Abstract
Aberrant DNA methylation of the CpG site is among the earliest and most frequent alterations in cancer. Detection of promoter hypermethylation of cancer-related genes may be useful for cancer diagnosis or the detection of recurrence. p16, an inhibitor of the cyclin D-dependent protein kinases, is a classical tumor suppressor gene, and its inactivation is closely associated with carcinogenesis. p16 hypermethylation could be detected in each stage, which is consistent with the finding that aberrant methylation of p16 is a very early event in carcinogenesis. We have developed an electrochemical procedure for detecting DNA methylation of the human p16Ink4a gene. The procedure is based on the coupling of DNA electrochemical sensors with linker-PCR- amplified DNA from human gastric tumor tissue and whole blood cells of healthy human. The synthesized oligonucleotide was immobilized on the modified gold electrode to fabricate a DNA biosensor. The hybridization reaction on the electrode surface was monitored by cyclic voltammogram (CV) and square wave voltammogram (SWV), using [Co(phen)3](ClO4)3 as a redox indicator. Methylation status of human p16Ink4a gene was detected and the results were validated by bisulfite DNA sequencing. A good reproducibility was observed in several parallel experiments. The coupling of DNA electrochemical sensors with PCR allowed quick detection and have the potential of the quantitative evaluation of the methylation status of the human p16Ink4a gene.
INTRODUCTION
The methylation of DNA is an epigenetic modification that can play an important role in the control of gene expression in mammalian cells. The epigenetic event has been observed in GC-rich regions, called CpG islands, frequently located in the promoter and the first exon regions of genes. CpG island hypermethylation is closely associated with transcriptional inactivation of the classic tumor suppressor genes, which is a common feature in human carcinomas (1). p16, an inhibitor of the cyclin D-dependent protein kinases, is a tumor suppressor gene, and its inactivation is closely associated with carcinogenesis. Hypermethylation on the CpG islands of the p16 gene has been proposed as an alternative mechanism for the loss of p16 expression. p16 hypermethylation could be detected in each stage, which is consistent with the finding that aberrant methylation of p16 is a very early event in carcinogenesis (2). Detection of aberrant promoter hypermethylation of cancer-related gene may be useful for cancer diagnosis or the detection of recurrence (3,4). Several methods have been developed to evaluate the methylation status of genes, such as Southern blot (5), bisulfite genomic DNA sequencing (6), restriction enzyme-PCR (7), MSP (methylation-specific PCR) (8), methylation-sensitive single nucleotide primer extension (MS-SNuPE) (9), DNA microarray based on fluorescence or isotope labeling (10,11) and so on. They have offered useful and powerful tools in studying the phenomenon of DNA methylation. However, the present methods are still laborious, time-consuming, less sensitive and not rigid enough for the clinical applications. It is of great importance to establish sensitive and reliable methods for the methylation detection of earlier cancer diagnosis.
In recent years, there has been considerable interest in developing a DNA electrochemical biosensor for rapid applications in genetic analysis. DNA electrochemistry biosensor plays an enormous potential for the detection of the presence of genes or mutant genes associated with inherited human diseases, because electrochemical ones provide simple, rapid and low-cost point-of-care detection of specific nucleic acid sequences (12–15). A DNA electrochemical biosensor generally is an electrode with an oligonucleotide immobilized on the surface. Recently, some reports have indicated that electrochemical techniques are well suited for measuring hybridization events (16–24). The electrochemical detections of DNA hybridization have been mainly oriented to the application of labeling or unlabeling target. A number of papers focused on the application of electroactive DNA intercalators such as daunomycin, [Co(phen)3](ClO4)3 and Co(bpy)33+ intercalates in the double strand to detect DNA hybridization (16–21). However, most of the present research was demonstrated only by using synthetic oligonucleotides as target gene. Only Marrazza et al. (17) have reported the experiments of DNA electrochemical biosensors for detection of PCR-amplified DNA products. We have not found any report to use the DNA electrochemical biosensor for methylation detection until now.
The aims of the present work are to develop a new and label-free method for detecting DNA methylation. We describe an electrochemical method to detect DNA methylation, using DNA fragments amplified by linker-PCR. The procedure involves the use of an electroactive indicator, [Co (phen)3](ClO4)3, which can intercalate the double strand DNA (25). We selected a segment of the 5′ untranslated region and the first exon of the p16Ink4a gene, as the investigated target, which includes the recognition site of BstUI restriction endonuclease. Genomic DNA is restricted with MseI from human gastric tumor tissue and whole blood cells of healthy human, and the cleaved ends of DNA are ligated to unphosphorylated linkers as universal PCR primers. Then the ligated DNA is digested with methylation-sensitive endonucleases BstUI. The digested products are amplified by PCR as targets. An oligonucleotide was synthesized and purified to completely match with the target DNA fragments. The oligonucleotide was immobilized on the modified gold electrode to fabricate a DNA biosensor. The DNA biosensor is hybridized with the target solution to determine methylation status of p16 gene according to peak current. We have successfully used the above electrochemical method to detect the methylated CpG islands of p16 gene from human gastric tumor tissue, and the results showed that the electrochemical monitoring of the DNA hybridization at the transducer surface would be reliable, low cost and the analysis would be rapid.
MATERIALS AND METHODS
2-Aminoethanethiol (AET), 1-ethyl-3 (3-dimethyl-aminopropyl)-carbodiimide (EDC) and N-hydroxysulfosuccinimide (NHS) were purchased from Sigma Chemicals. [Co(phen)3] (ClO4)3 was prepared according to the literature (26). Other chemicals were of analytical reagent grade.
Pretreatment and modification of the electrodes
The electrodes were pretreated and modified according to Ge et al. (26). Gold electrodes were polished firstly with w5 abrasive paper and subsequently with chamois followed by rinsing with water and sonification in ethanol and water for 2 min each, respectively. The extent of electrode pretreatment was evaluated by a cyclic voltammogram (CV) measurement in 1 mM K3Fe (CN)6 solution containing 0.1 M KCl. The peak-to-peak separation should be <70 mV at a scan rate of 100 mV/s.
Before chemisorptions, the electrodes were treated with ‘piranha solution’ (H2SO4/30% H2O2, 7:3 in v/v) for 5 min at room temperature and subsequently rinsed thoroughly with twice distilled water and, finally, ultrasonically cleaned with ethanol and twice distilled water for 3 min, respectively. The pretreated electrodes were immersed in 1 mM AET ethanol solution for 16 h resulting in the AET deposition on the electrodes. After that, the electrodes were thoroughly washed with ethanol and distilled water. The electrode was kept in distilled water till use and denoted as AET/Au.
DNA self-assembly on the modified gold electrodes
The phosphorylated oligonucleotide was synthesized and purified by Shenyou Inc. (Shanghai, China). The oligonucleotide sequence was 5′-ACGGCCGCGGCCCGGGGT-3′. A gold electrode modified with AET (denoted as AET/Au) was immersed in a 5 mM EDC and 8 mM NHS in PB buffer (pH 7.0) for 15 min containing DNA probes. The DNA modified electrode (denoted as DNA/AET/Au) was then soaked for 5 min in 0.1% SDS and distilled water, respectively, to remove any non-specially absorbed DNA.
Preparation of targets and hybridization
Whole blood cells of healthy human and gastric tumor tissue obtained from GuLou hospital (Nanjing, China). Genomic DNA was extracted from whole blood cells and gastric tumor tissue by standard methods using proteinase K digestion and phenol/chloroform extraction. The DNA derived from the blood cells of healthy human was divided into two parts. One aliquot was treated by methylase Sss I (SM) as a positive control with the conditions recommended by the supplier (New England Biolabs, Beverly, MA). Another was not treated by methylase Sss I (SM) as negative control. The positive control generated in this way had 100% methylated cytosine in the test CpG sites, whereas the negative control had all unmethylated cytosine residues in the test CpG sites.
The preparation process of targets is illustrated in Figure 1. Approximately 2 µg DNA was restricted to completion with 20 U of MseI following the conditions recommended by the supplier (New England Biolabs, Beverly, MA). This enzyme restricts bulk DNA into small fragments. As its recognition site (TTAA) rarely occurs in GC-rich regions, most CpG islands remain intact after the restriction. The digests were purified with QIAquick column (Qiagen) and mixed with 0.5 nmol of unphosphorylated linkers H-24 and H-12 in a DNA ligase buffer (New England Biolabs). The linker sequences were H-24, 5′-AGG CAA CTG TGC TAT CCG AGG GAT, and H-12, 5′-TAA TCC CTC GGA (27). Oligonucleotides were annealed by cooling the mixture gradually from 55°C to room temperature over 1 h and then ligated to the cleaved ends of the DNA fragments by incubation overnight with 400 U of T4 DNA ligase (New England Biolabs) at 16°C.
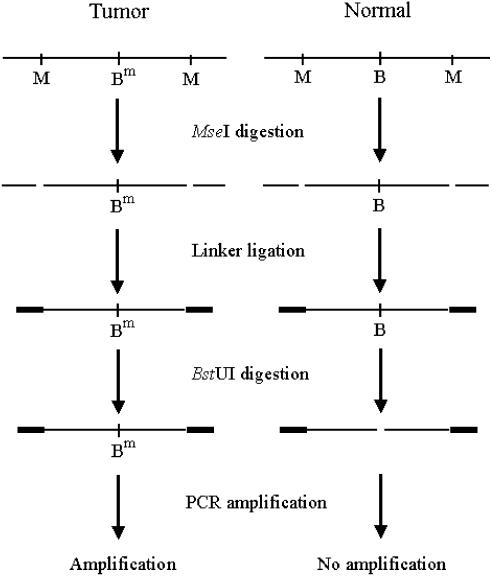
Figure 1.
A schematic outline of the procedure for the preparation of the DNA targets. M, B and Bm represent MseI, unmethylated and methylated BstUI recognition sites, respectively. Genomic DNA comes from whole blood cells of healthy human (normal) and gastric tumor tissue (tumor).
The ligated DNA was digested with the methylation-sensitive endonucleoase BstUI following the conditions recommended by the supplier (New England Biolabs). PCR reactions were performed in a 100 µl volume, containing 0.4 µM H-24 primer, 4 U Taq DNA polymerase, 5% DMSO and 200 µM dNTP in a buffer provided by the supplier. The tubes were incubated for 3 min at 72°C to fill in 5′-protruding ends of the ligated DNA and subjected to 20 cycles of amplification consisting of 1 min at 97°C and 3 min at 72°C in a PTC-225 thermocycler (MJ Research, Watertown, MA). The final extension was lengthened to 10 min.
The PCR products were suspended in unihybridization solution [1:4 dilution (v/v), Telechem]. The electrode hybridization was conducted in a moist hybridization chamber under a cover slip at 42°C for 2 h. After hybridization, the electrode was rinsed and washed at room temperature with 2× SSC–0.1% SDS and 0.1× SSC–0.1% SDS, respectively, for a total of 15 min, and dried under the flow of argon. The electrodes were denoted as positive control hybrid-DNA/AET/Au, negative control hybrid-DNA/AET/Au and sample hybrid-DNA/AET/Au, respectively.
Electrochemical measurement
CV and square wave voltammogram (SWV) experiments were performed on a CHI 660A workstation (American CH instrument). The electrochemical cell consisted of a three-electrode system with gold or modified gold electrode (0.5 mm in diameter) as the working electrode, a saturated calomel electrode (SCE) and a platinum wire as the reference and the counter electrode, respectively. The experimental temperature was controlled at 25 ± 1°C.
Bisulfite analysis
Bisulfite processing of DNA was performed in principle as described by Frommer et al. (6) and the modifications introduced by Clark et al. (28). Briefly, 1 µg of genomic DNA was digested by EcoRI and denatured in 0.35 M NaOH at 37°C for 20 min. The bisulfite reaction was carried out in 3.2 M sodium bisulfite and 0.5 mM hydroquinone (Sigma Chemical Co., USA) at 55°C for 16–24 h. DNA was recovered by a desalting column (DNA Clean-Up System, Promega Inc., USA) and desulphonated in 0.2 M NaOH at 37°C for 15 min, neutralized by ammonium acetate, alcohol precipitated, dried and then dissolved in 30 µl of deionized water. After bisulfite processing, the all unmethylated cytosine residues converted to uracil, whereas the methylated ones remained unchanged.
The 5′-CpG island regions of the p16Ink4a gene were amplified with primers for methylated and unmethylated DNA, respectively. The primer pairs are described in Table 1. The PCR reaction was performed in buffer containing 10 mM Tris–HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 5% DMSO, 1.75 mM MgCl2, 0.2 mM of each dNTP and 1 µl bisulfite-treated DNA. The amplification was carried out for 35 cycles (30 s at 95°C, 30 s at the annealing temperature listed in Table 1 and 30 s at 72°C), followed by a final 4 min extension at 72°C. The PCR products were gel purified and sequenced using an automated sequence analyzer (ABI377A, Applied Biosystem Inc.).
Table 1. PCR primers used for methylation-specific PCR.
| Primer set | Sense primer (5′→3′) | Antisense primer (5′→3′) | Size (bp) | Temperature (°C) | Genomic position |
|---|---|---|---|---|---|
| p16-M | TTATTAGAGGGTGGGGCGGATCGC | CCACCTAAATCGACCTCCGACCG | 234 | 65 | +167 |
| p16-U | TTATTAGAGGGTGGGGTGGATTGT | CCACCTAAATCAACCTCCAACCA | 234 | 60 | +167 |
Sequence differences between methylation/modified and unmethylated/modified are underlined. Primers were placed near the transcriptional start site. Genomic position is the location of the 5′ nucleotide of the sense primer in relation to the major transcriptional start site (GenBank accession no. X94154).
RESULTS AND DISCUSSION
Detection of methylation status of p16Ink4a gene with cyclic voltammogram
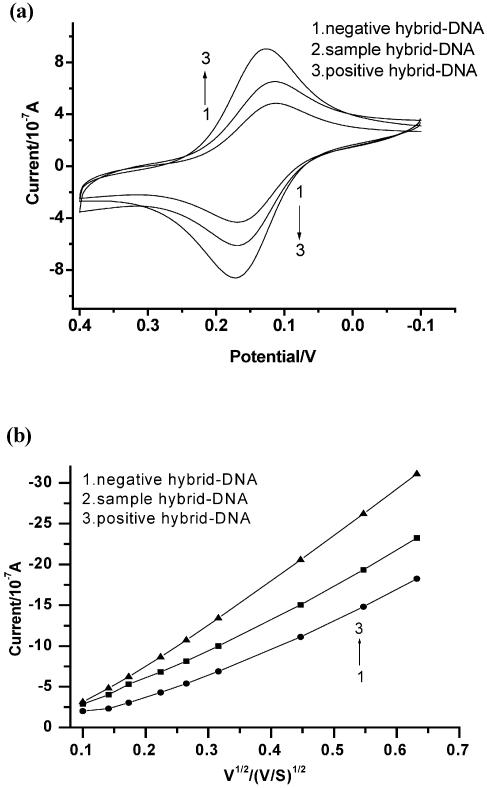
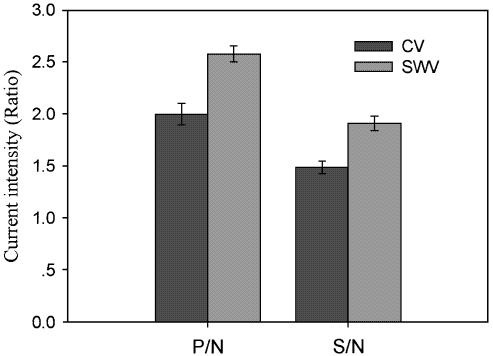
In the presence of water-soluble carbodiimide regent, the 5′-terminal phosphate of oligonucleotide probe formed a phosphoramidate bond with the primary amino group of aminoethanethiol monolayer on the gold electrode surface (29). These electrodes were reacted with positive control, negative control and sample in the hybridization buffer, respectively. The recognition of differently hybridized electrodes was justified by CV using 0.12 mM [Co(phen)3](ClO4)3 as a redox indicator that was added in the Tris–HCl buffer solution. Figure 2a and b shows the CV measurement at the scan rate of 50 mV/S and anodic peak currents versus root of the scan rate of CV v1/2 respectively for negative hybrid-DNA/AET/Au (curve 1), sample hybrid-DNA/AET/Au (curve 2) and positive hybrid-DNA/AET/Au (curve 3) in Tris–HCl buffer (pH 7.1) containing 0.12 mM [Co(phen)3](ClO4)3. Comprehensively, the following results were observed: (i) the peak current intensity reflected the methylation status of the p16Ink4a gene. Figure 2a shows that the redox peak currents of Co(phen)33+ species at the positive hybrid-DNA/AET/Au (curve 3) electrode (7.56 ± 0.21) were much larger than those at sample hybrid-DNA/AET/Au (curve 2) (5.62 ± 0.24) and negative hybrid-DNA/AET/Au (curve 1) (3.80 ± 0.31). Because the peak current intensity of negative hybrid-DNA/AET/Au (curve 1) was the smallest among three electrodes, a positive threshold could be established (30); therefore, methylation could be determined. The ratios of positive control/negative control and sample/negative control were calculated respectively and averaged for three parallel experiments. The positive control/negative control (P/N) and sample/negative control (S/N) were 1.989 and 1.479, respectively (Fig. 4). These data indicated that the CV could be used to detect methylation status of p16Ink4a gene. (ii) The peak potential differences (ΔEp = Epa – Epc) and the peak shape |Epc – Ep/2| were found to be between 60 and 70 mV, independent of potential sweep rate (10 ≤ v ≤ 100 mV/s), where ΔEp, Epa, Epc and Ep/2 represent the interspike interval, anodic spike potential, cathodic spike potential and half peak potential, respectively. These indicated that the reduction of [Co(phen)3]3+ at the surface of the electrode is a one electron transfer reversible process. A linear correlation between the oxidation peak current and the square root of the scan rate v1/2 was observed (Fig. 2b), as expected for a diffusion controlled electrochemical process. All electrodes are R ≥ 0.99 and P ≤ 0.001, respectively (Fig. 2b). On the other hand, at the positive hybrid-DNA/AET/Au and sample hybrid-DNA/AET/Au, the oxidation peak currents increase rapidly with the v1/2, which is consistent with the behavior of adsorption of the electroactive species that occurred at the surface of the electrode. According to the relationship of peak current ip with the scan rate v (25): ip = 2.69 × 105·n3/2·C0·D01/2·V1/2·A, where n, C0, D0 and A represent the number of electrons in redox reaction, concentration of electroactive species, diffusion coefficient and the area of electrode, respectively. The microscopic areas of 0.029 (±0.004) cm2 were determined by the integration of the cathode peak during the redox reaction of superficial gold in 1.0 M H2SO4 (26). For n = 1, the diffusion coefficients for differently hybridized electrodes with the measuring error were calculated from the slope of ip versus v1/2 (Fig. 2) and shown in Table 2.
Figure 2.
CV at the scan rate of 50 mV/S (a) and anodic peak currents versus root of the scan rate of CV v1/2 (b) for negative DNA/AET/Au (1), sample DNA/AET/Au (2) and positive DNA/AET/Au (3) in pH 7.1 Tris–HCl buffer containing 0.12 mM [Co(phen)3](ClO4)3.
Figure 4.
The plot of current intensity ratios of CV and SWV for differently hybridized electrodes. P/N, positive hybrid-DNA/AET/Au/negative hybrid-DNA/AET/Au; S/N, sample hybrid-DNA/AET/Au/negative hybrid-DNA/AET/Au.
Table 2. Diffusion coefficients for different hybridized electrodes for positive hybrid-DNA/AET/Au (P), sample hybrid-DNA/AET/Au (S), and negative hybrid-DNA/AET/Au (N) derived from CV.
| N | S | P | |
|---|---|---|---|
| ip/v1/2 (10–7A · S1/2/V1/2) | 29.7 | 34.5 | 47.7 |
| Δ(ip/v1/2) | 0.7 | 0.8 | 1.1 |
| R | 0.9988 | 0.9981 | 0.9996 |
| D0 (10–6cm2/S) | 10.1 | 13.5 | 25.9 |
| ΔD0 (10–6cm2/S) | 0.5 | 0.7 | 1.2 |
ip, A, v1/2, R and D0 represent respectively peak current, the area of electrode, square root of the scan rate, relativity and diffusion coefficient.
From Table 2, the diffusion coefficient (Do) for positive hybrid-DNA/AET/Au is as high as (25.9 ± 1.2) × 10–6 cm2/S, which is apparently higher than for sample hybrid-DNA/AET/Au [(13.5 ± 0.7) × 10–6 cm2/S] and negative hybrid-DNA/AET/Au [(10.1 ± 0.5) × 10–6 cm2/S)]. This result showed a reasonable agreement with the common conception that the Co (phen)33+ had the ability of strong interaction with dsDNA (31). Therefore, the CV could be used to detect methylation status of the p16Ink4a gene.
Detection of methylation status of p16Ink4a gene with square wave voltammogram
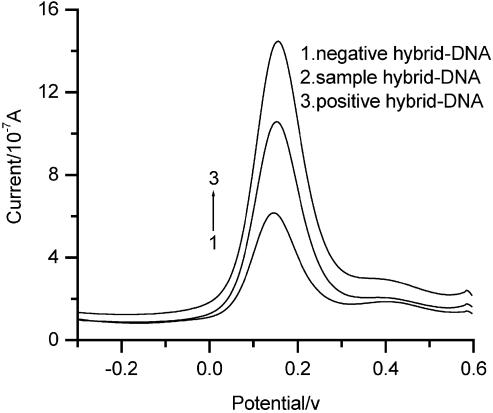
SWV is a sensitive method in electrochemical measurement. The different hybridized electrodes were also characterized with the square wave voltammetry with an amplitude of 25 mV, a pulse frequency of 30 Hz and step potential of 4 mV using 0.12 mM [Co(phen)3](ClO4) 3 as a redox indicator that was added in the Tris–HCl buffer solution. Figure 3 illustrates that the cathode peak current of of Co(phen)33+ species at the positive hybrid-DNA/AET/Au (curve 3) (12.22 ± 0.32) was much larger than at the sample hybrid-DNA/AET/Au (curve 2) (9.02 ± 0.22) and the negative hybrid-DNA/AET/Au (curve 1) (4.76 ± 0.24). The peak current intensity of negative hybrid-DNA/AET/Au (curve 1) was the smallest among three electrodes, which can be taken as a baseline of the hybridization signal. Therefore, the signal of the methylated samples could be compared with both negative and positive ones. We have calculated the positive control/negative control and sample/negative control and averaged from three parallel experiments. The ratios of P/N and S/N were 2.567 and 1.895, respectively (Fig. 4). The results show that SWV is suited to detect methylation status of CpG sites. The ratios of peak current (P/N and S/N) for SWV were larger than those for CV. It means that SWV and CV are sensitive in detecting methylation status of CpG sites. Moreover, the technology will have potential in the quantitative measurement for methylation samples.
Figure 3.
SWV for positive hybrid-DNA/AET/Au (1), sample hybrid-DNA/AET/Au (2) and negative hybrid-DNA/AET/Au (3) with an amplitude of 25 mV, a pulse frequency of 30 Hz and step potential of 4 mV, in pH 7.1 Tris–HCl buffer containing 0.12 mM [Co(phen)3](ClO4)3.
Methylation-specific PCR for methylation detection of p16 gene
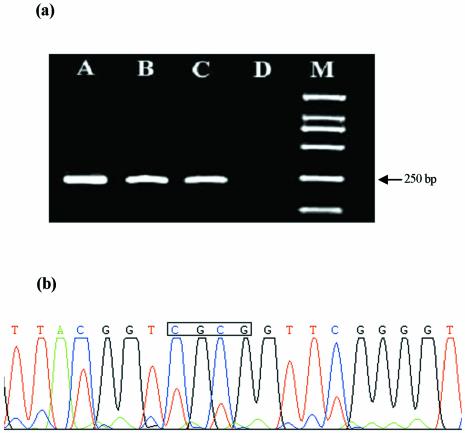
MSP, which can rapidly assess the methylation status of virtually any group of CpG sites within a CpG island, independent of the use of methylation-sensitive restriction enzymes. This assay entails initial modification of DNA by sodium bisulfite, converting all unmethylated, but not methylated, cytosines to uracil, and subsequent amplification with primers specific for methylated versus unmethylated DNA (8). Primer pairs were designed to discriminate between methylated and unmethylated alleles following bisulfite treatment. To accomplish this, primer sequences were chosen for regions containing frequent cytosines, and CpG pairs near the 3′-end of the primers (to provide maximal discrimination in the PCR between methylated and unmethylated DNA) (8). After bisulfite treatment, the amplification products were both detected with unmethylated primers (p16-U) in the genomic DNA from whole blood cells of healthy human and gastric tumor tissue. However, the amplification product was only obtained with methylated primers (p16-M) in the genomic DNA from the gastric tumor tissue (Fig. 5a). The results indicate that methylation was only detected in the gastric tumor tissue for p16Ink4a gene, which further confirmed the conclusion of CV and SWV. The results of MSP were validated by bisulfite DNA sequencing. Figure 5b illustrates that methylation was detected in the recognition site of BstUI (CGCG) for the gastric tumor tissue.
Figure 5.
(a) MSP of p16 gene. After treatment with bisulfate, amplification of DNA from gastric tumor tissue (A) and whole blood cells of healthy human (B) using unmethylated primer; amplification of DNA from gastric tumor tissue (C) and whole blood cells of healthy human (D) using methylated primer. Marker (M): from top to bottom, the bands are 2000, 1000, 750, 500, 250 and 100 bp, respectively. (b) Sequencing result of the PCR product with an ABI377A. The box indicates that the CpG sites in BstUI recognition sequence are methylated.
CONCLUSION
In this paper, we have described an electrochemical method to detect DNA methylation, using label-free DNA fragments amplified by linker-PCR. The results presented here confirm that the electrochemical methods have been successfully used to detect methylated CpG sites within the 5′-CpG islands of the p16Ink4a gene. A great advantage of the electrochemical monitoring of the DNA hybridization at the transducer surface has proved to be: (i) faster measurements, ≤10 s; (ii) eliminating the need for gel, radioisotopes and blotting methods; (iii) coupled with PCR and high sensitivity; (iv) label-free target; (v) low-cost point-of-care and reliable detection results; and (vi) potential in quantitative measurments. The possibility of detection methylation opens realistic prospects for a number of important genetic analyses.
Acknowledgments
ACKNOWLEDGEMENTS
The National Key Fundamental Research Foundation, the National Natural Science Foundation of China, and the National High-Tech Research Programme supported this work.
REFERENCES
- 1.Baylin S.B., Herman,J.G., Graff,J.R., Vertino,P.M. and Issa,J.P. (1998) Alteration in DNA methylation: a fundamental aspect of neoplasia. Adv. Cancer Res., 72, 141–196. [PubMed] [Google Scholar]
- 2.Belinsky S., Nikula,K.J., Palmisano,W.A., Michels,R., Saccomamno,G., Gabrielson,E., Baylin,S.B. and Herman,J.G. (1998) Aberrant methylation of p16INK4a is an early event in lung cancer and a potential biomarker for early diagnosis. Proc. Natl Acad. Sci. USA, 95, 11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valenzuela M.T., Galisteo,R., Zuluaga,A., Villalobos,M., Nunez,M.I., Oliver,F.J. and Ruiz,J.M. (2002) Assessing the use of p16INK4a promoter gene methylation in serum for detection of bladder cancer. Eur. Urol., 42, 622–630. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M., Montserrat,S.C., Rosell,R., Sidransky,D., Baylin,S. and James,G.H. (1999) Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res., 59, 67–70. [PubMed] [Google Scholar]
- 5.Bickle T.A. and Kruger,D.H. (1993) Biology of DNA restriction. Microbiol. Rev., 57, 434–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frommer M., McDonald,L.E., Millar,D.S., Collis,C.M., Watt,F., Grigg,G.W., Molloy,P.L. and Paul,C.L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA, 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kane M.F., Loda,M., Gaida,G.M., Lipman,J., Misbra,R., Goldman,H., Jessup,J.M. and Kolodner,R. (1997) Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human cell lines. Cancer Res., 57, 808–811. [PubMed] [Google Scholar]
- 8.Herman J.G., Graff,J.R., Myöhänen,S., Nelkin,B.D. and Baylin,S.B. (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA, 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuppuswamy M.N., Hoffmann,J.W., Kasper,C.K., Spitzer,S.G., Groce,S.L. and Bajaj,S.P. (1991) Single nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis genes. Proc. Natl Acad. Sci. USA, 88, 1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adorján P., Distler,J., Lipscher,E., Model,F., Müller,J., Pelet,C., Braun,A., Florl,A.R., Gütig,D., Grabs,G., Howe,A., Kursar,M., Lesche,R., Leu,E., Lewin,A., Maier,S., Müller,V., Otto,T., Scholz,C., Schulz,W.A., Seifert,H.H., Schwope,I., Ziebarth,H., Berlin,K., Pipenbrock,C. and Olek,A. (2002) Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res., 30, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou P., Ji,M.J., Liu,Z.C., Shen,J.Y., Cheng,L., He,N.Y. and Lu,Z.H. (2003) A microarray to analyze methylation patterns of p16Ink4agene 5′-CpG islands. Clin. Biochem., 36, 197–202. [DOI] [PubMed] [Google Scholar]
- 12.Wang J. (2000) From DNA biosensors to gene chips. Nucleic Acids Res., 28, 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J. (1999) Electroanalysis and biosensors. Anal. Chem., 71, 328R–332R. [DOI] [PubMed] [Google Scholar]
- 14.Palecek E. and Fojta,M. (2001) Detecting DNA hybridisation and damage. Anal. Chem., 73, 74A–83A. [DOI] [PubMed] [Google Scholar]
- 15.Kerman K., Ozkan,D., Kara,P., Meric,B., Gooding,J.J. and Ozsoz,M. (2002) Voltammetric determination of DNA hybridisation using methylene blue and self-assembled alkanethiol monolayer on gold electrodes. Anal. Chim. Acta, 462, 39–47. [Google Scholar]
- 16.Napler M.E. and Thorp,H.H. (1997) Modification of electrodes with dicarboxylate self-assembled monolayers for attachment and detection of nucleic acids. Langmuir, 13, 6342–6344. [Google Scholar]
- 17.Marrazza G., Chiti,G., Mascini,M. and Anichin,M. (2000) Detection of human apolipoprotein E genotype by DNA electrochemical biosensor coupled with PCR. Clin. Chem., 46, 31–37. [PubMed] [Google Scholar]
- 18.Millan K.M., Saraullo,A. and Mikkelsen,R.S. (1997) Voltammeteric DNA biosensor for cystic fibrosis based on a modified carbon paste electrode. Anal. Chem., 66, 2943–2948. [DOI] [PubMed] [Google Scholar]
- 19.Singhal P. and Kuhr,W.G. (1997) Ultrasensitive voltammetric detection of underivatized olignucleotides and DNA. Anal. Chem., 69, 4828–4832. [DOI] [PubMed] [Google Scholar]
- 20.Caruana D.J. and Heller,A. (1999) Enzyme-amplified amperometric detection of hybridisation and of a single base pair mutation in 18-base oligonucleotide on a 7-µm diameter microelectrode. J. Am. Chem. Soc., 121, 769–774. [Google Scholar]
- 21.Wang J., Xu,D.K., Kawde,A. and Polsky,R. (2001) Metal nanoparticle-based electrochemical stripping potentiometric detection of DNA hybridization. Anal. Chem., 73, 5576–5581. [DOI] [PubMed] [Google Scholar]
- 22.Cai H., Xu,C., He,P.G. and Fang,Y.Z. (2001) Colloid Au-enhanced DNA immobilization for the electrochemical detection of sequence-specific DNA. J. Electroanal. Chem., 510, 78–85. [Google Scholar]
- 23.Garcia M.B. and Garcia,A.C. (1995) Adsorptive stripping voltammetric behavior of colloidal gold and immunogold on carbon paste electrode. Bioelectrochem. Bioenerget., 38, 389–395. [Google Scholar]
- 24.Wang J., Polsky,R. and Xu,D.K. (2001) Silver-enhanced colloidal electrochemical striping detection of DNA hybridisation. Langmuir, 17, 5739–5741. [Google Scholar]
- 25.Bard A.J. and Faulkner,L.R. (2001) Electrochemical Methods, 2nd Edn. John Wiley & Sons, New York, NY, pp. 32–76. [Google Scholar]
- 26.Ge C.W., Liao,J.H., Yu,W. and Gu,N. (2003) Electric potential control of DNA immobilization on gold electrode. Biosens. Bioelectron., 18, 53–58. [DOI] [PubMed] [Google Scholar]
- 27.Yan P.S., Efferth,T., Chen,H.L., Lin,J., Rodel,F. Fuzesi,L. and Huang,H.M (2002) Use of CpG island microarrays to identify colorectal tumors with a high degree of concurrent methylation. Methods, 27, 162–169. [DOI] [PubMed] [Google Scholar]
- 28.Clark S.J., Harrison,J., Paulb,C.L and Frommer,M. (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res., 22, 2290–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun X.Y., He,P.G., Liu,S.H. and Fang,Y.Z. (1998) Immobilization of single-stranded deoxyribonucleic acid on gold electrode with self-assemble aminoethanthiol monolayer for DNA electrochemical sensor application. Talanta, 47, 487–495. [DOI] [PubMed] [Google Scholar]
- 30.Wojciechowski M., Sundseth,R., Moreno,M. and Henkens,R. (1999) Multichannel electrochemical detection system for quantitative monitoring of PCR amplification. Clin. Chem., 45, 1690–1693. [PubMed] [Google Scholar]
- 31.Carter M.T., Rodriguez,M. and Bard,A.J. (1989) Voltammetric studies of the interaction of metal chelates with DNA. 2. Tris-chelated complexes of cobalt (III) and iron (II) 1,10-phenanthroline and 2,2′-bipyridine. J. Am. Chem. Soc., 111, 8901–8911. [Google Scholar]