Chu,A.M., Giaever,G., Prokisch,H., Oefner,P.J. et al. (2002) Systematic screen for human disease genes in yeast. Nature Genet., 31, 400–404. [DOI] [PubMed] [Google Scholar]
- 5.Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]
- 6.Badarinarayana V., Estep,P.W., Shendure,J., Edwards,J., Tavazoie,S., Lam,F. and Church,G.M. (2001) Selection analyses of insertional mutants using subgenic-resolution arrays. Nat. Biotechnol., 19, 1060–1065. [DOI] [PubMed] [Google Scholar]
- 7.Mitra R. and Church,G. (1999) In situ localized amplification and contact replication of many individual DNA molecules. Nucleic Acids Res., 27, e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Hoek P., Modesti,A., Ramponi,G., Kotter,P., van Dijken,J.P. and Pronk,J.T. (2001) Human acylphosphatase cannot replace phosphoglycerate kinase in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek, 80, 11–17. [DOI] [PubMed] [Google Scholar]
- 9.Sherman F. (1991) Getting started with yeast. Methods Enzymol., 194, 3–21. [DOI] [PubMed] [Google Scholar]
- 10.Mumberg D., Muller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- 11.Burke D., Dawson,D. and Stearns,T. (2000) Methods in Yeast Genetics, 2000 Edn. Cold Spring Harbor Laboratory Press, Woodbury, NY, pp. 103–105. [Google Scholar]
- 12.Strathern J.N. and Higgins,D.R. (1991) Recovery of plasmids from yeast into Escherichia coli-shuttle vectors. Methods Enzymol., 194, 319–329. [DOI] [PubMed] [Google Scholar]
- 13.Edwards J.S. and Palsson,B.O. (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics and capabilities. Proc. Natl Acad. Sci. USA, 97, 5528–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherman M.A., Dean,S.A., Mathiowetz,A.M. and Mas,M.T. (1991) Site-directed mutations of arginine-65 at the periphery of the active-site cleft of yeast 3-phosphoglycerate kinase enhance the catalytic activity and eliminate anion-dependent activation. Protein Eng., 4, 935–940. [DOI] [PubMed] [Google Scholar]
- 15.Ladine J.R. and Cross,R.L. (1991) The adenine nucleotide-binding site on yeast 3-phosphoglycerate kinase—affinity labeling of Lys-131 by pyridoxal 5′-diphospho-5′-adenosine. J. Biol. Chem., 266, 7194–7198. [PubMed] [Google Scholar]
- 16.Mas M.T., Resplandor,Z.E. and Riggs,A.D. (1987) Site-directed mutagenesis of glutamate-190 in the hinge region of yeast 3-phosphoglycerate kinase—implications for the mechanism of domain movement. Biochemistry, 26, 5369–5377. [DOI] [PubMed] [Google Scholar]
- 17.Mas M.T., Bailey,J.M. and Resplandor,Z.E. (1988) Site-directed mutagenesis of histidine-388 in the hinge region of yeast 3-phosphoglycerate kinase—effects on catalytic activity and activation by sulfate. Biochemistry, 27, 1168–1172. [DOI] [PubMed] [Google Scholar]
- 18.Watson H.C., Walker,N.P.C., Shaw,P.J., Bryant,T.N., Wendell,P.L., Fothergill,L.A., Perkins,R.E., Conroy,S.C., Dobson,M.J., Tuite,M.F. et al. (1982) Sequence and structure of yeast phosphoglycerate kinase. EMBO J., 1, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oechsner U., Magdolen,V., Zoglowek,C., Hacker,U. and Bandlow,W. (1988) Yeast adenylate kinase is transcribed constitutively from a promoter in the short intergenic region to the histone H2a-1 gene. FEBS Lett., 242, 187–193. [DOI] [PubMed] [Google Scholar]
- 20.Forsburg S.L. (2001) The art and design of genetic screens: yeast. Nature Rev. Genet., 2, 659–668. [DOI] [PubMed] [Google Scholar]
- 21.Flaman J.M., Frebourg,T., Moreau,V., Charbonnier,F., Martin,C., Chappuis,P., Sappino,A.P., Limacher,J.M., Bron,L., Benhattar,J. et al. (1995) A simple P53 functional assay for screening cell-lines, blood and tumors. Proc. Natl Acad. Sci. USA, 92, 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia L.Q., Osada,M., Ishioka,C., Gamo,M., Ikawa,S., Suzuki,T., Shimodaira,H., Niitani,T., Kudo,T., Akiyama,M. et al. (1997) Screening the p53 status of human cell lines using a yeast functional assay. Mol. Carcinog., 19, 243–253. [DOI] [PubMed] [Google Scholar]
- 23.Chien C.T., Bartel,P.L., Sternglanz,R. and Fields,S. (1991) The 2-hybrid system—a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA, 88, 9578–9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brent R. and Finley,R.L. (1997) Understanding gene and allele function with two-hybrid methods. Annu. Rev. Genet., 31, 663–704. [DOI] [PubMed] [Google Scholar]
- 25.Tugendreich S., Bassett,D.E., McKusick,V.A., Boguski,M.S. and Hieter,P. (1994) Genes conserved in yeast and humans. Hum. Mol. Genet., 3, 1509–1517. [DOI] [PubMed] [Google Scholar]
- 26.Robinson A.S., Bockhaus,J.A., Voegler,A.C. and Wittrup,K.D. (1996) Reduction of BiP levels decreases heterologous protein secretion in Saccharomyces cerevisiae. J. Biol. Chem., 271, 10017–10022. [DOI] [PubMed] [Google Scholar]
- 27.Halushka M.K., Fan,J.B., Bentley,K., Hsie,L., Shen,N.P., Weder,A., Cooper,R., Lipshutz,R. and Chakravarti,A. (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nature Genet., 22, 239–247. [DOI] [PubMed] [Google Scholar]
- 28.Brookes A.J. (1999) The essence of SNPs. Gene, 234, 177–186. [DOI] [PubMed] [Google Scholar]
- 29.Kruglyak L. (1999) Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet., 22, 139–144. [DOI] [PubMed] [Google Scholar]
- 30.Jamshidi N., Wiback,S.J. and Palsson,B.O. (2002) In silico model-driven assessment of the effects of single nucleotide polymorphisms (SNPs) on human red blood cell metabolism. Genome Res., 12, 1687–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.