Abstract
The human ribonucleoprotein ribonuclease P (RNase P), processing tRNA, has at least 10 distinct protein subunits. Many of these subunits, including the autoimmune antigen Rpp38, are shared by RNase MRP, a ribonucleoprotein enzyme required for processing of rRNA. We here show that constitutive expression of exogenous, tagged Rpp38 protein in HeLa cells affects processing of tRNA precursors. Alterations in the site-specific cleavage and in the steady-state level of 3′ sequences of the internal transcribed spacer 1 of rRNA are also observed. These processing defects are accompanied by selective shut-off of expression of Rpp38 and by low expression of the tagged protein. RNase P purified from these cells exhibits impaired activity in vitro. Moreover, inhibition of Rpp38 by the use of small interfering RNA causes accumulation of the initiator methionine tRNA precursor. Expression of other protein components, but not of the H1 RNA subunit, is coordinately inhibited. Our results reveal that normal expression of Rpp38 is required for the biosynthesis of intact RNase P and for the normal processing of stable RNA in human cells.
INTRODUCTION
Biochemical purification of the human tRNA processing enzyme ribonuclease P (RNase P) has revealed that this ribonucleoprotein possesses at least 10 distinct protein subunits, associated with one RNA species, H1 RNA (1,2). These protein subunits are designated Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, hPop5 and hPop1 (3–9). Some of these protein subunits have homologs in yeast (10–15) and archaea (16).
The subunit composition of nuclear RNase P is shared, in part, with the ribonucleoprotein RNase MRP, processing rRNA (17,18). In yeast, RNase MRP is required for mitochondrial DNA replication (19), for processing of the internal transcribed spacer 1 (ITS1) of rRNA (17,20,21) and for the cell cycle progression at the end of mitosis (17,22). The involvement of these two ribonucleoprotein enzymes in stable RNA processing or in the cell cycle, however, has not been demonstrated in human cells. One of the subunits shared by RNase P and RNase MRP is Rpp38, a protein with a major Th/To autoantigenic determinant (23). Rpp38 has sequence similarity with the Haloarcula marismortui L7Ae and it may bind the K-turn structure in the RNA component of RNase MRP (24). Rpp38 is primarily localized in the nucleolus and Cajal bodies of mammalian cells and appears to have a role in coordinating the intranuclear localization and assembly of RNase P and RNase MRP (25,26). Recent findings suggest that Pop3, the yeast homolog of Rpp38, is dispensable for tRNA substrate recognition and catalysis by nuclear RNase P (27).
We show here that constitutive expression in HeLa cells of an exogenous Rpp38 protein causes shut-off in expression of the endogenous Rpp38 and affects the biosynthesis of an intact RNase P. A decrease in the processing of the initiator methionine precursor tRNA, as well as miscleavage and rapid degradation of the 3′ sequences of ITS1 rRNA, are observed. Moreover, by using small interfering RNA (siRNA) directed against Rpp38 mRNA, we demonstrate that inhibition of expression of Rpp38 affects processing of precursors for tRNA and 5.8S rRNA. The results presented in this study reveal that normal expression of a single protein subunit of RNase P affects its structure and function in human cells.
MATERIALS AND METHODS
Cell culture and transfection
Stable transfection of HeLa S3 cells was performed using the calcium phosphate method and individual cell lines obtained were maintained in a selective medium that contained 0.4 mg/ml G418. For transfection of cells with synthetic siRNA (Dharmacon Research Inc., Lafayette CO) directed against Rpp38, Oligofectamine reagent was used, following the instructions of the manufacturer (Invitrogen). When siRNA and plasmids were introduced simultaneously into cells, Lipofectamine plus (Invitrogen) was utilized.
Gene constructs and probes
Two primers, one encompassing the first 22 nt of the 5′-untranslated region (5′-UTR) of Rpp38 and the other containing the last 29 nt of the Rpp38 open reading frame and an extra 18 nt that correspond to six histidine residues followed by a stop codon, were used to amplify Rpp38 cDNA (4). This cDNA was first subcloned into pBluescript, sequenced and then the cDNA was released by EcoRI and XbaI (located in the 3′ primer) and inserted into a pCI-neo vector (Promega) digested with EcoRI and XbaI, thereby generating pCIRpp38H.
For RNase protection analysis, probe B (Fig. 1C) was prepared by linearizing pCIRpp38H with NcoI located in the Rpp38 C-terminus (4) and transcribed using the T3 RNA polymerase promoter located downstream of the Rpp38H cDNA. Probe A (Fig. 1C) was generated by transcribing with T7 RNA polymerase a HindIII–PstI Rpp38 cDNA fragment subcloned in pBluescript. This fragment contains the 5′-UTR and the first 9 nt of the Rpp38 open reading frame.
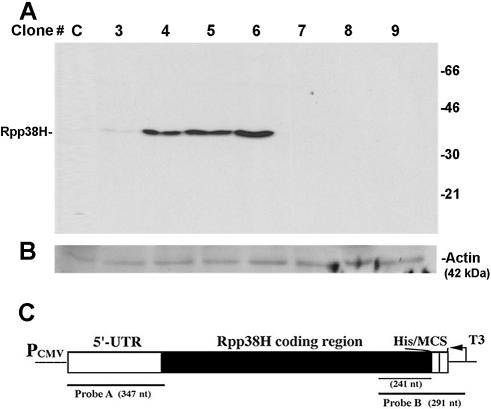
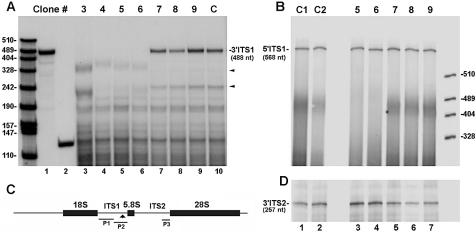
Figure 1.
Constitutive expression of Rpp38H in transfected HeLa S3 cells. (A) S100 crude extracts (100 µg) obtained from eight individual HeLa cell clones transfected with pCI-neo (clone C) or pCIRpp38H (clones 3–9) were subjected to western blot analysis using a monoclonal anti-polyhistidine antibody to test for Rpp38H expression. Proteins were separated by 12% SDS–PAGE, and positions of Rpp38H and protein size markers are indicated. (B) The membrane in (A) was analyzed using a monoclonal antibody against actin, 42 kDa, which serves as an internal control. (C) Schematic map of pCIRpp38H (not drawn to scale) containing a Rpp38 cDNA that has the 5′-UTR and the entire coding region fused to six histidine (His) residues. The cDNA (Rpp38H) is subcloned in the multiple cloning site (MCS) of pCI-neo and transcribed from an upstream cytomegalovirus immediate-early promoter/enhancer (PCMV). Probe A (347 nt) and probe B (291 nt), used for RNase protection analysis, are shown. T3 indicates T3 RNA polymerase used to transcribe probe B.
pTZ18R/ITS1 and pTZ18R/ITS2 were kindly provided by Jean-Pierre Bachellerie (CNRS, Toulouse, France). A 568 bp NarI–KpnI fragment of human genomic rDNA, encompassing positions 31–598 of the 1095 bp ITS1, was subcloned in pTZ18R digested by AccI and KpnI. This plasmid was linearized by cleavage at the HindIII site located in the multiple cloning site of the plasmid and transcribed by T7 RNA polymerase to yield an antisense RNA complementary to 568 nt of the 5′ region of ITS1 RNA. A 937 bp MluI–HinfI segment of human genomic rDNA, covering positions 176–1112 of the 1155 bp ITS2, was inserted into pTZ18R that was digested with SmaI. pTZ18R/ITS2 was linearized with NotI located within the ITS2 rDNA segment and transcribed by T7 RNA polymerase to yield an antisense RNA covering positions 855–1112. A pT3/T7α19 plasmid that harbors an EcoRI–HindIII PCR product that covers positions 606–1094 of the human ITS1 rDNA was kindly provided by David Tollervey (University of Edinburgh, Edinburgh, UK). The construct was linearized with EcoRI and the insert was transcribed by T7 RNA polymerase to yield a 488 nt antisense probe.
The human tRNAiMet gene (clone T54T60) subcloned in pT3/T7α, kindly provided by Uttam L. RajBhandary (MIT, Cambridge, MA), was linearized with EcoRI and an antisense RNA probe was synthesized using T3 RNA polymerase, while a clone for the human tRNASer gene was transcribed by T7 RNA polymerase.
Antisense oligonucleotides 5′-TCCTGCAATTCACATTAATTCTCGCAGCTAGC-3′, 5′-AAAGCCTACAGCACCCGGTATTCCC-3′ and 5′-TCCTCCGAGCCGGATTCGAA-3′ directed against human 5.8S rRNA, 5S rRNA and tRNATyrGUA respectively, were 5′ end labeled by T4 polynucleotide kinase using [γ-32P]ATP.
A custom-designed siRNA, which corresponds to positions 158–180 (5′-AAGCUAUUGGACUUCAGAAGAUU-3′) of the open reading frame of Rpp38 mRNA (4) was used for targeting Rpp38 in cells. The complementary synthetic strands of siRNA38 used in this study were as follows: 5′-GCU AUUGGACUUCAGAAGAUUdTdT-3′ and 3′-dTdTCGA UAACCUGAAGUCUUCUAA-5′.
RNA hybridization analyses
Total RNA was extracted from 0.5–1 × 107 cells with the use of Trizol reagent (Invitrogen) or by the acid/phenol RNA extraction method. RNA (30 µg) was subjected to RNase protection analysis using 0.5 × 106 c.p.m. of antisense RNA probe internally labeled with [α-32P]GTP in the presence of cold GTP. For rRNA analysis, 2–5 µg of total RNA was analyzed. Hybridization was performed in 40 µl of buffer A (80% deionized formamide, 40 mM PIPES, pH 6.7, 400 mM NaCl and 1 mM EDTA) for 14 h at 42–46°C. RNA was digested for 15 min at 22–25°C by adding 10 vol of buffer B (10 mM Tris–HCl, pH 7.5, 5 mM EDTA, 300 mM NaCl) that contained 0.8 µg/ml RNase A and 60 U/ml RNase T1. RNases were then eliminated by proteinase K/SDS treatment and the protected RNA was extracted with phenol:chloroform and ethanol precipitated in the presence of tRNA as a carrier. The pellet of RNA was resuspended in loading buffer (95% formamide, 10 mM Tris–HCl, pH 7.5, 1 mM EDTA), heat denatured and separated on a 4–6% polyacrylamide/7 M urea gel. For single-stranded DNA size markers, pBluescript DNA was digested with MspI and labeled with [α-32P]dCTP using Klenow enzyme.
Northern blot hybridization analysis was performed essentially as described (5). RNA was quantitated by PhosphorImager or by Scion Image (Scion Corp.).
Purification of human RNase P from HeLa cells
RNase P from HeLa S3 cells or from stably transfected cell lines was purified as previously described (7). Briefly, 1–2 × 109 cells were pelleted, disrupted and the whole cell extract was centrifuged at 20 000 g, followed by another centrifugation at 100 000 g to obtain S100 crude extract (7). This extract was loaded on a DEAE–Sepharose anion exchange chromatography column and the eluted fractions were tested for RNase P activity using the yeast suppressor precursor tRNASer (pSupS1) or Escherichia coli precursor tRNATyr as substrate. When crude extracts were assayed for RNase P, 5–20 U RNasin and 10 µg poly(I:C) were added to the reaction buffer to minimize precursor tRNA degradation. The specific activity of RNase P was determined as described (4). Each assay of RNase P was repeated using crude cell extracts obtained from a series of similar transfection experiments.
Analysis of RNase P RNA and protein subunits
For protein analysis, western blotting was performed using monoclonal mouse antibody directed against polyhistidine tag (HIS-1), γ-tubulin, actin (Sigma), C23 and B23 (Santa Cruz Biotechnology Inc.) or with affinity-purified polyclonal rabbit antibodies directed against Rpp subunits as described (5,6).
RNase P from crude extracts was immunoprecipitated with HIS-1 or polyclonal anti-Rpp antibodies and RNA in the immunoprecipitate was extracted with phenol:chloroform, precipitated by ethanol and then subjected to 3′ end labeling with [32P]cytidine phosphate or analyzed by northern blotting using antisense RNA probes transcribed from pBluescript(SK) harboring cDNAs of H1 RNA (340 nt) and MRP RNA (265 nt).
RESULTS
Selection of HeLa cells that constitutively express histidine-tagged Rpp38
HeLa S3 cells were transfected with pCIRpp38H, a eukaryotic expression vector that contains a gene that confers resistance to neomycin (G418) and a cDNA coding for a histidine-tagged Rpp38 protein (Rpp38H) (Fig. 1C). G418-resistant cell foci obtained after 2–3 weeks were selected and established as separate cell lines (see Materials and Methods). After eight passages in tissue culture, S100 crude extracts were obtained from several selected cell lines and were subjected to western blot analysis using a monoclonal anti-polyhistidine antibody (HIS-1) to test for expression of Rpp38H. A protein of Mr ≈ 38 kDa that corresponds to Rpp38H was expressed in cell clones 3–6 (Fig. 1A, clones 3–6) but not in cells stably transfected with pCI-neo (Fig. 1A, clone C). The level of Rpp38H expression was variable in these cell lines, the highest being in clones 5 and 6. Cell clones 7–9 were found to be resistant to G418, but expression of Rpp38H was not detectable (Fig. 1A, clones 7–9). This might be due to disruption of the Rpp38H cDNA as a result of random chromosomal DNA integration of pCIRpp38H.
Cell clones 3–6 exhibited slow growth rates (a 2-fold decrease in doubling time) when compared to those of cell clones 7–9 or to parental HeLa cells (data not shown). Furthermore, out of 15 individual cell clones that were selected for this study, only those that produced detectable levels of Rpp38H, including cell clones 3–6 described here, died after 11–14 passages in tissue culture. Therefore, high expression of Rpp38 is deleterious for survival of cells.
Lack of expression of endogenous Rpp38 in HeLa cells that produce Rpp38H
We next tested for expression of Rpp38H and Rpp38 mRNA in cell clones 3–9. To discriminate between these two mRNAs, which have similar sizes, total RNA extracted from clones 3–9 was subjected to quantitative RNase protection analysis using probe B. This 291 nt RNA probe covers the 3′ end of the Rpp38 coding region (241 nt), the hexahistidine coding sequence, the stop codon and an extra 29 nt sequence spanning the multiple cloning site of the vector (map in Fig. 1C). A 241 nt RNA was protected by Rpp38 mRNA in untransfected cells (Fig. 2A, clone C1) and in cells transfected with pCI-neo (Fig. 2A, clone C2). This mRNA, however, was barely detected in cell clones 3–6, which express Rpp38H protein (Fig. 2A, clones 3–6). Instead, larger RNAs of 250–260 nt that correspond to Rpp38H mRNAs were obtained (Fig. 2A, clones 3–6). Since these Rpp38H mRNAs encoded intact protein products (Fig. 1A), the 3′ ends of the protected Rpp38H mRNAs differ by some nucleotides as a result of variations in the RNase protection assays. The triplet repeats enriched with AU (5′-CAU CAU CAU CAU CAU CAU-3′), which code for the C-terminal six histidine residues, may expose the probe annealed to the Rpp38H mRNAs to differential RNase A attacks. The use of probe A (map in Fig. 1C) revealed the presence of intact 5′-UTR sequences in Rpp38H mRNAs, an indication that there was no extensive elimination of these mRNA molecules in clones 3–6 (data not shown).
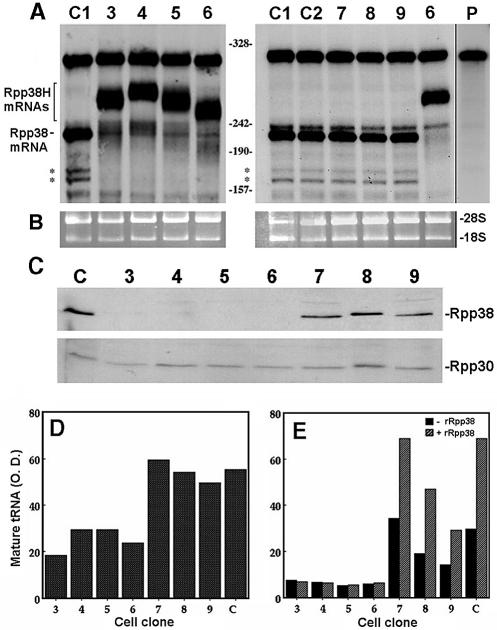
Figure 2.
Shut-off of Rpp38 mRNA and protein expression and impaired RNase P activity in cells expressing Rpp38H. (A) Total RNA extracted from untransfected HeLa cells (clone C1), cells transfected with pCI-neo (clone C2) and cell clones 3–9 (clones 3–9) was subjected to RNase protection analysis using uniformly labeled antisense probe B. Rpp38 mRNA protects 241 nt from the entire 291 nt probe, while Rpp38H mRNAs protect 250–270 nt. Since the probe has sequences derived from the plasmid and a 291 nt RNA was still seen in untransfected cells (clone C1), this RNA represents free probe. The shorter RNA bands indicated by asterisks represent Rpp38 mRNA species. The right-most lane represents undigested probe B (weaker exposure is presented). Numbers between panels indicate single-stranded DNA size markers generated by MspI digestion of pBlueScript. (B) Ethidium bromide staining of 18S and 28S rRNAs, used as internal controls for total RNA analyzed in (A). (C) S100 crude extracts from untransfected cells (clone C) and from cell clones 3–9 were subjected to western blot analysis using affinity purified antibodies against Rpp38 (upper panel) or Rpp30 (lower panel). (D) A representative assay of RNase P activity in S100 crude extracts with equal amounts of protein from clones 3–9 and from untransfected cells using precursor tRNASer as a substrate. Cleavage products were resolved in 8% polyacrylamide/7 M urea and mature tRNA (3′) was quantitated as described in Materials and Methods. (E) Extracts were tested for RNase P activity in the absence (–rRpp38) or presence (+rRpp38) of 8 pmol of highly purified recombinant Rpp38. This recombinant protein has a histidine tag at its N-terminus (5). The optical density values of tRNA are presented in arbitrary units.
The lack of Rpp38 mRNA expression in cell clones 3–6 was accompanied by very low expression of endogenous Rpp38 protein (Fig. 2C, upper panel), but strikingly also of Rpp38H (Fig. 2C, upper panel). Because in this western blot analysis we used polyclonal rabbit antibodies raised against a histidine-tagged recombinant Rpp38 (5), and therefore they presumably exhibit similar binding affinities to Rpp38H and Rpp38, cell clones 3–6 seem to intrinsically produce low levels of Rpp38H, even though their Rpp38H mRNA levels were comparable to those of Rpp38 mRNA expressed in parental cells or in clones 7–9 (see Fig. 2A). In fact, the variations in the levels of expression of Rpp38H in clones 3–6, as revealed by the HIS-1 antibody (Fig. 1A), are below the normal levels of Rpp38 in control cells (Fig. 2C). No change in Rpp30 levels has been observed in the various clones (Fig. 2C, lower panel).
Our findings demonstrate that overproduction of Rpp38H in HeLa cells causes shut-off of the expression of endogenous Rpp38 at the mRNA level. The observation that Rpp38H mRNA can be detected (Fig. 2A) while the Rpp38H protein level is low (Fig. 2C) suggests a rapid degradation of the exogenous protein in the cell or weak translation of the Rpp38H mRNAs that lack authentic 3′-UTR sequences.
Impaired activity of RNase P in extracts of cells expressing Rpp38H
The results described above raised the possibility that RNase P activity may also be affected in cell lines 3–6. Accordingly, S100 crude extracts described above were tested for enzyme function. A 2- to 3-fold reduction in the specific activity of RNase P, defined by the rate of substrate cleavage divided by total protein in the extract (4), in processing of precursor tRNASer was measured in clones 3–6 when compared to extracts of clones 7–9 or of control cells (Fig. 2D and E). The addition of a highly purified recombinant Rpp38 protein, fused to a histidine tag (5), to the extracts obtained from clones 7–9 and control cells, but not from clones 3–6, enhanced RNase P activity by 2- to 3-fold (Fig. 2E). This enhancement is specific for recombinant Rpp38, because the addition of recombinant, histidine-tagged Rpp30 protein (5) did not stimulate enzyme activity in any extract tested (data not shown). Hence, recombinant, histidine-tagged Rpp38 is functional in terms of its ability to enhance RNase P activity (by binding and/or replacing Rpp38), but only in extracts from clones 7–9 and control cells. However, the reduced activity of the enzyme in clones 3–6 is not related to a reduction in total protein. This impaired activity is partly due to a reduction in the steady-state level of both Rpp38 and Rpp38H in cell clones 3–6 (Fig. 2C). Additionally, RNase P produced in cells expressing low levels of Rpp38 may have defects in other subunits that cannot be alleviated by recombinant Rpp38 (see below).
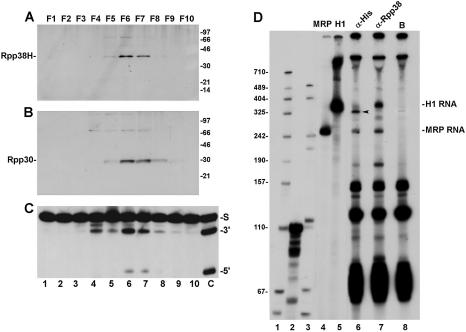
Rpp38H is part of a defective RNase P complex
S100 crude extracts from cell clone 6 described above were fractionated on a DEAE–Sepharose anion exchange chromatography column (7). Active RNase P was eluted from this column at 260–340 mM KCl (Fig. 3C, lanes 6 and 7), a concentration of salt shown previously to elute RNase P ribonucleoprotein from untransfected HeLa S3 cells from the same column (4). Fractions F6 and F7 contained the peak of enzymatic activity (Fig. 3C). Both Rpp38H (Fig. 3A) and Rpp30 (Fig. 3B), as judged by western blot analysis using their corresponding antibodies described above, were found in F6 and F7. As with the S100 crude extracts (Fig. 2D and E), the activity of RNase P in fractions F6 and F7 was significantly lower than that exhibited by DEAE-purified RNase P obtained from untransfected HeLa cells (Fig. 3C, lane C). Qualitative immunoprecipitation analysis using monoclonal anti- polyhistidine antibody (HIS-1) revealed that Rpp38H was associated with active RNase P (data not shown) that has an H1 RNA smaller than 340 nt (Fig. 3D, lane 6, arrow), the normal size of H1 RNA brought down by anti-Rpp38 antibody from untransfected HeLa cells (Fig. 3D, lane 7). In contrast, the MRP RNA co-immunoprecipitated by HIS-1 or anti-Rpp38 antibodies was intact (265 nt) and no general degradation of other RNAs was seen (Fig. 3D, lane 6 versus 7).
Figure 3.
Rpp38H is associated with active RNase P that has truncated H1 RNA. (A) S100 crude extracts from cell clone 6 (Fig. 1A) were loaded on a DEAE–Sepharose chromatography column and all the eluted fractions obtained (lanes 1–10) were tested for the presence of Rpp38H as described in Figure 1A. (B) The membrane in (A) was analyzed for Rpp30 using affinity purified antibodies against Rpp30. Proteins were separated by 12% SDS–PAGE, and the positions of Rpp38H, Rpp30 and protein size markers are shown. The protein of Mr ≈ 72 kDa in (A) represents dimers of Rpp38H. (C) RNase P activity in the DEAE eluted fractions as determined by processing of precursor tRNATyr (S) to mature tRNA (3′) and leader sequence (5′). The right-most lane shows a control assay using a DEAE purified RNase P from untransfected cells. The non-specific cleavage of precursor tRNA found in fractions that preceded RNase P activity is commonly seen during fractionation of S100 extracts on a DEAE column (2). (D) 3′ End labeling of RNAs immunoprecipitated with RNase P from whole crude extract of cell clone 6 expressing Rpp38H using anti-polyhistidine antibody (lane 6) or beads alone (lane 8). As a control, anti-Rpp38 antibodies were used to bring down RNase P from normal HeLa cells (lane 7). Positions of the intact 340 nt H1 RNA and 265 nt MRP RNA are shown. The arrow points to truncated H1 RNA. Labeled precursor tRNASer (110 nt) (lane 2) and a MspI digest of pBluescript (lane 1) or pGEM-3 (lane 3) were used as size markers.
Reduced processing of tRNA in HeLa cells that express Rpp38H
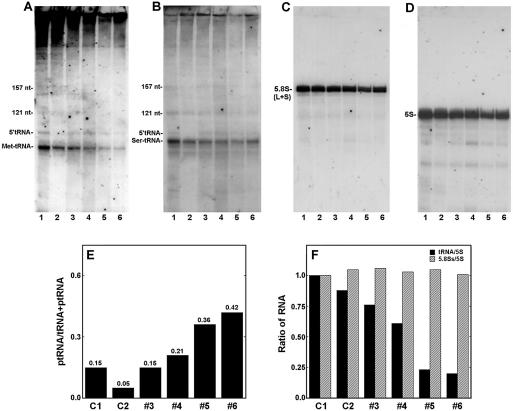
To produce evidence that RNase P has impaired activity in Rpp38H-producing cells, the steady-state level of the initiator methionine tRNA in cell clones 3–6 was determined by northern blot hybridization analysis using an antisense RNA probe transcribed from the human tRNAiMet gene (see Materials and Methods). A 6- to 8-fold decrease in the level of the 75 nt tRNAiMet was measured in cell clones 4–6 (Fig. 4A, lanes 4–6) when compared to the level in untransfected HeLa cells (Fig. 4A, lane 1) or to cells transfected with vector alone (Fig. 4A, lane 2). In contrast, only a 2-fold decrease in mature tRNAiMet was measured in clone 3 (Fig. 4A, lane 3), which expressed lower levels of Rpp38H compared to clones 4–6 (Fig. 1A). Moreover, a marked increase of 6- to 8-fold in the ratio of 5′-tRNAiMet precursor containing the 5′ leader sequence to mature tRNAiMet was pronounced in clones 5 and 6 when compared to the ratio obtained in control cells (Fig. 4A and E). Accordingly, processing of precursor tRNAiMet by RNase P is impaired in clones 3–6, which produce Rpp38H.
Figure 4.
Expression of tRNAs and rRNAs in HeLa cells that express Rpp38H. (A) Total RNA extracted from untransfected cells (lane 1), pCI-neo- transfected cells (lane 2) and clones 3–6 expressing Rpp38H (lanes 3–6) was analyzed by northern blot hybridization analysis using a uniformly labeled antisense RNA probe for precursor tRNAiMet. This probe detects the mature tRNAiMet, 75 nt in length, and the precursor tRNAiMet (ptRNAiMet, 95 nt). (B–D) The blot in (A) was rehybridized with antisense RNA probes against human precursor tRNASer, the 157 nt 5.8S rRNA (large and small species) or the 121 nt 5S rRNA, respectively. The signal seen at the top of (A) is non-specific hybridization, which was not seen in panels (B)–(D). (E) The ratio of ptRNAiMet to ptRNAiMet + tRNAiMet in each lane of (A) was determined after quantitation of RNA. (F) tRNAiMet, 5S rRNA and 5.8SS rRNA in (A), (C) and (D) were quantitated and the ratios tRNAiMet:5S rRNA and 5.8SS rRNA:5S rRNA are plotted.
It has been shown that processing of tRNAiMet precursors is more sensitive than other tRNA precursors to RNase P inhibition in gcd14 yeast mutants (28). The GCD14 gene codes for a protein essential for maturation of a subset of RNA polymerase III transcripts and cooperates with the La antigen (28). Indeed, the reduction in the steady-state levels of another precursor tRNA, tRNASer, is less pronounced in these cells. When the same RNA blot in Figure 4A was rehybridized with an antisense RNA probe against human tRNASer only a 2-fold decrease in the level of this tRNA was seen in clones 5 and 6 (Fig. 4B). Moreover, the ratio of precursor tRNASer to mature tRNASer was less pronounced when compared to that measured for tRNAiMet (Fig. 4A versus B). Likewise, only a slight decrease in the steady-state level of tRNALeu was observed in clones 3–6 (data not shown).
The above results indicate that tRNA processing is impaired in cells expressing Rpp38H, but processing of the initiator methionine tRNA precursor is particularly sensitive to the reduced activity of RNase P (see below).
Incorrect cleavage and reduced steady-state level of internal transcribed spacer 1 of rRNA in cells expressing Rpp38H
The yeast RNase MRP has been shown to cleave ITS1 rRNA at site A3, thus triggering the removal of the remaining downstream sequences by exoribonuclease activity to yield the 5′-end of 5.8SS rRNA (29,30). An RNase MRP-independent pathway produces the 5.8SL rRNA (29,30). A similar pathway for rRNA processing may exist in other eukaryotes (31). Therefore, we checked for a defect in RNase MRP function by examining the steady-state level of 5.8S rRNA in cells expressing Rpp38H. We found no significant change in the levels of 5.8S rRNAs in clones 3–6 when compared to those in control cells (Fig. 4C and F). There was no alteration in the ratio of 5.8SS to 5.8SL rRNAs either (see below). Moreover, the steady-state level of mature 5S rRNA (121 nt) remained unaffected in these clonal cell populations (Fig. 4D and F). However, quantitative RNase protection analysis of 3′ ITS1 rRNA sequences in cells expressing Rpp38H revealed that their processing is actually not normal (Fig. 5A). A 488 nt RNA that corresponds to positions 606–1094 of the 3′ sequence of ITS1 rRNA was protected by the P2 probe when total RNA from cell clones 7–9 (Fig. 5A, lanes 7–9) and from control HeLa cells (Fig. 5A, lane 10) was first analyzed. RNA of ∼320 nt in length, which was generated from endonucleolytic cleavage of ITS1 rRNA (32), was also protected in clones 7–9 and in control cells (Fig. 5A, lanes 7–9 and 10, respectively, arrow). This finding is consistent with previous S1 nuclease mapping analysis, in which it was shown that ITS1 rRNA is endonucleolytically cleaved in a U-tract region located 161–163 nt upstream of the 5.8S rRNA (33). In addition, another minor RNA of ∼250 nt was also seen in these cells (Fig. 5A, lanes 7–10, arrow). However, none of the three protected RNAs just described were detected when total RNA from clones 3–6 was analyzed (Fig. 5A, lanes 3–6). Instead, rRNA fragments of 230–350 nt were obtained (Fig. 5A, lanes 3–6).
Figure 5.
Alterations in the processing pattern and steady-state level of 3′ ITS1 rRNA in cells expressing Rpp38H. (A) Total RNA extracted from cell clones 3–6 (lanes 3–6), clones 7–9 (lanes 7–9) and untransfected HeLa cells (lane 10) was subjected to RNase protection analysis using probe P2 (lane 1). This probe protects 488 nt of the 3′ sequence of ITS1 rRNA. The two other protected RNAs of ∼230 and 350 nt seen in control cells (lane 10, small arrows), but not in clones 3–6 (lanes 3–6), represent specific cleavage sites in the 3′ ITS1 rRNA sequence. Lower bands may be generated from partial digestion of the probe as a result of forming intramolecular stem–loop rRNA structures. Labeled precursor tRNATyr (131 nt, lane 2) and MspI-digested pBlueScript were used as size markers. Protected RNAs were separated in 5% polyacrylamide/7 M urea gels. (B) RNA from untransfected cells (lane 1), pCI-neo-transfected cells (lane 2), cell clones 5 and 6 (lanes 3 and 4) and clones 7–9 (lanes 5–7) was analyzed by RNase protection analysis using probe P1 that protects 568 nt of the 5′ sequence of ITS1 rRNA. (C) The antisense RNA probes P1, P2 and P3, which cover different regions in ITS1 and ITS2 that separate 18S, 5.8S and 28S rRNAs, are indicated. The arrow points to a putative cleavage site in the 3′ end of ITS1. (D) Total RNA seen in (B) was analyzed using probe P3 that protects 257 nt of the 3′ end of ITS2 rRNA.
The above analyses reveal that processing of 3′ ITS1 rRNA is aberrant in clones 3–6, thereby implicating RNase MRP in rRNA processing in human cells. Work is in progress to elucidate the enzymatic basis for processing of ITS1 rRNA by RNase MRP in vitro.
To verify whether the lack of 3′ ITS1 rRNA sequences in clones 3–6 is the result of specific degradation, 5′ sequences of ITS1 rRNA were quantitated through the use of the P1 probe that covers positions 31–598 of this spacer (Fig. 5C). A 568 nt RNA was protected when total RNAs from cell clones 5 and 6 (Fig. 5B, lanes 3 and 4) and clones 7–9 (Fig. 5B, lanes 5–7), as well as from control cells (Fig. 5B, lanes 1 and 2), were analyzed. Moreover, analysis of the 3′ end of ITS2 rRNA, using the P3 probe (Fig. 5C) that covers positions 855–1112 of this downstream spacer, showed that this sequence existed in the cell clones expressing Rpp38H (Fig. 5D). Accordingly, the absence of 3′ ITS1 rRNA is due to enhanced degradation in cells that produce Rpp38H.
Inhibition of expression of Rpp38 by the use of small interfering RNA
We also examined the effect of inhibition of expression of endogenous Rpp38 on stable RNA processing in HeLa cells by the use of siRNA, designated siRNA38 (see Materials and Methods), which inhibited the expression of Rpp38 in a dose-dependent manner as seen after transfection of cells for 24 (Fig. 6A, left panel) and 48 h (Fig. 6A, right panel). A reduction of ∼50% in Rpp38 protein level was measured in cells transfected with 5.2 µg/ml of siRNA38, when compared to mock-treated cells and relative to γ-tubulin, which served as an internal control (Fig. 6A, numbers below panels). The efficiency of cell transfection, as determined by a reporter gene, pEGFP-C1, was 70–80% (data not shown). Cells treated with siRNA38 showed a slower growth rate (between 12 and 72 h after transfection), when compared to that of mock-treated cells (data not shown), but without affecting viability, as determined by FACS analysis (Fig. 6B). No effect on Rpp38 expression was observed in the presence of siRNA directed against lamin A/C (data not shown).
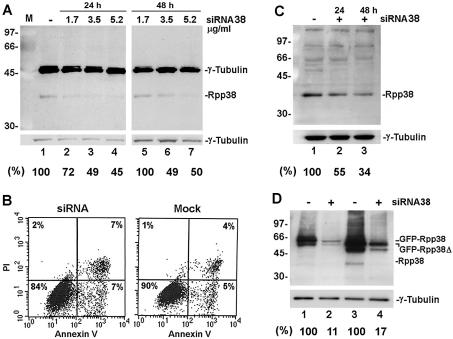
Figure 6.
Inhibition of expression of Rpp38 in HeLa cells transfected with siRNA38. (A) Equal amounts of proteins in S20 crude extracts of HeLa cells transfected for 24 (left panel) or 48 h (right panel) with 1.7, 3.5 or 5.2 µg/ml siRNA38 were separated by 12% SDS–PAGE and then subjected to western blot analysis using anti-Rpp38 and anti-γ-tubulin antibodies. Positions of Rpp38, γ-tubulin and protein size markers are shown. Numbers below panels represent percent inhibition of Rpp38, as determined by optical density scans of the band of Rpp38 and normalized to that of γ-tubulin. Percentage inhibition is relative to the control cells. (B) FACS analysis of siRNA38-treated (left) and mock-treated (right) HeLa cells using an Annexin V and propidium iodide (PI) kit (from Roche Molecular Biochemicals, Germany). Percentage of living (lower left quadrant), apoptotic (lower right) and necrotic (upper right) cells are shown. (C) Equal amounts of proteins in S20 crude extracts of 293 HEK cells transfected for 24 (lane 2) or 48 h (lane 3) with 5.2 µg/ml siRNA38 were separated by 12% SDS–PAGE and then subjected to western blot analysis using anti-Rpp38 antibody (upper panel) or anti-γ-tubulin antibody (lower panel). Positions of Rpp38, γ-tubulin and protein size markers are indicated. Numbers below the panel represent percent inhibition of Rpp38, as determined by optical density scans of the band of Rpp38 normalized to that of γ-tubulin. (D) HeLa cells were transfected for 48 h with pEGFP-Rpp38 or pEGFP-Rpp38Δ in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 5.2 µg/ml siRNA38. Equal amounts of protein in S20 crude extracts of the cells were subjected to western blot analysis as described in (A). Numbers below the panel represent percentage inhibition of the fused proteins.
Inhibition of Rpp38 expression by siRNA38 is not cell type specific, as destruction of Rpp38 by siRNA38 was also observed in 293 human embryonic kidney (HEK) cells (Fig. 6C, lane 1 versus 3). A reduction of ∼66% in the Rpp38 level was measured in cells treated with 5.2 µg of siRNA38 (Fig. 6C). The efficiency of cell transfection as determined by pEGFP-C1 was 70–90% (data not shown).
To obtain more information as to the targeting specificity of the siRNA38 described above, we co-transfected HeLa cells with siRNA38 and pEGFP-Rpp38, an expression vector that contains the open reading frame of the Rpp38 cDNA fused to green fluorescent protein (GFP) (25). A significant reduction of ∼90% in expression of GFP–Rpp38 has been measured in cells co-transfected with siRNA38, when compared to the control (Fig. 6D, lane 1 versus 2). Similar results have been obtained with pEGFP-Rpp38Δ (Fig. 6D, lane 3 versus 4), in which GFP was fused to Rpp38 lacking the last 36 amino acids that constitute a nucleolar localization domain (25). Thereby, siRNA38 efficiently leads to the destruction of exogenous Rpp38 expressed in human cells. This discrepancy in the targeting efficiency of exogenous and endogenous Rpp38 by siRNA38 is not related to transfection efficiency. We are currently testing cell lines expressing siRNA38 and combinations of siRNAs directed against different regions in the Rpp38 mRNA.
The results described above show that siRNA38 specifically targets endogenous and exogenous Rpp38 for destruction in human cells.
Reduced activity of RNase P purified from cells transfected with siRNA38
Whole crude extracts obtained from HeLa cells treated with various concentrations of siRNA38 were tested for RNase P activity using precursor tRNATyr (Fig. 7A). A significant reduction of ∼82% in tRNA processing has been measured in cells treated with 6.4 µg/ml siRNA38, when compared to control (Fig. 7B). Expression of Rpp38 in these cells was inhibited by ∼84%, when compared to mock-treated cells (Fig. 7C, lane 1 versus 5). A slight increase in the expression of the nucleolar phosphoprotein B23 (Fig. 7D), but not of C23 (nucleolin) (Fig. 7E), was observed in siRNA38-treated cells.
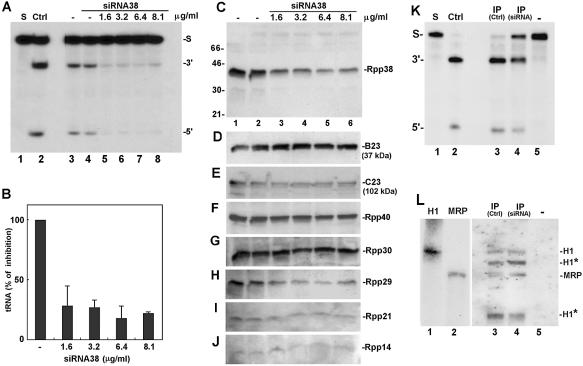
Figure 7.
Inhibition of RNase P activity in extracts of HeLa cells transfected with siRNA38. (A) Whole crude extracts of HeLa cells transfected for 48 h with 1.6, 3.2, 6.4 or 8.1 µg/ml siRNA38 (lanes 5–8) were tested for the activity of RNase P in processing of labeled precursor tRNATyr. Control extracts of cells mock-treated for 24 and 48 h are shown in lanes 3 and 4. Cleavage was for 4 min at 37°C so as to be in the linear range (<30% cleavage of substrate in control extracts). The positions of substrate (S) and cleavage products, the 5′ leader sequence (5′) and tRNA (3′) are indicated. Ctrl represents enzymatic assay of DEAE-purified HeLa RNase P (lane 2). (B) Quantitation of processing of precursor tRNATyr seen in (A). Values are the mean of three independent experiments whose standard deviation is shown by the bars. (C–J) Whole crude extracts of cells seen in (A) were subjected to western blot analyses using antibodies against Rpp38 (C), B23 (D), C23 (E), Rpp40 (F), Rpp30 (G), Rpp29 (H), Rpp21 (I) and Rpp14 (J). Positions of protein subunits and size markers are indicated. (K) Processing of precursor tRNATyr by RNase P immunoprecipitated by anti-Rpp30 antibody from S20 crude extracts of HeLa cells treated with 8.1 µg/ml siRNA38 (lane 4) or left untreated (lane 3). Lane 5 indicates beads not coupled to antibody and lane 3 shows activity of RNase P brought down by anti-Rpp30 antibodies from equal amounts of proteins in S20 crude extracts of untreated HeLa cells. (L) Northern blot analysis of RNA that was extracted from the immunoprecipitates described in (K) (lanes 3–5). Labeled, antisense H1 RNA and MRP RNA were used as probes (see Materials and Methods). The RNA bands indicated by asterisks represent truncated H1 RNA molecules (∼310 and 170 nt), which can also be detected by the use of an antisense H1 RNA alone (5; data not shown). As a control, sense H1 and MRP RNAs were analyzed (lanes 1 and 2) and a shorter exposure of the blot (lanes 1 and 2) is attached.
We also tested the expression of other protein subunits of human RNase P in siRNA38-treated cells and found no substantial changes in the levels of Rpp40, Rpp30 and Rpp14 (Fig. 7F, G and J, respectively). In contrast, reductions of ∼54 and 73% in expression of the subunits Rpp21 and Rpp29 were measured (Fig. 7I and H, respectively).
RNase P immunoprecipitated from S20 protein extracts of siRNA38-treated cells by polyclonal antibodies against the subunit Rpp30 (5) showed an ∼50% reduction in processing of substrate (Fig. 7K, lane 3 versus 4). This weak activity of RNase P, however, was not accompanied by any decrease in the steady-state level of H1 RNA, as judged by northern blot hybridization analysis of RNA precipitated with the anti-Rpp30 antibody (Fig. 7L, lane 3 versus 4). Shorter H1 RNAs were seen both in control and siRNA38-treated cells (Fig. 7L, lane 3 versus 4, asterisks).
The above observations reveal that the level of H1 RNA, MRP RNA and some protein subunits remained unchanged in cells that have an ∼80% reduction in the level of Rpp38, but some other subunits (Rpp21 and Rpp29) concomitantly decreased.
Processing of tRNA precursors in siRNA38-treated cells
To directly assess the effect of siRNA38 on RNase P activity in cells, total RNA was extracted from the siRNA38-treated cells and subjected to northern blot hybridization analysis using the antisense RNA probe derived from the gene encoding the initiator methionine tRNA (Fig. 8A). Two precursors, 5′-tRNA and 5′-tRNA-3′, accumulated in a dose-dependent manner to increasing concentrations of siRNA38 (Fig. 8A and B, lanes 2–5). 5′-tRNA-3′ represents the tRNAiMet primary transcript that has a 5′ leader sequence of 7 nt and 3′ trailer of 2–4 nt in length (34). The level of 5′-tRNA increased by about 5-fold at 7.8 µg/ml siRNA38, when compared to the level in mock-treated cells (Fig. 8B, lane 5 versus 1, and C). However, no concomitant decrease in the level of the mature tRNAiMet was detected (Fig. 8A and B, lanes 2–5). No noticeable decrease in mature tRNATyr, tRNASer or tRNALeu was observed (data not shown).
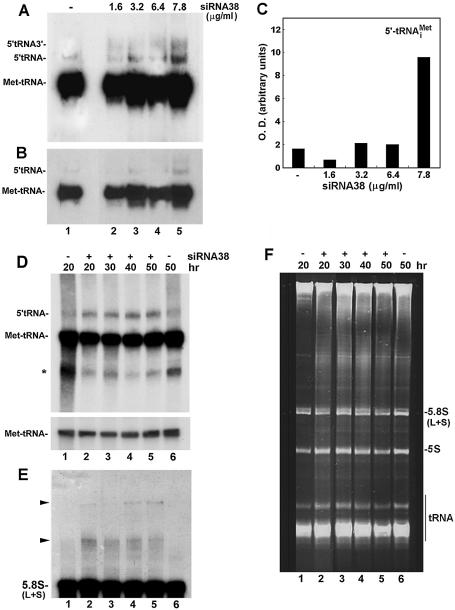
Figure 8.
Processing of tRNA and 5.8S rRNA in siRNA38-treated HeLa cells. (A) Total RNA was extracted from HeLa cells treated with siRNA38 and then subjected to northern blot hybridization analysis using an antisense RNA probe against tRNAiMet. 5′-tRNA-3′ represents the primary transcript with 5′ leader sequence and 3′ trailer. 5′-tRNA represents a precursor with 5′ leader sequence. (B) A shorter exposure of the gel seen in (A). (C) Quantitation of the 5′-tRNA seen in (A). (D) Cells were transfected with 7.8 µg/ml siRNA38 for the indicated time points and total RNA was extracted and analyzed as in (A). The asterisk may represent uncharged or truncated tRNA. The lower panel depicts shorter exposure of mature tRNAiMet. (E) The membrane shown in (D) was rehybridized with an antisense oligonucleotide against 5.8S rRNA. Arrows point to large RNA transcripts containing 5.8S rRNA sequences. (F) Ethidium bromide staining of total RNA separated in denaturing 8% polyacrylamide/7 M urea gels. tRNAs, 5S rRNA and small and large 5.8S rRNAs are indicated.
Kinetic analysis revealed that high levels of 5′-tRNAiMet were observed after 20 h of cell transfection with 7.8 µg/ml siRNA38 and persisted for up to 50 h (Fig. 8D, lanes 2–5). A concomitant, but slight, decrease in tRNAiMet was seen in these cells (Fig. 8D, lower panel).
Processing of 5.8S rRNA in siRNA38-treated cells
We found no reduction in the levels of mature 5.8S rRNAs in siRNA38-treated cells, when compared to those in control cells (Fig. 8E and F, lanes 1 and 6 versus 2–5). The ratio of 5.8SS to 5.8SL rRNA remained unchanged (Fig. 8F). However, two large rRNA transcripts have accumulated in the presence of siRNA38 (Fig. 8E, lanes 2–5, arrows). The larger rRNA species accumulated at later time points during transfection (40–50 h) when compared to the smaller one (Fig. 8E, lanes 4 and 5). These transcripts may represent precursors containing 5.8S rRNA sequences but their accumulation did not alter the final production of 5.8S rRNAs in response to inhibition of expression of Rpp38.
DISCUSSION
Role of Rpp38 in processing of tRNA precursors in human cells
We have shown that inhibition of expression of Rpp38 in human cells, whether by the use of the RNA interference approach or by constitutive expression of an exogenous, histidine-tagged Rpp38 protein, alters the activity of nuclear RNase P in tRNA processing. Processing of the initiator methionine tRNA precursor is particularly sensitive to inhibition of RNase P activity, when compared to processing of other tRNA precursors. Inhibition of expression of Rpp38 is accompanied by a decrease in the levels of some protein subunits, such as Rpp21 and Rpp29, but not of Rpp14, Rpp30, Rpp40 or H1 RNA. Therefore, the intracellular level of Rpp38 influences the expression of certain protein subunits of RNase P and affects the processing of tRNA precursors.
Rpp38H expressed in cells is physically associated with RNase P that exhibits decreased specific activity, when compared to that of RNase P purified from untransfected cells. This tagged ribonucleoprotein is assembled with a truncated H1 RNA that may affect its enzymatic activity. Additionally, the integration of Rpp38H into RNase P (Fig. 3) may change the overall structure or composition of the complex. Previous biochemical purification analyses have indicated that the positively charged Rpp38 of human RNase P can be trapped on a cation exchange FPLC chromatography column, thus resulting in active, albeit unstable, holoenzyme that lacks this subunit (4,7). The addition of recombinant, histidine-tagged Rpp38 to this partially purified holoenzyme can increase its activity. Moreover, Rpp38 is not required for substrate cleavage by RNase P in vitro (H. Mann and N. Jarrous, unpublished data). However, RNase P purified from cells with reduced levels of Rpp38 exhibits very weak activity in processing of synthetic precursor tRNAs in vitro. Therefore, purified complexes of nuclear RNase P do not necessarily reflect those in living cells in terms of catalysis or catalytic efficiency. Similar results have been reported for tRNA processing in HeLa cells in which Rpp38 was targeted by an external guide sequence (35). In this latter study, a total reduction of 25% in tRNA levels has been reported (35). Since other protein subunits were also inhibited along with Rpp38, the possibility exists that RNase P does not require all its subunits to efficiently process certain tRNA precursors in living cells. Alternatively, processing of tRNA in human cells can take place in the presence of trace amounts of intact RNase P. RNase P may utilize its distinct protein subunits to deal with the large number of tRNA isotypes expressed in growing cells. In support of these findings, it has been shown that Pop3, the yeast homolog of Rpp38, can be depleted from Saccharomyces cerevisiae without abolishing RNase P activity and that this subunit is added at a late stage to a precursor RNase P ribonucleoprotein complex that is catalytically active (27).
Function of Rpp38 in rRNA processing
In addition to the alteration in tRNA processing, defects in the cleavage and degradation of ITS1 rRNA are observed in HeLa cells that constitutively express Rpp38H. Although the enzymatic basis of processing of rRNA in human cells is not known in detail, our observations reveal that the 3′ region of human ITS1 rRNA contains two endonucleolytic cleavage sites that have been affected by the constitutive production of Rpp38H. Because the 3′ ITS1 rRNA in yeast serves as a substrate for RNase MRP (29) and the physical association of Pop3 (the homolog of Rpp38) with RNase MRP has been demonstrated by biochemical means (11), our results implicate human RNase MRP in processing of ITS1 rRNA. This non-essential role of RNase MRP in rRNA processing is further supported by the finding that precursor RNAs containing 5.8S rRNA sequences accumulated in siRNA38-treated cells. It remains to be shown if human RNase MRP can cleave model substrates of ITS1 rRNA.
One of the unexpected findings of our study is the complete lack of 3′ ITS1 rRNA sequences in cells expressing Rpp38H. Although processing of ITS1 rRNA is an early step in rRNA maturation and the half-life of rRNA intermediates with 3′ ITS1 sequences is very short (29,31), the results indicate that these sequences, whether they exist as part of large rRNA transcripts or as free by-product molecules, are rapidly and efficiently eliminated in cells that produce Rpp38H. It has been shown that yeast 3′ ITS1 rRNA sequences are degraded by exoribonucleases that functionally interact in synthetic lethal screens with RNase MRP (30,36). Since similar degradation events exist in higher eukaryotes (31,37), the lack of 3′ ITS1 rRNA may involve excessive exonucleolytic activity in cells producing Rpp38H.
A role of Rpp38 in transcription?
The intracellular level of Rpp38, which is encoded by an intronless RPP38 gene candidate (2) is tightly regulated and selectively affects the expression of other RNase P protein subunits. The lack of normal expression of endogenous Rpp38 in HeLa cells that constitutively produce Rpp38H appears to involve a negative feedback mechanism that regulates RPP38 gene expression. Because the synthesis of both protein and mRNA for Rpp38 were inhibited in these cells, the molecular mechanism(s) responsible for this inhibition may act at the transcriptional level, possibly by Rpp38 itself. Of note, Rpp38 has a conserved domain, spanning positions 183–216, found in the retinoblastoma-associated proteins RbAp46 and RbAp48, both of which are components of chromatin remodeling complexes.
Acknowledgments
ACKNOWLEDGEMENTS
Part of this study was initiated in the laboratory of Professor Sidney Altman at Yale University. This research was supported by the Israel Science Foundation (grant no. 549/01) and by the United States–Israel Binational Science Foundation (grant no. 2001017) to N.J. N.J. is the recipient of a Kahanoff Foundation fellowship.
REFERENCES
- 1.Altman S., Gopalan,V. and Vioque,A. (2000) Varieties of RNase P: a nomenclature problem? RNA, 6, 1689–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarrous N. (2002) Human ribonuclease P: subunits, function and intranuclear localization. RNA, 8, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lygerou Z., Pluk,H., van Venrooij,W.J. and Seraphin,B. (1996) hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J., 15, 5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 4.Eder P.S., Kekuda,R., Stolc,V. and Altman,S. (1997) Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl Acad. Sci. USA, 91, 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarrous N., Eder,P.S., Guerrier-Takada,C., Hoog,C. and Altman,S. (1998) Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA, 4, 407–417. [PMC free article] [PubMed] [Google Scholar]
- 6.Jarrous N., Eder,P.S., Wesolowski,D. and Altman,S. (1999) Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA, 5, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarrous N. and Altman,S. (2001) Human ribonuclease P. Methods Enzymol., 342, 93–100. [DOI] [PubMed] [Google Scholar]
- 8.van Eenennaam H., Lugtenberg,D., Vogelzangs,J.H., van Venrooij,W.J. and Pruijn,G.J. (2001) hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J. Biol. Chem., 276, 31635–31641. [DOI] [PubMed] [Google Scholar]
- 9.Guerrier-Takada C., Eder,P.S., Gopalan,V. and Altman,S. (2002) Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA, 8, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lygerou Z., Mitchell,P., Petfalski,E., Seraphin,S. and Tollervey,D. (1994) The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev., 8, 1423–1433. [DOI] [PubMed] [Google Scholar]
- 11.Dichtl B. and Tollervey,D. (1997) Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J., 16, 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu S., Zengel,J.M. and Lindahl,L. (1997) A novel protein shared by RNase MRP and RNase P. RNA, 3, 382–391. [PMC free article] [PubMed] [Google Scholar]
- 13.Stolc V. and Altman,S. (1997) Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev., 11, 2926–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chamberlain J.R., Lee,Y., Lane,W.S. and Engelke,D.R. (1998) Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev., 12, 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stolc V., Katz,A. and Altman,S. (1998) Rpp2, an essential protein subunit of nuclear RNase P, is required for processing of precursor tRNAs and 35S precursor rRNA in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 95, 6716–6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall T.A. and Brown,J.W. (2002) Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA, 8, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton D.A. (2001) A big development for a small RNA. Nature, 410, 29–31. [DOI] [PubMed] [Google Scholar]
- 18.Xiao S., Scott,F., Fierke,C.A. and Engelke,D.R. (2002) Eukaryotic ribonuclease P: a plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem., 71, 165–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D.Y. and Clayton,D.A. (1997) RNase mitochondrial RNA processing correctly cleaves a novel R loop at the mitochondrial DNA leading-strand origin of replication. Genes Dev., 11, 582–592. [DOI] [PubMed] [Google Scholar]
- 20.Lindahl L., Archer,R.H. and Zengel,J.M. (1994) Alternate pathways for processing in the internal transcribed spacer 1 in pre-rRNA of Saccharomyces cerevisiae. Nucleic Acids Res., 22, 5399–5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chu S., Archer,R.H., Zengel,J.M. and Lindahl.L. (1994) The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc. Natl Acad. Sci. USA, 91, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai T., Aulds,J., Gill,T., Cerio,M. and Schmitt,M.E. (2002) The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics, 161, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Eenennaam H., Vogelzangs,J.H., Lugtenberg,D., Van Den Hoogen,F.H., Van Venrooij,W.J. and Pruijn,G.J. (2002) Identity of the RNase MRP- and RNase P-associated Th/To autoantigen. Arthritis Rheum., 46, 3266–3272. [DOI] [PubMed] [Google Scholar]
- 24.Klein D.J., Schmeing,T.M., Moore,P.B. and Steitz,T.A. (2001) The kink-turn: a new RNA secondary structure motif. EMBO J., 20, 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jarrous N., Wolenski,J.S., Wesolowski,D., Lee,C. and Altman,S. (1999) Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J. Cell Biol., 146, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Eenennaam H., van der Heijden,A., Janssen,R.J., van Venrooij,W.J. and Pruijn,G.J. (2001) Basic domains target protein subunits of the RNase MRP complex to the nucleolus independently of complex association. Mol. Biol. Cell, 12, 3680–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srisawat C., Houser-Scott,F., Bertrand,E., Xiao,S., Singer,R.H. and Engelke,D.R. (2002) An active precursor in assembly of yeast nuclear ribonuclease P. RNA, 8, 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvo O., Cuesta,R., Anderson,J., Gutierrez,N., Garcia-Barrio,M.T., Hinnebusch,A.G. and Tamame,M. (1999) GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4167–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lygerou Z., Allmang,C., Tollervey,D. and Seraphin,B. (1996) Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science, 272, 268–270. [DOI] [PubMed] [Google Scholar]
- 30.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borovjagin A.V. and Gerbi,S.A. (1999) U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in Xenopus oocytes. J. Mol. Biol., 286, 1347–1363. [DOI] [PubMed] [Google Scholar]
- 32.Hadjiolova K.V., Nicoloso,M., Mazan,S., Hadjiolov,A.A. and Bachellerie,J.-P. (1993) Alternative pre-rRNA processing pathways in human cells and their alteration by cycloheximide inhibition of protein synthesis. Eur. J. Biochem., 212, 211–215. [DOI] [PubMed] [Google Scholar]
- 33.Hadjiolova K.V., Georgiev,O.I., Nosikov,V.V. and Hadjiolov,A.A. (1984) Mapping of the major early endonuclease cleavage site of the rat precursor to rRNA within the internal transcribed spacer sequence of rDNA. Biochim. Biophys. Acta, 782, 195–201. [DOI] [PubMed] [Google Scholar]
- 34.Drabkin H.J. and RajBhandary,U.L. (1985) Site-specific mutagenesis on a human initiator methionine tRNA gene within a sequence conserved in all eukaryotic initiator tRNAs and studies of its effects on in vitro transcription. J. Biol. Chem., 260, 5580–5587. [PubMed] [Google Scholar]
- 35.Kovrigina E., Wesolowski,D. and Altman,S. (2003) Coordinate inhibition of expression of several genes for protein subunits of human nuclear RNase P. Proc. Natl Acad. Sci. USA, 100, 1598–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dichtl B., Stevens,A. and Tollervey,D. (1997) Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J., 16, 7184–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peculis B.A. and Steitz,J.A. (1993) Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell, 73, 1233–1245. [DOI] [PubMed] [Google Scholar]