Abstract
Successful use and reliability of microarray technology is highly dependent on several factors, including surface chemistry parameters and accessibility of cDNA targets to the DNA probes fixed onto the surface. Here, we show that functionalisation of glass slides with homemade dendrimers allow production of more sensitive and reliable DNA microarrays. The dendrimers are nanometric structures of size-controlled diameter with aldehyde function at their periphery. Covalent attachment of these spherical reactive chemical structures on amino-silanised glass slides generates a reactive ∼100 Å layer onto which amino-modified DNA probes are covalently bound. This new grafting chemistry leads to the formation of uniform and homogenous spots. More over, probe concentration before spotting could be reduced from 0.2 to 0.02 mg/ml with PCR products and from 20 to 5 µM with 70mer oligonucleotides without affecting signal intensities after hybridisation with Cy3- and Cy5-labelled targets. More interestingly, while the binding capacity of captured probes on dendrimer-activated glass surface (named dendrislides) is roughly similar to other functionalised glass slides from commercial sources, detection sensitivity was 2-fold higher than with other available DNA microarrays. This detection limit was estimated to 0.1 pM of cDNA targets. Altogether, these features make dendrimer-activated slides ideal for manufacturing cost-effective DNA arrays applicable for gene expression and detection of mutations.
INTRODUCTION
DNA microarrays are produced by in situ synthesis of oligonucleotides (1) or by mechanical deposition of pre-fabricated nucleotides onto chemically reactive support by means of ink jetting or spotting robot devices (2,3). The activated glass plates (e.g. microscope slides) are readily available, easy to handle and commonly used to deposit DNA, peptides, antibodies, low-molecular-weight molecules, etc. (4,5), because of their great chemical resistance against solvents, their mechanical stability and low intrinsic fluorescence properties. The performance of functionalised glass surface, e.g. signal intensity, signal-to-noise ratio, spot homogeneity, is directly influenced by the amount of DNA probes attached to the surface, by the probe length and by the accessibility of the labelled targets to the probes. Taking into account these requirements, an ideal support should have an excellent surface chemistry allowing covalent binding of DNA with assurance that this attachment is stable, a high binding capacity obtained by increasing the number of reactive sites to which the probes can be covalently bound, and a low background. Furthermore, probes should not be spread on the support, as this could induce problems of accessibility during the hybridisation step. Shchepinov et al. (6) suggested that an optimal spacer should possess at least 40 atoms in length, to not contain positive or negative charges, in order to space away the probe from the surface.
To try to fulfil these needs, two major achievements have been reported in recent years, which have progressively replaced the low-efficiency non-covalent binding of DNA on poly-l-lysine-coated glass slides (7,8). The first progress was the covalent binding of DNA probes onto silanised glass slides via a spacer of four to 12 carbon units, with an epoxy, aldehyde or isothiocyanate reactive function at its extremity (9–11). The attachment of the DNA probes occurs by their 5′-termini, which must be amino or thiol-modified, depending on the chemical reactive group of the linkers. While good quality, accuracy and reliability of these microarrays were demonstrated (11), the linear structure of the spacer still leaves a planar surface structure on the glass-slide, which neither increases by very much the accessibility of the targets to the probes nor the loading capacity of the support. To overcome in part these failures, a second major advancement has been to tentatively build up a pseudo-three-dimensional structure using the so-called ‘dendrimeric’ linker system. A significant contribution to this achievement was provided by the work of Beier and Hoheisel (12) who generated a pseudo-dendritic linker by direct chemical reaction on the glass slide or on polypropylene surfaces. With this structure, they could show an increased binding capacity of the activated-support for the probe beyond that of the glass slide and a high stability of the bonding of the dendrimer to the support. However, a major disadvantage of this product is that the linker system must be done in situ by eight time-consuming chemical steps, precluding any quality control of the linkers during the process, and limiting strongly a potential industrial development of this methodology. An alternative chemical process was the p-aminophenyltrimethoxysilane (ATMS)/diazotisation chemistry of the glass slides (13). However, major drawbacks of this grafting technology are that the ATMS-activated glass plates must be kept at 4°C throughout the process and that the covalent binding of DNA cannot be controlled as it could occur by any base of the probe. Another work described the use of prefabricated polyamino-functionalised ‘PAMAM starbust’ macromolecules for the manufacture of biochips, which includes an activation of the dendrimer layer surface prior to coupling with the DNA probes (14,15). Though this technology likely provides the best quality for DNA chips in terms of spots homogeneity, high sensitivity, low background and possible reutilisation, the chemical procedure to functionalise glass slides is time-consuming and leads to low thermal stability of the PAMAM dendrimers. Moreover, the support must be grafted with probes immediately upon activation, which limits the large-scale development of PAMAM-activated slides.
In the past 10 years, we have developed in our laboratory the production of dendrimers (16–18) based on an iterative reaction sequence that originates from a central core of hexachlorocyclo-triphosphazene (N3P3Cl6) linked to external ramifications to give rise to various generations (Table 1). The dendrimers are nanometric spherical structures whose diameter depends on the number of ramifications and which possess at their periphery a reactive function that can be aldehyde, thiol, epoxy, etc. In this work, we used dendrimers of generation 4 with aldehyde function to generate on amino-silanised glass surface a stable layer onto which NH2-modified DNA probes are covalently fixed by their 5′-ends. Advantages of this dendrimer-based technology and corresponding DNA microarrays over commercially available activated surface and DNA chips are illustrated, as well as their validation in biological applications.
Table 1. Size of dendrimers and number of aldehyde reactive functions according to the dendrimer generation.
| Generation | Number of reactive CHO functions | Size (Å) |
|---|---|---|
| 1 | 12 | 30 |
| 2 | 24 | 45 |
| 3 | 48 | 60 |
| 4 | 96 | 75 |
| 5 | 192 | 90 |
| 6 | 384 | 105 |
| 7 | 768 | 120 |
MATERIALS AND METHODS
Materials
All chemical and solvents were purchased from Fluka/Sigma (Gillingham, UK) and were of the highest analytical grade. Untreated microscope glass slide (Gold Seal Microslides) and Sigma Screen APS were purchased from Sigma/Aldrich (Gillingham, UK), QMT Amino-slide from Quantifoil Micro Tools GmbH (Jena, Germany), SAL-1 and SA-1 GENORAMA™ Microarray slides from Asper Biotech (Tartu, Estonia), SuperAldehyde and SuperAmine from Arraylt™ (Telechem, USA), Ez-rays amino-slide from Apogent Discoveries (Hudson, USA) CMT-Gap, CMT-Gap2 Ultragap from Corning (Kentucky, USA), EasySpot® Oligo Microarray slide from U-Vision Biotech (Taipei, Taiwan) and 3D-Link™ from Surmodics/Amersham Biosciences (Minnesota, USA). The 70mer oligonucleotides package corresponding to the 6200 yeast ORFs was purchased from Qiagen/Operon (Germany). PCR products that corresponded to yeast ORFS (see Figures 4 and 5 for details) were amplified using the commercially available set of primers (GenePair, Invitrogen Corp.).
Figure 4.
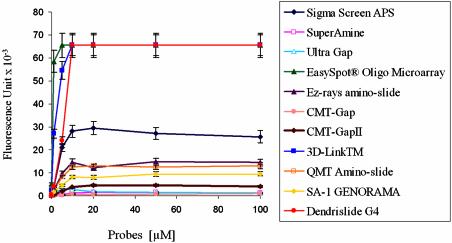
Immobilisation efficiency of G4-dendrislides compared with other commercially available activated-glass slides. Single-stranded 35mer oligonucleotide carrying a 3′-amino function and a 5′-Cy5 fluorophore (5′-Cy5-TTAGCGCATTTTGGCATATTTGG GCGGACAACTT-NH2-3′) was spotted at concentrations from 0.01 to 100 µM, on the different functionalised-glass slides. The value of fluorescence intensity (fluorescent signal – background) was measured at 635 nm with the Genepix scanner at 500 PMT.
Figure 5.
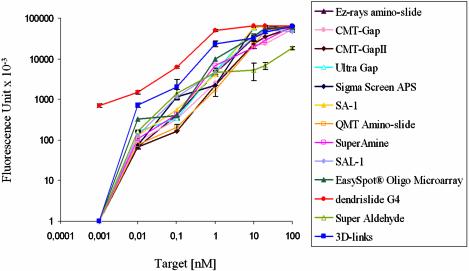
Detection sensitivity of dendrislide compared with commercially available activated glass slides. A single-stranded 35mer amino-oligonucleotide (5′-NH2-GTGATCGTTGTATCGAGGAATACTCCGATACCATT-3′) at a concentration of 10 µM was spotted on the different functionalised glass slides. The target Cy5-labelled 15mer oligonucleotides (5′-Cy5-AATGGTATCGGAGTA-3′) were hybridised at concentrations ranging from 0.001 to 100 nM. Image capture was measured at 635 nm with GenePix 4000A at 500 PMT. The value of fluorescence intensity reported is the fluorescent signal – background.
Production of the dendrimer-activated glass slides
Step 1: glass cleaning. Microscope glass slides were cleaned by immersion for 2 h in 60% ethanol containing NaOH 100 g/l (19). The slides were rinsed three times with MilliQ-filtered water and were then submerged in 95% ethanol.
Step 2: silanisation of the slides. Cleaned microscope slides were immersed in a solution consisting of 3-aminopropyltriethoxysilane 10% made in 95% ethanol for 12–15 h at room temperature under constant agitation. The slides were washed twice in 95% ethanol, twice in MilliQ-filtered water, dried under a stream of nitrogen and finally baked for 3 h at 120°C.
Step 3: bonding of dendrimers on the silanised slides. The silanised glass slides were immerged in an aqueous solution of KOH (8%) for 5 min. They were rinsed three times in MilliQ-filtered water and dried by centrifugation (8 min at 500 r.p.m.) or under a nitrogen stream. These activated slides were then incubated in a solution containing phosphorus dendrimers [0.1%, w/v, generation 4 (17)] in dichloromethane for 7 h at room temperature under gentle agitation. The slides were then washed twice with dichloromethane then with 95% ethanol and finally with absolute ethanol. They were dried by centrifugation or under a nitrogen stream. The dendrislides were kept at room temperature in plastic bags before use.
Spotting DNA probes onto the activated support surfaces
Unless otherwise stated, a 35mer 5′-labelled Cy5-oligonucleotide-3′NH2 (5′Cy5-TTAGCGCATTTTGGCATATTTGGGC GGACAACTT-NH2-3′) or the PCR products corresponding to the yeast ORFs were dissolved in the phosphate buffer 0.3 M, pH 9. All capture probes were synthesised with a modified primer bearing an amine at their 5′- or 3′-end. The probes were printed on glass slides by mechanical spotting with a robot (Eurogridder from Virtek/Bio-Rad). The print head on the arrayer uses ‘quill’ pins, whose contact on the surface delivers ∼2 nl of DNA solution per spot, with a medium size diameter of ∼80 µm under this condition. The quality of the printing was optimal at 45% relative humidity and a constant temperature of 22°C. After spotting, the slides were allowed to dry overnight at room temperature. The reduction of the imines function formed between probes and dendrimer was carried out by immersion of the slides into a solution containing NaBH4 at 3.5 mg/ml for 3 h at room temperature under agitation. The DNA slides were washed in boiling water for 2 min, twice in room-temperature water and then dried under a stream of N2. The DNA arrays were then stored in a plastic container at 4°C until use.
Preparation of labelled targets
In experiments shown in Figures 2B and 5, the target was a 15mer 5′-labelled Cy5 oligonucleotide (Cy5-AATGGTAT CGGAGTA) that is complementary to the 35mer probes (5′NH2-GTGATCGTTGTATCGAGGAATACTCCG ATACCATT). Other targets were prepared from total yeast RNA as a template by incorporation of fluorescent-labelled Cy5 or Cy3-deoxyribonucleotides during first-strand cDNA synthesis or by using the post-labelling (amino-allyl) method (20). The reaction was carried out with 15 µg total RNA using the CyScribe™ First Strand cDNA Labelling Kit or CyScribe Post-Labelling Kit (Amersham Bioscience) and purified on a GFX column (Amersham Bioscience).
Figure 2.
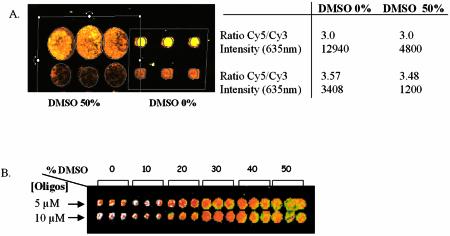
Images of hybridisation of a DNA probe spotted on activated glass slides with dendrimer generation 4 in the absence or in the presence of DMSO. (A) PCR products from two yeast ORFs (CDC19/YAL038w and CLN3/YAL040c) were resuspended in Na2 phosphate buffer 0.3 M, pH 9 in the absence or in the presence of DMS0 50%. (B) A 5′-NH2 35mer oligonucleotide was prepared in the same buffer at two different concentrations (5 and 10 µM) in the absence or presence of increasing concentration of DMSO. DNAs were spotted in triplicate on the activated dendrislides and hybridisation was carried out with yeast labelled cDNA (A) or with a complementary Cy5-labelled 15mer (5′-Cy5-AATGGTATCGGAGTA-3′) (B).
Hybridisation to the DNA-arrays, image detection and stripping procedures
Prehybridisation of the printing area was carried out with 20 µl solution containing 18 µl Dig Easy buffer (Roche Diagnostic) and 2 µl of denaturated salmon sperm DNA over 1 h at 42°C. The slide was washed with 2× SSC and then hybridised at 42°C for 12–14 h in a similar solution to which was added labelled cDNA targets that was boiled for 5 min prior to addition. In initial experiments, the hybridisation was carried out in a coverslip hybridisation cassette (Corning Inc), which was kept humid by addition of 20 µl water in the chamber. Experiments reported in Figures 4B and 6 were carried out with an automatic hybridisation chamber (Discovery from Ventana Medical System, Inc). Prehybridisation was carried out with a freshly prepared solution of 1% BSA, 2× SSC, 0.2% SDS over 30 min at 42°C. After automatic washing according to manufacturer’s instructions, the slides were hybridised for 10 h in 200 µl of ChipHybe™ buffer (Ventana Medical System, Inc) containing 10 µl of labelled cDNA purified on the GFX column. After hybridisation, the slides were washed for 2 min in 2× SSC/0.2% (v/v) SDS, for 2 min in 0.2× SSC/0.2% (v/v) SDS and for 2 min in 0.2× SSC, and then dried under a stream of nitrogen. Detection of the fluorescence signals was captured with a laser scanner (GenePix 4000A from Axon Instrument, CA) and analysed with the GenePix 3.01 software. Data were further processed by statistical methods using our homemade BioPlot Software (S.Sokol, C.Declercq and J.François, unpublished data, see http://bio71.gba.insa-tlse.fr/). For re-use, DNA-chips were incubated in a stripping solution [2.5 mM Na2HPO4 and 0.1% (v/v) SDS] for 10 min at 95°C. They were then washed with ddH2O, dried under a stream of N2 and checked for complete loss of fluorescence by scanning with the laser scanner (GenePix 4000A from Axon Inc) at appropriate sensitivity levels of photomultiplier (PMT).
Figure 6.
Use of dendrislides to detect base mutations. Thirteen 20mer oligonucleotides were spotted in triplicate at 10 µM in Pi buffer 0.3 M, pH 9.0. These oligonucleotides corresponded to a part of the yeast HSP12 gene, and varied from each other by a single or two mutations proximal to the 5′- or 3′-end or in the middle of the sequence. The 20mer sequences from HSP12 were as follows: 1: NH2 5′-AATATGTTTCCGGTCGTGTC-3′; 2: NH2 5′-AATATGTTTCGGGTCGTGTC-3′; 3: NH2 5′-AATATGTT TCAGGTCGTGTC-3′; 4: NH2 5′-AATATGTTTCtGGTCGTGTC-3′; 5: NH2 5′-AATATGTTTCCGGTCGTGTA-3′; 6: NH2 5′-AATATGTTTCC GGTCGTGTG-3′; 7: NH2 5′-AATATGTTTCCGGTCGTGTT-3′; 8: NH2 5′-AATATGATTCCGGaCGTGTC-3′; 9: NH2 5′-AATATGGTTCCGG GCGTGTC-3′; 10: NH2 5′-AATATGCTTCCGGcCGTGTC-3′; 11: NH2 5′-AATAAGTTTCCGGTCGTGTC-3′; 12: NH2 5′-AATAGGTTTCCGGT CGTGTC-3′; 13: NH2 5′-AATACGTTTCCGGTCGTGTC-3′. Hybridisation was carried out with Cy5-labelled cDNA from yeast total RNA. At the top of the corresponding spots is given the mean value of fluorescent signal (intensity at 635 nm – background).
RESULTS AND DISCUSSION
Properties of the dendrimer-activated support
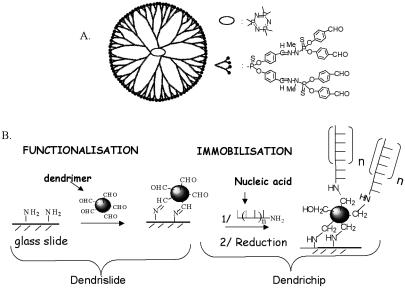
We have developed a chemical process for the covalent immobilisation of DNA probes (either PCR fragments or oligonucleotides) to microscope slides using a poly-functional polymer called a dendrimer. The dendrimers are nanometric spherical structures that originate from a central core from which emerge external ramifications (16), giving rise to different generations of dendrimers, according to the number of ramifications. They harbour at their outer sphere a specific reactive function (i.e. aldehyde, epoxide or thiol, etc.), the number of which is proportional to the ramification units (Table 1). The synthesis of these dendrimers has been described in detail by Launay et al. and Slomkowski et al. (16–18), and the chemical process for their covalent binding on the glass-slide has been patented (21). Figure 1 briefly outlines the protocol of manufacturing dendrimer-activated glass slides (dendrislide) and the covalent attachment of DNA probes on these dendrislides (dendrichip). This procedure to yield dendrimer-activated glass slides is very simple, effective and much faster than a previous method using ‘PAMAM starburst’ macromolecules (14). Moreover, the slides can be stored for months at room temperature before use which provides a serious technological advantage over PAMAM-dendrimer-activated glass surface. As the dendrimers differ in size and number of ramifications, different generations from G1 to G7 (Table 1) have been evaluated for their ability to reduce the signal-to-background after hybridisation of a 15mer Cy5-labelled complementary strand to a 35mer DNA probe. Since the same values for signal-to-background ratio were found with dendrimers G4–G7 (data not shown), we decided to use generation 4 merely because the next generation is dependent on the previous one due to the iteration process for dendrimer synthesis (16). Dendrimer G4 has a diameter size of about 75 Å, and displays 96 aldehyde functions at its periphery. We also verified that the binding of DNA probes on the aldehyde function of dendrimers occurs specifically via the 5′-amino modified terminal of the probes because no fluorescent signal was recorded after processing of the dendrimer-activated glass surface with an unmodified 15mer Cy3-labelled DNA probe (data not shown).
Figure 1.
Scheme of dendrimer ‘generation 4’ (A) and description of surface activation to fix the dendrimers and immobilisation method for covalent binding of nucleic acids (DNA probes) on the activated glass slide surface (B).
Choice of spotting conditions
As suggested by Diehl et al. (22), the buffer into which DNA probes are dissolved prior to spotting must be adequate for the chemical function of the activated-glass surface. It was also shown that the quality of the spotting on the nylon membrane was improved when the nucleic acids are dissolved in 50% dimethylsulfoxide (DMSO) (23). The advantage of this organic solvent is its denaturing property and its effect of strongly reducing evaporation. Both properties improve the homogeneity of the spots: they remain circular without a ‘doughnut’ effect. We also successfully applied this protocol for the spotting of DNA probes on amino-silanised glass slides purchased from different companies (amino-silane surface CMT-GAP™ from Corning; Telechem SuperAmine; Asper Biotech amino-silane SA, data not shown). However, this protocol did not work appropriately with dendrimer-functionalised glass slides. As shown in Figure 2, the spot’s size increases as the concentration of DMSO in the DNA probes increases, and reached ∼200 µm in the presence of 50% DMSO. Under this condition, the fluorescent signal of hybridisation with a mixture of Cy5-/Cy3-labelled targets obtained from total yeast RNA was 2- to 4-fold lower. This result suggests that DMSO may either reduce the actual amount of DNA fixed on the dendrimeric-activated support, affect the interaction between probes and targets or may even partially modify the backbone of the dendrimer [likewise a transformation of the P(S) groups into P(O) groups, see Fig. 1].
To circumvent this problem and to find a solvent capable of reducing evaporation during the spotting process, we tried the methodology of Diehl et al. (22) who showed that the addition of 1.5 M betaine to DNA solution prior to spotting on amino-silanised glass slides increased binding efficiency and fluorescence signal, and reduced evaporation. In agreement with this, we also found that betaine offered the same advantages with the dendrimer-activated glass slide, and increased the signal intensities after hybridisation by 2–3 times (Fig. 3A). However, we noticed a precipitation of the DNA probes dissolved in 1.5 M betaine after storage at –20°C. Thus, it is recommended to use the same DNA preparation for spotting within a short period of time.
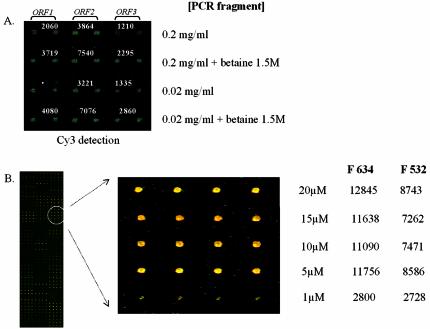
Figure 3.
Image capture from hybridisation of arrays manufactured with dendrislide bearing DNA probes spotted at decreasing concentrations. (A) PCR from yeast ORF (ORF1: MET14/YKL001c, ORF2: SPC42/YKL042w, ORF3: MRP17/YKL003c) was spotted in duplicate (column) in phosphate buffer 0.3 M, pH 9 at 200 and 20 µg/ml (rows) in the absence or in the presence of 1.5 M betaine. (B) A 70mer oligonucleotide corresponding to the gene SSA1/YAL005c was spotted in quadruplet in the same buffer at decreasing concentration ranging from 20 to 1 µM. The hybridisation was carried out with Cy3-labelled cDNA (A) or a mix of Cy3-/Cy5-labelled cDNA (B) obtained from 10 µg of yeast total RNA. In (A), the value of fluorescence intensity (fluorescent signal – background) measured at 532 nm is given on the top of the corresponding spots. The fluorescence intensity was measured with the Genepix scanner at 500 PMT.
Immobilisation efficiency
The binding capacity of the activated support is an important criterion for high density coupling of DNA probes, which can increase the signal-to-noise ratio and lower the detection limit upon hybridisation analysis. However, high surface densities and high hybridisation signal might not necessarily be related, since steric interferences between immobilised probes could hinder access to target DNA strand and reduce the hybridisation efficiency (15,24,25). Therefore, we compared the immobilisation capacity of G4-dendrimer slides with 10 conventional/commercial available activated glass slides (Materials and Methods). This was performed by spotting a 35mer oligonucleotide (Cy5-labelled at its 5′-end and containing a NH2 group at its 3′-end for covalent binding to supports) four times at concentrations ranging from 0.01 to 100 µM, which correspond to probe densities ranging from 0.02 to 250 fmol/mm2. The fluorescence signal, which estimates the immobilisation efficiency, was recorded with a laser scanner (GenePix 4000A) at PMT 600, and the mean value for the four data points was calculated for each concentration of spotted DNA probes. Using our dendrislides, the fluorescence intensity reached saturation at 10 µM DNA probes (Fig. 4). Apparently, this chemistry did not offer a better binding capacity since saturation was already attained at this concentration with two other commercial arrays that are functionalised for covalent attachment of the probes (EasySpot® Oligo Microarray slide from U-Vision Biotech and 3D Links from Surmodics/Amersham Biosciences). Interestingly, this mode of attachment resulted in a 2- to 3-fold higher fluorescent intensity than that from amino-silanised slides (as for instance the very commonly used ultra Gap slides from Corning or Sigma screen APS, see Fig. 4). This result suggests that the dendrimer-modified surface has a higher surface coverage and reactivity in comparison to amino-silanised glass slides. The lower reactivity of these latter glass slides might be due to steric constraints or by the fact that the probes are spread over the surface, therefore reducing accessibility of other molecules for binding.
We carried out two other experiments by lowering the concentration of probes for spotting. In the first experiment, unlabelled PCR products ranging from 400 to 600 bp from three yeast ORFs were spotted in duplicate at 0.2 mg/ml and at a 10-fold lower concentration. The binding capacity was indirectly estimated by hybridisation of these DNA probes with Cy5-labelled cDNA from total yeast RNA. Figure 3A shows that the hybridisation signal was the same at both concentrations of the spotted probes, and as indicated above, the signal intensities were 2-fold higher when the probes were prepared in the presence of 1.5 M betaine prior to spotting. The second experiment was carried out with 70mer oligonucleotides from a set of 96 yeast genes kindly provided by Operon (Array-Ready Oligo Set™ Qiagen/Operon). They were spotted in quadruplets at decreasing concentrations from 20 to 1 µM. The arrays were hybridised as described above but using a mixture of Cy5-/Cy3-labelled cDNA targets from total yeast RNA (Fig. 3B). The hybridisation image of the 96 oligonucleotides microarrayed on the dendrimer-activated slides shows that signal intensities were the same from 20 to 5 µM, and dropped by three times at 1 µM DNA probe (see enlarged area of Fig. 3B). This suggests that at concentrations lower than 5 µM, the DNA probe was no longer in excess over the target DNA. From this experiment, we could estimate a minimal concentration of oligonucleotides probes of ∼5 µM for efficient target/probe hybridisation on our dendrichips. As a consequence, the high surface coverage together with an efficient target capture at low oligonucleotide probe concentration offer several advantages for the dendrislides in both technological and economical terms.
Comparison of sensitivity and detection limit of dendrichips with other microarrays
The experiments reported above have already indicated that glass slides functionalised with spherical dendrimeric structure not only have higher immobilisation efficiency, but also confer an excellent accessibility to the target DNA. To quantify the target/probe hybridisation sensitivity, arrays from dendrislides, as well as from 11 available activated slides from commercial sources, were manufactured with a fixed amount of a 35mer oligonucleotide (spotted at 10 µM solution) and were hybridised with increasing concentration of a Cy5-labelled 15mer oligonucleotide complementary to the probe from 0.001 to 100 nM. As shown in Figure 5, the fluorescent intensity as recorded with the laser scanner set at PMT 600 reached a plateau at a concentration of 10 nM target DNA for almost all manufactured arrays. However, the logarithmic expression of signal intensities versus concentration of targets illustrated a clear advantage of the dendrimer-activated slides over all other microarrays at target concentrations <1 nM. A fluorescent signal, which corresponded to total fluorescence backgound of 1500 ± 30, was measured at a concentration of 0.01 nM of cDNA target, which is about 10-times higher than for other commercially available slides, and at 0.001 nM of DNA target, a signal (fluorescence background) was still easily quantifiable only using dendrichips. This result indicates a detection sensitivity of dendrichips that is ∼10- to 100-fold higher than arrays made with most of other functionalised glass slides currently available on the market (13,19,22,23). The high hybridisation sensitivity is particularly interesting when measuring low abundance transcripts in cells and for studies involving very low amounts of biological material, such as biopsies. As an indication of this high sensitivity, we produced Cy3-labelled cDNA from 10 µg total yeast RNA by reverse transcription, and used these labelled cDNA targets at 1- to 100-fold dilution to hybridise dendrichips made with 70mer oligonucleotides from a set of randomly chosen 96 yeast genes provided by Operon. A quantifiable fluorescent signal on almost all spots could be recorded even with the lowest concentration of cDNA targets (data not shown).
Caution about re-usability of DNA microarrays
The possibility to re-use DNA arrays could be an advantage in transcriptomic studies, as it allows reproducibility of experiments on the same technical support. It also reduces the cost, while improving accuracy of the genomic data. Thus, we investigated the re-usability of the DNA arrays generated from dendrimer-activated support. The first approach that has been reported in several previous papers (12,13,20,22) was to spot a few sets of PCR or oligonucleotide probes. In our case, we spotted a 35mer oligonucleotide several times on dendrislides, and hybridised this prototype of microarrays with a Cy5-labelled target complementary to the probe. After reading the fluorescence intensity, the slides were regenerated by ‘stripping’ using phosphate buffer (pH 7.5) containing SDS (0.1%, w/v) for 5 min at 95°C. After each stripping the slides were scanned to guarantee the complete removal of the target and subjected to rehybridisation. By application of this strategy, the arrays manufactured with dendrislides were re-used at least 10 times without significant loss of the signal intensity. The same advantages were claimed with many other activated-glass slides that allow covalent linkage of the probes (12,15,20). We therefore wanted to verify this performance, using arrays bearing the whole yeast genome spotted as PCR products (300–600 bp length) or 70mer oligonucleotides (purchased from Qiagen/Operon) (see our website http://bio71.gba.insa-tlse.fr for details on yeast microarrays construction). Contrary to expectation, the stripping procedure for these arrays was much less effective; leaving patches of faint colour on the slides that could not be removed even after two or three successive strippings. Similar data were obtained with yeast and rat oligo microarrays from commercial sources (not shown). This lack of complete removal of the targets from the arrays may be due to several reasons, including the heteroduplex interactions which involve secondary structures of the nucleic acids (26), as well as residual unspecific binding of dyes on the glass slides.
Biological application to mutations detection
Search for a single mutation in a given gene (SNPs) is a research field where the DNA chip technology should bring a revolution (27–30). Although methods to identify SNPs already exist and most of them rely on the Affymetrix technology (1,31), the development of an alternative, inexpensive, cost-effective high throughput method to identify point mutations in DNA sequences is still awaited in the context of medical diagnosis. Search of mutations in a given gene must use dedicated chips that contain all possible sequences of the DNA in which one wishes to seek these mutations. We examined the potential application of our dendrimer-activated support in such an endeavour. To this end, we spotted on dendrislides 13 20mer oligonucleotides from HSP12 encoding a protein chaperone in yeast (30), which differed from each other by a single or a double base mutation at positions proximal to 5′, 3′ or in the middle of the sequence. These DNA probes were then hybridised with Cy5-labelled cDNA obtained from reverse transcription of yeast total RNA. Figure 6 shows the image capture of this experiment. As expected, the highest fluorescent signal was recorded with probe 1, which exhibits the perfect complementary DNA sequence of the yeast HSP12. Interestingly, a lower but measurable fluorescence intensity was recorded with oligonucleotides bearing mismatch mutation at the 3′-end (i.e. probes 5–7). This result is an indirect indication that the probes attached to a dendrislide are highly accessible to cDNA targets.
Conclusion
The explosion of genome sequences from microbes, plants, animals and humans forces academic and industrial laboratories to investigate functional genomics with the use of DNA chips in a massive way. So far, these tools have remained too expensive for most of the academic laboratories, and hence, efforts must be carried out to propose DNA arrays that are more reliable, cheaper and easy to use for these customers. To this end, we have generated a very effective chemically activated glass slide based on the use of homemade neutral phosphorus dendrimers. These G4-dendrimer-coated slides display most of the suitable criteria that one should expect for accurate, reliable and cost-effective DNA arrays. Since the production of dendrimer and the chemistry for activation of the surface are relatively simple and inexpensive, this process could be reproduced at an industrial scale. Moreover, we found that dendrislides were still active after 3–5 months of storage at room temperature. Since other functions, for example thiol, epoxy, etc., can be introduced at the periphery of the dendrimer structure, this renders these polymers particularly attractive for the immobilisation of other macromolecules such as peptides, proteins, antibodies, lipids, etc.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Drs C. Bergaud (LAAS-CNRS) and G. Pratviel (LCC-CNRS) for stimulating discussion during this work. This work was supported in part by grants from Région Midi-Pyrénées (01002664) and Génopole Toulouse Midi-Pyrénées.
REFERENCES
- 1.Lipshutz R.J., Fodor,S.P.A., Gingeras,T.R. and Lockhart,D.J. (1999) High density synthetic oligonucleotide arrays. Nature Genet., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 2.Hughes T.R., Mao,L.M., Jones,A.R., Burchard,J., Marton,M.J., Shannon,K.W. et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol., 19, 342–347. [DOI] [PubMed] [Google Scholar]
- 3.Cheung V.G., Morley,M., Auilar,F., Massimi,A., Kucherlapati,R. and Childs,G. (1999) Making and reading microarrays. Nature Genet., 21, (suppl.), 15–19. [DOI] [PubMed] [Google Scholar]
- 4.MacBeath G., Koehler,A.N. and Schreiber,S.L.(1999) Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J. Am. Chem. Soc., 121, 7967–7968. [Google Scholar]
- 5.Mac Beath G. and Schreiber,S.L. (2000) Printing proteins as microarrays for high-throughput function determination. Science, 289, 1760–1762. [DOI] [PubMed] [Google Scholar]
- 6.Shchepinov M.S., Case-Green,S.C. and Southern,E.M. (1997) Steric factors influencing hybridisation of nucleic acids to oligonucleotide arrays. Nucleic Acids Res., 25, 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 8.De Risi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 9.Joos B., Kuster,H. and Cone,R. (1997) Covalent attachment of hybridisable oligonucleotides to glass supports. Anal. Biochem., 247, 96–101. [DOI] [PubMed] [Google Scholar]
- 10.Rogers Y.-H., Jianf-Baucon,P., Huang,Z.-J., Bogdanov,V., Anderson,S. and Boyce-Jacino,M.T. (1999) Immobilization of oligonucleotides onto a glass support via disulfide bonds: a method for preparation of DNA microarrays. Anal. Biochem., 266, 23–30. [DOI] [PubMed] [Google Scholar]
- 11.Guo Z., Gatterman,M.S., Hood,L., Hansen,J.A. and Petersdorf E.W. (2002) Oligonucleotide arrays for high throughput SNPs detection in the MHC class I genes: HLA-B as a model system. Genome Res., 12, 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beier M. and Hoheisel,J.D. (1999) Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dolan P.L., Wu,Y., Ista,L.K., Metzenberg,R.L., Nelson,M.A. and Lopez,G.A. (2001) Robust and efficient synthetic method for forming DNA microarrays. Nucleic Acids Res., 29, e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benters R., Niemeyer,C.M. and Wohrle,D. (2001) Dendrimer-activated solid supports for nucleic acid and protein microarrays. Chem. Biochem., 2, 686–694. [DOI] [PubMed] [Google Scholar]
- 15.Benters R., Niemeyer,C.M., Drutschmann,D., Blohm,D. and Wohrle,D. (2002) DNA microarrays with PAMAM dendritic linker systems. Nucleic Acids Res., 30, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Launay N., Caminade,A.M., Lahana,R. and Majoral,J.P (1994) A general synthetic strategy for neutral phosphorus-containing dendrimers. Int. Ed. Engl., 33, 1589–1592. [Google Scholar]
- 17.Launay N., Caminade,A.M. and Majoral,J.P (1997) Synthesis of bowl-shaped dendrimers from generation 1 to generation 8. J. Organomet. Chem., 529, 51–53. [Google Scholar]
- 18.Slomkowski S., Miksa,B., Chehimi,M.M., Delamar,M., Cabet-Deliry,M., Majoral,J.P. and Caminade,A.-M. (1999) Inorganic-organic systems with tailored properties controlled on molecular, macromolecular and microscopic level. Reactive Functional Polymers, 41, 45–57. [Google Scholar]
- 19.Zammatteo N., Jeanmart,L., Hamels,S., Courtois,S., Louette,P., Hevesi,L. and Remacle,J. (2000) Comparison between different strategies of covalent attachment of DNA to glass surfaces to build DNA microarrays. Anal. Biochem., 280, 143–150. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X., Nampalli,S., Serino,A.J. and Kumar,S. (2001) Immobilization of oligonucleotides with multiple anchors to microchips. Nucleic Acids Res., 29, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trévisiol E., Leclaire,J., Pratviel,G., Caminade,A.M., Majoral,J.P., François,J. and Meunier,B. (2002) Supports solides fonctionalisés par des dendrimères phosphorés, leurs procédés de préparation et applications. French patent no. 0205049 (April 23nd).
- 22.Diehl F., Grahlmann,S., Beier,M. and Hoheisel,J.D. (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res., 29, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegde P., Qi,R., Abernathy,K., Gay,C., Dharap,S., Gaspard,R., Hughes,J.E., Snesrud,E., Lee,N. and Quackenbush,J. (2000) A concise guide to cDNA microarray analysis. Biotechniques, 29, 548–562. [DOI] [PubMed] [Google Scholar]
- 24.Eisen M.B. and Brown,P.O. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 25.Henke W., Herdel,K., Jung,K. Schnorr,D. and Loening,S.A. (1997) Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res., 25, 3957–3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peterson A.W., Heaton,R.J. and Georgiadis,R.M. (2001) The effect of surface probe density on DNA hybridisation. Nucleic Acids Res., 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hacia J. (1999) Resequencing and mutational analysis using oligonucleotide microarrays. Nature Genet., 21, 42–47. [DOI] [PubMed] [Google Scholar]
- 28.Hacia J.G. and Collins,F.S. (1999) Mutational analysis using oligonucleotide microarrays. J. Med. Genet., 36, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastinen T., Raitio,M., Lindroos,K., Tainola,P., Peltonen,L. and Syvanen,A.C. (2000) A system for specific, high-throughput genotyping by allele-specific primer extension on microarrays. Genome Res., 10, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Praekelt U.M. and Meacock,P.A. (1990) HSP12, a new small heat shock gene of Saccharomyces cerevisiae: analysis of structure, regulation and function. Mol. Gen. Genet., 223, 97–106. [DOI] [PubMed] [Google Scholar]
- 31.Maskos U. and Southern,E.M. (1992) Parallel analysis of oligodeoxyribonucleotide (oligonucleotide) interactions. I. Analysis of factors influencing oligonucleotide duplex formation. Nucleic Acids Res., 20, 1675–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]