Abstract
A statistical model is proposed for the analysis of errors in microarray experiments and is employed in the analysis and development of a combined normalisation regime. Through analysis of the model and two-dye microarray data sets, this study found the following. The systematic error introduced by microarray experiments mainly involves spot intensity-dependent, feature-specific and spot position-dependent contributions. It is difficult to remove all these errors effectively without a suitable combined normalisation operation. Adaptive normalisation using a suitable regression technique is more effective in removing spot intensity-related dye bias than self-normalisation, while regional normalisation (block normalisation) is an effective way to correct spot position-dependent errors. However, dye-flip replicates are necessary to remove feature-specific errors, and also allow the analyst to identify the experimentally introduced dye bias contained in non-self-self data sets. In this case, the bias present in the data sets may include both experimentally introduced dye bias and the biological difference between two samples. Self-normalisation is capable of removing dye bias without identifying the nature of that bias. The performance of adaptive normalisation, on the other hand, depends on its ability to correctly identify the dye bias. If adaptive normalisation is combined with an effective dye bias identification method then there is no systematic difference between the outcomes of the two methods.
INTRODUCTION
Microarrays are a technology for measuring the expression levels of genes simultaneously, allowing the complete transcriptome of a cell, tissue or organ to be defined. However, microarray experiments generate data that contain errors, which arise from various sources, such that the noise may significantly distort the real signal. Experimental error can be classified into two categories: systematic error and random error. The former reflects the accuracy of measurements, while the latter reflects the precision of measurements. Obtaining data with satisfactory precision and accuracy has been one of the biggest challenges in the application of microarray technology.
In a typical spotted slide microarray experiment, two mRNA samples are compared by reverse transcribing them into cDNA, labelling using red and green fluorescent dyes, respectively, and then hybridising these labelled targets simultaneously to denatured PCR product or cDNA probes spotted on a glass slide. The relative level of gene expression in the two samples is then measured by determining the ratio of fluorescence intensity of the two dyes. This technique is very effective in overcoming the weak point of microarray experiments, which is that their reproducibility in measurement of absolute (as opposed to relative) expression level is poor. However, associating two samples with two different fluorescent dyes introduces dye bias into the measurements. Dye bias is a systematic error that should be removed before any further analysis of microarray data is performed. To remove systematic errors from microarray data one can employ a normalisation method. A number of normalisation approaches have been introduced for microarray data analysis, which include the housekeeping gene approach (1), total RNA approach (2), global normalisation (3), ANOVA (4), the centralisation method (5), self-normalisation (6) and adaptive normalisation involving regression techniques such as LOESS (3).
The housekeeping gene approach builds on the assumption that there are sets of genes in any genome that are constitutively expressed at a relatively constant level under any set of conditions. The total RNA approach is based on the assumption that each cell carries the same amount of total RNA at different times. Unfortunately, there is substantial evidence that these assumptions are incorrect in many cases (7,8). Global normalisation is based on two assumptions. Firstly, that the centre (i.e. the mean or median) of the distribution of gene expression ratios on a log scale is zero. Secondly, that the systematic error makes the central line move vertically, but otherwise leaves the distribution invariant. Therefore, a global normalisation shifts the centre of the log ratio distribution to zero. However, systematic error in microarray data is often not constant (3,9). In this study, we will show that the log ratio distribution is not always centred to zero. Therefore, the performance of global normalisation is not satisfactory. Kerr et al. (4) proposed an ANOVA approach for microarray data analysis, but this is also global in nature. The dye bias contained in microarray data usually not only varies from spot to spot over a slide, but also from replicate to replicate for a given spot. Therefore, the interaction between slide and spot should be considered, but this cannot be achieved using the ANOVA approach. The centralisation method of Zien et al. (5) was shown to provide improved results; however, they discarded the data points whose intensity was below 10 or above 1000 in their study. This operation excluded 487 out of 1189 genes in their data sets. In addition, the number of replicates used was very large and so their approach may be difficult to apply in practice.
The self-normalisation method (6) assumes that experimentally introduced error is multiplicative and that, for corresponding spots in replicated measurements, it is consistent. Based on this, the error on a log scale is additive and a subtract operation applied to the data sets from two replicates will remove this systematic error. However, this approach requires the association of a dye-flip technique, as otherwise the biological difference between two samples being measured will also be removed by the operation. The dye-flip (also known as dye swap or reverse labelling) technique generates paired slides where, on the first slide, one mRNA sample is labelled by Cy5 and the other mRNA sample is labelled by Cy3, while, on the second slide, the labels for the two samples are exchanged. Based on self-normalisation, the normalised result (logged ratio of expression) for a measured spot is half the difference between logged ratios measured from a pair of dye-flipped replicates for this spot (6). Therefore, self-normalisation has the property that it corrects feature-specific (i.e. probe- or spot-specific) differences so that, for a feature measuring the expression level of a given gene, the normalised result only measures the relative abundance of the gene itself and is not influenced by the measurement of any other features. Compared to global normalisation, in which the normalised result for a given gene spot will depend on the measurements of the whole set of genes, the self-normalisation approach goes to the other extreme and ignores all other measurements. Self-normalisation typically performs much better than global normalisation when dye-flip replicates are available (6).
Adaptive normalisation is an approach that falls somewhere between global normalisation and self-normalisation. The approach employs the assumption that the bias introduced by the experiment is dependent on a number of factors (spot intensity, print tips, spot position, etc.) and employs regression techniques to obtain a fit of the specific relationship, and then makes the correction. Because it takes systematic error as neither a constant nor spot-specific, the method has the advantages of both the global normalisation and the self-normalisation approaches, but without their disadvantages. Adaptive normalisation may perform differently for the different regression techniques employed. Generally, the regression can be either global or local. For global regression, one can employ either a linear or a non-linear function for the regression. For local regression, the LOESS (LOWESS) regime (10) is currently the most popular (3), although some basic knowledge of the method is required for the analyst to choose appropriate values for the parameters involved in the method.
Both self-normalisation and adaptive normalisation have been shown to provide advantages over global normalisation. Furthermore, adaptive normalisation may have advantages over self-normalisation. Firstly, self-normalisation can only be applied to dye-flip paired microarray slides. It cannot be applied to single slides, in contrast to the adaptive approach. Secondly, for spots whose systematic errors are not consistent on a slide pair, adaptive normalisation may produce better results because the correction of any gene spot is dependent on the bias of all gene spots that have the same value of spot intensity (i.e. the same value of the dependent variable used in the regression). However, self-normalisation is much easier to apply since it can be used without identifying the nature of the experiment-introduced bias or the genuine difference between two samples. In contrast, one must know, or estimate, at least one of these two components in order to apply an adaptive normalisation. Furthermore, for non-self-self slides, both of these components are unknown.
Given the above complexity, researchers often ask which currently used normalisation method performs better. Does the dye-flip technique play an indispensable role in obtaining better data quality? How does one improve the data quality through more effective normalisation operations? This study tries to answer these questions by a comparison of data quality before and after normalisation. This has permitted an investigation of which type of replication (dye-flip or non-dye-flip) will lead to better data quality and what normalisation techniques can generate data with higher accuracy. In order to compare the data quality obtained by different experimental designs and different normalisation regimes under a given number of replicates, we propose a method for assessing the accuracy and precision of normalised data. The former depends on our ability to remove the systematic error contained in microarray data. For self-self hybridisations, the data points should centre to the zero line on an M–A plot (6) and this can be used for the assessment of the accuracy of normalised data. In general, for non-self-self data, it is not known to what line the data spots should centre. However, self-normalisation has the intrinsic capacity to remove dye bias without detection of that bias. Hence, accuracy can be assessed by the systematic difference between normalised data from self-normalisation and from other normalisation techniques. The precision of normalised data can be assessed by the data consistency, which is represented by the difference of normalised data from replicate experiments, since this difference is only related to experimental noise and the performance of the normalisation regime employed. Therefore, comparison of this difference between a range of normalisation techniques applied to the same data sets can answer the question of which normalisation approach works better. Similarly, comparison of data consistency corresponding to different experimental designs shows which experimental design generates data of higher quality.
In this study, a statistical model for microarray experiments is proposed and used to investigate how systematic error is removed by different normalisation operations under certain simplifying assumptions. Deductions derived from the model are tested on real data from a set of microarray experiments. From analysis of the model and experiments, we make a number of useful conclusions. Firstly, we clarify the role of dye-flip replicates for improving microarray data quality. Secondly, we introduce a high performance normalisation approach that involves up to three stages in the regime. Details of the normalisation protocols and derivations of the results are contained in the Appendix.
ANALYTICAL METHODS
A statistical model is employed to investigate the main sources of systematic error and the effects of different normalisation approaches on these terms. In the following discussion, we consider log transformed data. Unless otherwise specified, the ratio (M) and spot intensity (A) are actually the ratio and mean spot intensity on a log scale, i.e. M = logR/G and
where R and G are measurements from the red and green channels, respectively, and we use base 2 logarithms.
A statistical model of experimental error
Let n represent the number of replicates of an experiment, g represent the number of the features (probes) on the slide, mj (j = 1, 2, …, g) represent the true ratio of expression levels for the gene measured by feature j, and Mjk (j = 1, 2, …, g; k = 1, 2, …, n) represent the measured ratio of expression levels for feature j on replicate k. The measurement Mjk will be modelled as:
Mjk = mj + c + ck + e(Fj) + ek(Ajk) + ek(Pj) + εjk 1
with
where c represents the expected global measurement bias between two channels, ck represents the variation of global measurement bias shown on replicate k, e(Fj) represents feature-specific bias for feature j, ek(Ajk) represents spot intensity-dependent bias for feature j on replicate k, ek(Pj) represents spot location-related bias for feature j on replicate k and εjk represents zero mean random error introduced to feature j on replicate k.
The global measurement bias c is a constant for the given data sets of n replicates and ck is a constant which represents the difference between the global bias of the kth replicate and mean global bias of n replicates. The function e(Fj) depends on the feature only, but the functions of spot intensity ek(Ajk) and the spot position ek(Pj) depend on both the feature and replicate indices. The global measurement bias has been included in c and ck and we therefore take e(Fj), ek(Ajk) and ek(Pj) to be centred to zero. We make simplifying assumptions that the three systematic error terms are independent. This is most likely sensible because their dependent factors are expected to be unrelated or only very weakly related. The spot intensity is mainly dependent on the expression of genes. The spot position-specific term is mainly related to the heterogeneity of the experiment over a slide surface and the feature-specific term mainly reflects the different performance of different print tips and related factors. We also assume that the function of intensity and function of spot position are continuous functions of their independent variables, but that the function of feature changes abruptly from feature to feature.
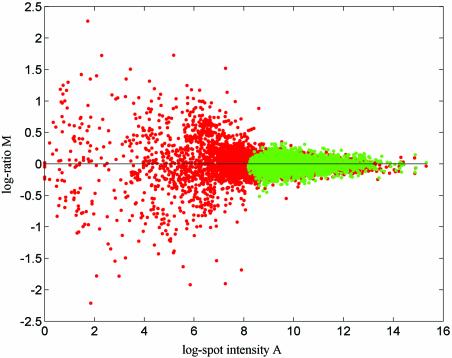
The proposed model contains only multiplicative items and is therefore additive on a log scale. Such a multiplicative model appears to work very well on microarray data without background correction. The use of background correction introduces an additive error item into the data that can be significant for low expression levels and may make this model unsuitable. Data that contain a mixture of additive and multiplicative terms can be modelled using, for example, the model introduced by Rocke and Durbin (11). For the microarray data studied here we believe it to be most likely that naïve background subtraction has more negative effects than positive effects on microarray data quality and it is therefore not used here. This point is illustrated by examining some of our normalised self-self data (data measured from self-self hybridisation chips). For self-self data, the genuine log ratio of each data point should be zero and normalised self-self data reflects the data quality of the corresponding microarray experiment. A comparison of the outcomes from applying the normalisation to background-corrected self-self data and to the same data without background correction will show the impact of background correction clearly. We have done a few of these kinds of comparisons based on self-self data from different microarray experiments and all of them show that background correction decreases the data quality. The plots presented in Figure 1 show normalised yeast data from self-self hybridisation microarray slides (for protocols, see 12,13). After normalisation, the background-corrected data is much noisier than the data without background correction.
Figure 1.
Plots of the estimated log ratio (base 2) after normalisation of four self-self replicates from a yeast microarray experiment. Red spots show results from data sets with background correction and green spots show results from data sets without background correction.
Normalisation approaches and procedures
Our model contains three systematic error items which are factor-specific [ek(Ajk), ek(Pj) and e(Fj)]. We employed an approach that involves a number of normalisation stages in order to remove these systematic effects wherever possible. We outline our normalisation approach below and provide further details in the Appendix.
If the data comes from an experiment with dye-flip replicates, then normalisation can be conducted in two or three steps. The first step is to correct the intensity-dependent dye bias. This task can be carried out by adaptive normalisation or self-normalisation. Self-normalisation simply involves subtracting the results of each pair of dye-flipped replicates. Adaptive normalisation involves the fitting of a regression function to the M–A plot for each slide and correcting accordingly. In our experiments, we used a generalisation of a standard polynomial fit that includes fractional powers (regression function M = β1 + ∑4i = 1βi + 1A1/(i + 1) + βi + 5Ai + ε). In some cases, it will be advantageous to identify the dye bias using a pooled set of dye-swap replicates. The parameterised regression function from the pooled data set is then applied to each data set in the pool so as to correct its dye bias. We call this technique ‘improved adaptive normalisation’ (iAN) (see Appendix for details).
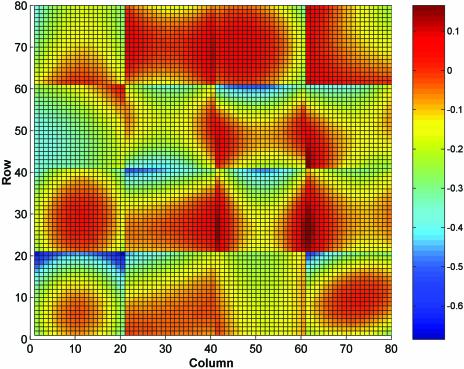
The second step is to remove the spot location-specific error using regional normalisation. A two-dimensional regression technique can be used to identify and correct the error in this case. The regression can be applied to each block of a replicate separately or can be applied to an entire replicate. Block-based regression is used in this study and is recommended for the situation where different blocks are printed by different print tips or are printed at different times. Figure 2 provides an example of the outcome of applying a two-dimensional quadratic polynomial regression [regression function f(x,y) = α1 + α2x + α3y + α4x2 + α5xy + α6y2 + ε, where x, y are spot coordinates in a block on a microarray slide] to each block of a yeast microarray data set (12,13). The plot shows that the difference in print tips and printing time may cause an abrupt change in spot position-dependent error (see border area of adjacent blocks). However, if a border of two adjacent blocks is in an area where the position-dependent error is dominated by the heterogeneity of the experiment over a slide surface, then the impact of difference in print tips and printing time may not be seen clearly. If a two-dimensional fit is applied to an entire replicate, then the fitted spot position-dependent error will change smoothly across any borders of two adjacent blocks, and this is usually not the case.
Figure 2.
We show the spot position-dependent error contribution from one of the yeast microarray chips used in this study. The data set has 6400 (80 × 80) data points grouped as 16 (4 × 4) blocks. Graduated colours are used to show the spatially averaged log ratio (base 2) obtained by fitting a 2D quadratic regression function to the log ratios from each block. Calibration of the colour scheme is shown by the bar on the right in log ratio units.
If we use self-normalisation as the first step, then the normalisation process only requires the two steps described above. However, if we use adaptive normalisation then a third normalisation step is required in order to remove any remaining feature-specific bias. This can be done using feature normalisation, which is essentially identical to self-normalisation. We simply subtract the values (after adaptive and regional normalisation) of each dye-flipped pair. The only difference between this and self-normalisation is that we carry out feature normalisation after the other normalisation methods have been applied.
If dye-flip replicates are not available, then self-normalisation and feature normalisation are not possible. In this case, we can only carry out adaptive and regional normalisation. If the intensity-dependent bias contains genuine biological signal, then there is no effective way to identify these two items separately and it is difficult to achieve satisfactory normalisation results, as we will demonstrate in the forthcoming sections.
To summarise, we will consider several normalisation methods involving combinations of the following normalisation steps: AN, adaptive normalisation; iAN, improved adaptive normalisation; SN, self-normalisation; RN, regional normalisation; FN, feature normalisation.
Combinations of these abbreviations will be used to denote different normalisation regimes. For data sets with dye- swap replicates we will use SN+RN, AN+RN, iAN+RN, AN+RN+FN and iAN+RN+FN, as outlined above. For data sets without dye swap replicates we will use AN+RN.
ANALYTICAL DEDUCTIONS
The analytical deductions given in this section are based on the statistical model previously described (1). The analysis is applied to four different situations: (i) data from self-self replicates (where they are treated as replicates without dye-flip); (ii) data from self-self replicates (where they are treated as replicates with dye-flip); (iii) data from non-self-self replicates without dye-flip; (iv) data from non-self-self replicates with dye-flip. We should emphasise that treating self-self replicates as dye-flipped replicates or as non-dye-flipped replicates is only a technique of data analysis and not a real experimental technique, because in a self-self microarray experiment the dye-flip is meaningless.
In the following analysis, we make the simplifying assumption that the dependent terms can be identified by regression without error. This is a fairly gross assumption, but we believe that the main differences between normalisation approaches can be more easily illustrated in this way. Identifying error introduced by the particular regression technique will be more dependent on details of the methods used. We are assuming that this source of error is less significant than the terms identified below. In the remainder of this section, only the results and the conclusions based on those results are provided, while details of the analyses are contained in the Appendix.
Self-self replicates without dye-flip
For this situation, the model is:
Mjk = c + ck + e(Fj) + ek(Ajk) + ek(Pj) + εjk. 2
The genuine log ratio for each spot on each replicate is zero, so that we have set mj = 0 in equation 1. The normalisation process for these data involves two steps: adaptive correction of dye bias and regional normalisation (AN+RN). Let m¯j represent the estimated log ratio for feature j after normalisation, which is given by equation 3 below
Self-self replicates with dye-flip
For this situation, we can take the n replicated measurements as s pairs (n = 2s). The statistical model can then be shown to be:
Mjk = c + ck + e(Fj) + ek(Ajk) + ek(Pj) + εjk
Mjk + s = c + ck + s + e(Fj) + ek + s(Ajk + s) + ek + s(Pj) + εjk + s
for k = 1, 2, …, s. 4
Normalisation of this data set can be carried out by two different approaches. The first approach includes the adaptive correction of dye bias, regional normalisation and the correction of feature-specific error (AN+RN+FN). The second approach involves self-normalisation and regional normalisation (SN+RN). After AN+RN+FN the estimated log ratio m¯j can be shown to be:
where the random error term ɛ̃j has equal variance to ɛmacr; and has an identical distribution if the error is symmetrically distributed. After SN+RN the estimated log ratio m¯j can be shown to be:
Non-self-self replicates without dye-flip
For this situation the genuine log ratio mj may contain a spot intensity-dependent bias that will be removed by intensity-dependent regression. Therefore, we split mj into two parts: a spot intensity-independent term, mj*, and a spot intensity-dependent component, m(Aj), where Aj can be thought of as the expectation value of the spot intensity for gene j over many replicates. We substitute mj = mj* + m(Aj) into equation 1 and get the appropriate statistical model below:
Mjk = mj* + m(Aj) + c + ck + e(Fj) + ek(Ajk) + ek(Pj) + εjk. 7
Self-normalisation is unusable since no dye-flipped measurement is available. Furthermore, when the dye bias is corrected by employing regression to the M–A plot, the fit will contain m(Aj) and the dye bias cannot be singled out from the fit. The most common current practice is to assume that m(Aj) is negligible, take the whole fit as dye bias and remove it from the data. In this case, we apply adaptive and regional normalisation (AN+RN), and m¯j can be shown to be:
m¯j = mj – m(Aj) + e(Fj) + ɛ̄j. 8
Here, we have made a simplifying approximation that m(Aj) would be completely removed by regression against Ajk. The main point of equation 8 is that the expression ratio estimate will be biased if the true expression ratio includes an intensity-dependent contribution, and this will hold in general.
Non-self-self replicates with dye-flip
In this situation, we can take the n replicated measurements as s pairs (n = 2s) and the statistical model is:
Mjk = mj* + m(Aj) + c + ck + e(Fj) + ek(Ajk) + ek(Pj) + εjk
Mjk + s = mj* – m(Aj) + c + ck + s + e(Fj) + ek + s(Ajk + s) + ek + s(Pj) + εjk + s, 9
where k = 1, 2, …, s. In this case, we consider three normalisation approaches. After AN+RN+FN, the estimated log ratio m̄j can be shown to be:
m¯j = mj – m(Aj) + ɛ̃j, 10
after iAN+RN+FN, m̄j can be shown to be:
m¯j = mj + ɛ̃j, 11
and after SN+RN, m̄j can be shown to be:
Conclusions based on analytical deductions
From the analytical deductions given by equations 3, 5, 6, 8 and 10–12, we make the following conclusions. (i) Equations 3 and 8 show that the normalised results from non-dye-flip replicates contain the feature-specific error term, while normalised results from dye-flip replicates (equations 5, 6, 11 and 12) do not. Therefore, dye-flip replication is an effective way of removing feature-specific bias introduced by the experiment and we can make use of it to achieve better data quality. (ii) Compared to the normalised results from approaches using adaptive normalisation (equations 5 and 11), the results from approaches using self-normalization (equations 6 and 12) contain a term which arises from differences in spot intensity and dye response between replicates. Hence adaptive normalisation is usually more effective in removing dye bias than self-normalisation. (iii) In the right-hand side of equations 8 and 10 there is a –m(Aj) term which reflects the intensity-dependent biological bias. This means that the dye bias will be overcorrected by a normalisation approach that includes adaptive normalisation if there is an intensity-dependent biological bias between two samples. In this case, the traditional adaptive normalisation technique is not suitable and iAN is required. (iv) From the normalised results (equations 10–12), we can see clearly that there will be an intensity-dependent systematic difference, m(Aj), between the results from the AN methods and the results from the SN method. However, there is no such bias between the normalised results from the iAN method and the results from the SN method. This provides a way to identify the intensity-dependent biological difference between two mRNA samples.
EXPERIMENTAL METHODS
To test conclusions 1 and 2 (above), we will compare the outcome of normalising self-self data sets using the different methods introduced. It is worth remembering that we can take self-self replicates as replicates with or without dye-flip. For testing conclusions 3 and 4, we will use data from reference treatment replicates with and without dye-flip. In order to determine that the biological bias can be identified and retained during normalisation, we compare the outcomes from traditional adaptive normalisation methods and from self-normalisation. Our model predicts that the systematic difference between the outcomes of these two approaches is just equal to the spot intensity-dependent biological bias between the two samples. The reason is that self-normalisation removes the dye bias but not the biological bias, while traditional adaptive normalisation removes both the dye bias and the biological bias. The difference between them is the biological bias between two measured samples. Finally, we compare the outcomes from the improved adaptive method of dye bias correction with self-normalisation so as to demonstrate which method produces results with higher accuracy.
Data and analysis
Data from a microarray study on yeast was employed for our analysis (12,13). The microarray study included four microarray slides and on each slide there were two sections, which were actually two replicates. In each replicate, there are 16 blocks with 400 spots per block. Among 6400 spots in a replicate, 6277 are spotted with gene products and the remaining 123 are empty spots. Of the four slides, two are self-self hybridisation slides, which provide four replicated measurements. The remaining two are hybridisation slides of reference versus heat shock samples. Each of the two slides provides two replicates without dye-flip and, at the same time, the two slides constitute two dye-flipped replicates. Though the number of slides is small, they contain data from self-self slides, reference heat shock slides, replicates in dye-flip form and replicates in non-dye-flip form. This means that all experimental combinations required for our analysis are available from this set of slides.
So far, we have assumed that a slide contains only one set of measurements. However, the microarray experiments used for this test have two duplicate sets of measurements on each slide and, effectively, we treat these sets as separate slides. Therefore, we have eight sets of measurements from these slides. We denote the eight data sets as SS1A, SS1B, SS2A, SS2B, RT1A, RT1B, RT2A and RT2B, where SS stands for self-self slides, RT stands for reference treatment slides, 1 stands for the first of two replicated slides and 2 stands for the second of two replicated slides, A stands for the first section on one slide and B stands for the second section on one slide.
Analysis of SS1 and SS2
We first take the two slides as non-dye-flip replicates and employ AN+RN normalisation. We then take the two slides as dye-flip replicates and use the AN+RN+FN and SN+RN normalisation approaches. Because the genuine log ratio is zero (mj = 0) for self-self microarray data, the normalised results provide a measure of the error contained in the data. If a normalisation method can remove all systematic errors contained in the data, then the normalised data contains only the random error inherent in the corresponding microarray experiment. However, if a normalisation approach cannot remove all the systematic errors, then this will result in increased errors, and if the systematic errors are not constant, then the normalised data will show greater variation. Therefore, a comparison of normalisation approaches can be conducted by comparing the variation (standard deviation and range) of self-self microarray data after normalisation. We also measure the correlation between replicated data sets after different normalisation approaches, since this also provides an indication of any systematic variation. This correlation should become weaker if a better normalisation approach is employed.
Analysis of RT1 and RT2
Four different normalisation approaches are employed to normalise the four data sets. They are iAN+RN, SN+RN, AN+RN+FN and iAN+RN+FN. The outcomes are compared to evaluate the performance of the approaches. It is necessary to emphasise that we do not know the genuine log ratio (mj) and, therefore, we cannot assess the performance of a normalisation approach by observing the variation of a normalised data set. For this reason, we used a method different from that used in the analysis of SS1 and SS2. In this experiment, we have four replicated measurements and each replicate contains both random noise and systematic errors. The random error is related to uncontrollable factors in the microarray experiment and systematic error may be removed by normalisation. If the systematic error can be removed completely by a normalisation approach, then the consistency (difference between two normalised results from two replicates) reflects the random noise of the experiments. However, if there is still a part of the systematic error in the normalised data, then the consistency of the data will deteriorate. Therefore, the performance of the normalisation approaches can be assessed by comparison of the consistency of replicated data after being normalised by different normalisation approaches. In practice, the consistency of replicated data should be equivalent to the random error contained in the data set. We measure consistency using the standard deviation and range of the difference between normalised replicates. We also visually compare normalised results from different normalisation approaches in order to identify any systematic differences between normalisation approaches.
Additional data
The yeast data set contains all of the different types of experiment required to test our theoretical predictions; however, this data set is small and analysis of some additional data is therefore desirable in order to confirm our results. A set of chips comparing tissue type in carp (mixed versus gill tissue), with dye-flip biological replicates, was obtained from A. Cossins, A. Gracey and J. Fraser of the University of Liverpool. The data consist of eight chips comprising four dye-flip biological replicate pairs, which we refer to as A, B, C and D. Each pair provides results from the same fish, while the four pairs are also biological replicates in the sense that they are comparing the same tissue types. Each slide contains 14 112 spots, of which 13 440 are labelled with gene products. The spots are arranged in 32 blocks, each containing 441 spots. The slides do not contain a large number of replicated spots and therefore we cannot use within-chip consistency as a measure of error for this data set. However, we would expect a successful normalisation scheme to reduce the difference between results derived from different replicate pairs.
EXPERIMENTAL RESULTS
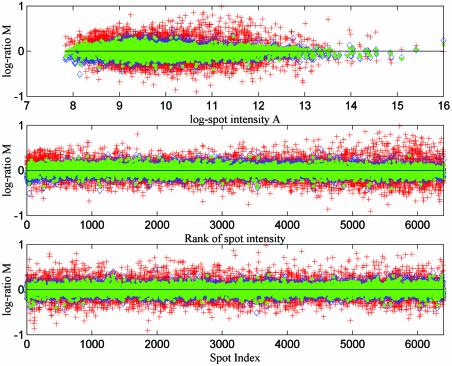
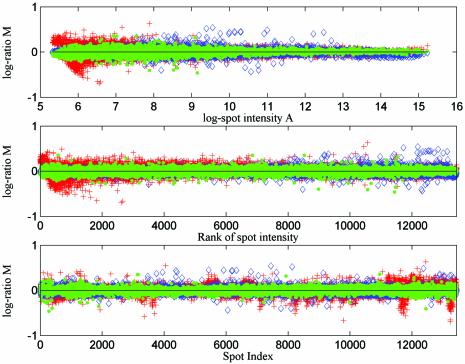
The outcomes of normalisation from self-self slides SS1 and SS2 are plotted in Figure 3. The three subplots in Figure 3 show the same results using different values on the x-axis: the log intensity, the rank of intensity and the spot index. Spots are indexed block by block and row by row, left to right and then top to bottom in each case, e.g. the top left spot of the second block from the left at the top of the slide would be indexed 401. In each subplot, results from three different normalisation methods are plotted (AN+RN, AN+RN+FN and SN+RN). The legend indicates which normalisation approach has been used in each case. The data sets are measurements of self-self data, so that the true log ratio for every spot should be zero and the three plots show the experimental error after data normalisation.
Figure 3.
We plot the estimated log ratios for every spot after applying different normalisation approaches to self-self replicated microarray data sets from yeast experiments (AN+RN, red crosses; SN+RN, blue diamonds; AN+RN+FN, green dots). In the top subplot the estimated log ratios (base 2) are plotted against mean log intensity. In the middle and bottom subplots the estimated log ratios are plotted against spot index and rank, where the rank of spot intensity is obtained by sorting the data set by mean log intensity.
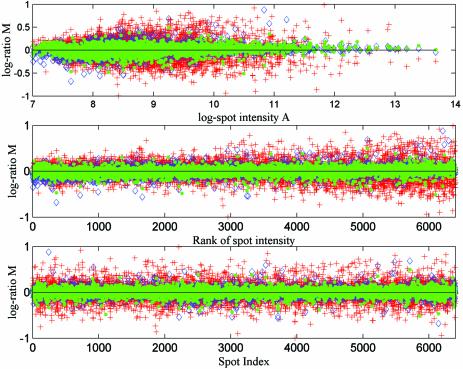
The normalised results of slides RT1 and RT2 are plotted as Figures 4 and 5. Figure 4 shows the error contained in the results from the three different normalisation approaches iAN+RN, SN+RN and iAN+RN+FN. The error here is defined as the difference in log ratio between the replicated spots on each slide after normalisation. Figure 5 shows the mean and the difference between the results (log ratios) from the three normalisation approaches AN+RN+FN, iAN+RN+FN and SN+RN. Figure 6 shows the same normalisation methods as in Figure 4 applied to the carp data set. In this case the error is defined as the difference between the estimated log ratios from different dye-flip pairs.
Figure 4.
We plot the estimated error for every spot after applying different normalisation approaches to reference treatment replicated yeast microarray data sets (iAN+RN, red crosses; SN+RN, blue diamonds; iAN+RN+FN, green dots). The error is defined as the difference in estimated log ratio (base 2) between replicated spots. In the top subplot the errors are plotted against mean log intensity. In the middle and bottom subplots the errors are plotted against spot index and rank, where the rank of spot intensity is obtained by sorting the data set by mean log intensity.
Figure 5.
Comparison of the results (estimated log ratios, base 2) after using three different normalisation approaches on reference treatment replicated yeast microarray data sets (AN+RN+FN, res1; iAN+RN+FN, res2; SN+RN, res3). The mean of each pair of results are plotted as red crosses and the difference between each pair of results are plotted as green dots.
Figure 6.
We plot the estimated error for every spot after applying different normalisation approaches to the carp microarray data sets (iAN+RN, red crosses; SN+RN, blue diamonds; iAN+RN+FN, green dots). The error is defined as the difference in estimated log ratio (base 2) from two distinct dye-flip pairs after normalisation. Other details are as in Figure 4.
Statistics of the normalised yeast data are presented in Tables 1 and 2. Table 1 lists the range and standard deviation of the normalized self-self replicated data using AN+RN, SN+RN and AN+RN+FN. Table 2 lists the range and standard deviation of errors after applying the AN+RN, SN+RN, AN+RN+FN and iAN+RN+FN approaches to reference treatment samples, where the error is defined as the difference in log ratio between the replicated spots on each slide after normalisation. In Table 3 we show results for these four normalisation methods applied to all possible pairings of dye-flip replicates available in the carp data set.
Table 1. Statistics measuring the experimental error of self-self yeast slides after three different normalisation methods.
| AN+RN | SN+RN | AN+RN+FN | |
|---|---|---|---|
| Range | 1.9200 | 1.1219 | 0.7805 |
| Standard deviation | 0.2124 | 0.1046 | 0.0865 |
We show the range and standard deviation of estimated log ratios (base 2) of all spots.
Table 2. Statistics measuring the experimental error of reference heat shock yeast slides after four different normalisation methods.
| AN+RN | SN+RN | AN+RN+FN | iAN+RN+FN | |
|---|---|---|---|---|
| Range | 2.0263 | 1.5939 | 0.9725 | 0.9964 |
| Standard deviation | 0.1970 | 0.0838 | 0.0682 | 0.0758 |
We show the range and standard deviation of the difference between log ratios (base 2) estimated from replicated spots.
Table 3. Statistics measuring the difference between estimated results from distinct dye-flip pairs of carp slides after four different normalisation methods.
| Pair A–Pair B | Pair C–Pair D | Pair A–Pair C | Pair B–Pair D | Pair A–Pair D | Pair B–Pair C | ||
|---|---|---|---|---|---|---|---|
| AN+RN | Range | 1.4917 | 1.2257 | 1.5175 | 1.3812 | 1.3124 | 1.4804 |
| SD | 0.0994 | 0.0891 | 0.0983 | 0.0864 | 0.0972 | 0.0942 | |
| SN+RN | Range | 1.3352 | 0.9798 | 1.4896 | 1.1591 | 1.1345 | 1.1549 |
| SD | 0.0652 | 0.0475 | 0.0783 | 0.0618 | 0.0726 | 0.0549 | |
| AN+RN+FN | Range | 1.0460 | 0.8125 | 0.8994 | 0.9876 | 0.6851 | 1.0098 |
| SD | 0.0524 | 0.0455 | 0.0534 | 0.0547 | 0.0502 | 0.0464 | |
| iAN+RN+FN | Range | 1.0526 | 0.8245 | 1.0571 | 1.0515 | 0.8896 | 1.0982 |
| SD | 0.0549 | 0.0468 | 0.0556 | 0.0482 | 0.0521 | 0.0481 |
We show the range and standard deviation of the difference between log ratios (base 2) estimated from the same spot in different dye-flip pairs.
We also calculated the correlation of the normalised self-self results. For the four self-self results (estimated log ratios) after using the AN+RN approach, we found that the mean value of the correlation coefficients of replicated spots is 0.62. Taking the four replicates as two pairs and conducting feature normalisation, we obtained two sets of results with a mean correlation coefficient of 0.22.
DISCUSSION
There are three important points revealed by Figures 3 and 4. The first is that, through normalisation, we can get more accurate results from dye-flip replicated experiments than from non-dye-flip replicates. This point can also be demonstrated by the summary statistics of the resulting data sets (see Tables 1 and 2). In addition, based on our statistical model of experimental errors and the corresponding analytical deductions, it is easy to understand why this conclusion is reasonable, because the feature-specific bias introduced by experiment cannot be removed if there is no dye-flip replicate available. Furthermore, comparing the correlation of normalised results shows the existence of feature-specific bias. For self-self replicated data sets, our model predicts that there may be a strong correlation among replicates after the AN+RN normalisation approach where the feature-specific error is not removed. In contrast, a much smaller correlation will be shown after applying the AN+RN+FN approach to paired replicates.
The second point revealed by Figures 3 and 4 is that adaptive normalisation can achieve higher precision than self-normalisation. This agrees with the inference based on our model, and the statistics in Tables 1 and 2 also support this point.
The third point revealed by these two figures is that the random errors introduced by microarray experiments may be different from spot to spot, but these differences can reasonably be taken as independent of the spot intensity and spot position (on a log scale). This can be seen clearly from the subplots showing error versus spot index and error versus the rank of spot intensity. At first glance, the error versus log intensity subplot seems to contradict this conclusion. However, in this subplot, the spots are not plotted evenly over the horizontal axis. It is clear that the error plotted in a region will appear to show a larger range of variation if there are more spots. Plotting against spot rank provides a better visualisation of the range and variability of the data.
Figure 5 demonstrates an important point. Compared to the reference sample, the up-regulation and down-regulation of genes in the treatment sample may not be balanced. Furthermore, the direction and degree of this imbalance may relate to the spot intensity. If the AN method is employed in this case, then the intensity-dependent imbalance between up- and down-regulation will be removed. Therefore, the systematic difference between the outcome from the AN+RN+FN and SN+RN methods (shown by the middle subplot) reflects the intensity-dependent biological difference between two samples. This demonstrates the necessity of identifying the experimentally introduced bias and the intensity-dependent biological bias [m(Aj) in our model]. Based on our analysis, if we conduct a fit of M versus A of a replicate, then both these biases (as well as other global biases) will be contained in the fitting. The biological bias should be retained and the other items should be removed, but this fit cannot single out the biological bias. To achieve this, we proposed an improved adaptive normalisation method. The top subplot in Figure 5 shows that the iAN method behaves differently from AN. The bottom subplot provides strong evidence that this method performs well as it shows no systematic difference from the SN+RN method. Therefore, we conclude that the iAN+RN+FN method removes systematic bias effectively, while retaining the improved dye-bias correction displayed by the adaptive method.
Results from the additional carp data set are shown in Table 3 and Figure 6 and are consistent with our results using the yeast data set. The iAN+RN+FN approach provides a greater reduction in variation in log ratio after normalisation when compared to the AN+RN, iAN+RN and SN+RN approaches. We have also confirmed that there is no systematic difference between results from the SN+RN and iAN+RN+FN methods, which shows that the improved adaptive normalisation method is not introducing any bias into the results.
From the above discussion, we can see that the experimental results strongly support all the analytical deductions based on our statistical model of experimental error. However, we have also made a few observations from the experimental results that cannot be obtained through model-based analyses. Firstly, from comparison of Tables 1 and 2, we can see that the data quality of reference heat shock replicates may be as good as the data quality of self-self replicates. In fact, the random error of RT (reference treatment) replicates has a slightly smaller standard deviation than that of SS (self-self) replicates. In contrast, the former has a larger range of variation; this means it has more notable outliers. Secondly, we can see from Figure 5 that heat shock treatment makes a number of genes become differentially expressed. Furthermore, it appears that more highly expressed genes are more highly regulated since they tend to show a systematic increase in log ratio on average as intensity increases. It would be interesting to see whether this is true of other organisms. Thirdly, the feature-specific error is the dominant error item in normalised yeast microarray data from the AN+RN approach (see Tables 1 and 2). This is clear evidence of the impact of feature-specific error and it also reveals that using dye-flip replicates is crucial for improving the performance of microarray data normalisation. Finally, Figure 4 shows that the error contained in the final results from the iAN+RN+SN method is bounded in the interval (–0.5, 0.5). This means that we can generate microarray data with such a level of quality comfortably (only two dye-flip pairs were used in the experiment) if dye-flip replicates and proper normalisation approaches are employed. Therefore, the genes with 2-fold or larger changes in expression level can be identified very confidently (we employed base 2 log transformations in this study so that a 2-fold change corresponds to a log ratio of 1 or –1).
SUMMARY AND CONCLUSIONS
Microarray data contains different sources of variation, which can be classified as systematic and random errors. Removal of systematic error items is a key issue for the improvement of microarray data quality. Though increasing the number of replicates can reduce random error in the data set, it can do nothing to reduce the systematic error. From consideration of the statistical model proposed in this study, we demonstrate that removing systematic error is not only the business of the data analyst but is also greatly influenced by experimental design. Employment of dye-flip replicates is critical for obtaining better results through normalisation. In addition, dye-flips provide the precondition for the analyst to identify the experiment-introduced systematic bias effectively. In comparison to non-dye-flip replicates, the use of dye-flip replicates associated with a suitable normalisation method can generate much improved results.
The statistical model of experimental errors introduced in this study provides useful guidance for microarray data analysis. Based on the model, we know why better results can be obtained from dye-flip replicates and we know why adaptive normalisation can achieve higher precision than self-normalisation. We also developed an improved method for identifying the experiment-introduced bias correctly. Using the model, we adopted suitable methods for removal of error items from different sources and so achieved very promising results. In turn, all the inferences from the model have been confirmed experimentally by analysis of real data sets.
This study provides a sound basis for microarray data analysis. The approach proposed can be used to perform not only a data normalisation, but also a data quality assessment. The data quality assessment method is potentially useful for data filtering and experimental quality control. For data filter purposes, we suggest that one could classify the replicated measurements of a given gene spot into good measurement and poor measurement subsets, based on the error contained in each of the measurements, and then discard the poor measurements of the gene spot, while retaining the good replicates of the gene spot for use in further analysis. To discard any gene spot being measured is a method that has been adopted by some researchers (5,9); however, there is typically no evidence that the gene spots being discarded are not of biological importance. We leave the testing of this approach and the details of the method for microarray data quality control for further study.
The main conclusions of our study appear to be robust with respect to the particular regression method used, e.g. using locally weighted regression in our adaptive normalisation method gives very similar errors after normalisation (results not shown). However, the analytical deductions presented in this paper are based on the assumption that all the normalisation processes involved can be carried out perfectly (the relevant error item can be identified and removed exactly), and this is clearly an idealisation. For example, in the adaptive normalisation process, the dye bias may not be identified and removed exactly. One problem with doing standard regression on an M–A plot is that both variables contain experimental error. In this case, it may be better to use a total least squares method in which both variables are modelled as variables that depend on some latent independent variable. Since the noise in M and A is correlated, it may be better to carry out this analysis using the original channel intensities. We leave the analysis of improved regression methods for future study.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Andrew Cossins, Andrew Gracey and Jane Fraser for providing the carp microarray data. This work was supported by a COGEME grant made to S.G.O. and A.B. under the ‘Investigating Gene Function’ Initiative of the BBSRC, a contract to S.G.O. within the frame of the GARNISH network of FP5 of the European Commission (contract no. QLK3-2000-00174) and by grants to A.B. and S.G.O. from the North-West Science Initiative’s Bioarray programme. Y.F. was supported by an ARK-Genome grant to A.B. and M.R. under the ‘Investigating Gene Function’ initiative of the BBSRC.
APPENDIX: AN ANALYSIS OF THE PERFORMANCE OF NORMALISATION APPROACHES
In adaptive normalisation (AN), we conduct a regression to an M–A plot of a replicate and then take the fitting as the dye bias and remove it from the data. The fitting is obtained by taking the spot intensity A as the independent variable and log ratio M as the dependent variable. The method for obtaining this fit can be either global linear regression or local regression. In this study, we used a non-linear regression method which is a generalisation of a standard polynomial fit. We followed the model in equation 1 and used fk(Ajk) to represent the fitted value for spot j on slide k after carrying out regression, i.e. fk(.) was the fitted function from the kth replicate and Ajk was the intensity of the jth spot on the kth replicate. The correction was achieved by subtracting the fitted value from Mjk,
Mjk ← Mjk – fk(Ajk). A1
It is easy to see that AN performs a slide-specific correction, i.e. for different slides, the correction may be different. The method can be applied to any type of microarray data set.
Improved adaptive normalisation (iAN) is similar to AN. However, a general function of dye bias is used for all n replicates. Although the function for n replicates stays unchanged, the correction for a given gene spot may be different from replicate to replicate because the spot intensity on different replicates will be measured as a different value. We propose a technique for obtaining this general function for replicates with dye-flip. The technique includes four steps: Firstly, we conduct a scale and location adjustment of spot intensity so that the intensity distribution of n replicated data sets has the same range and location. Secondly, we pool the data from all replicates together into one big data set. Thirdly, we make an M–A plot of this pooled data. Finally, we carry out regression on the M–A plot and obtain the parameterised regression function. The correction is then carried out for each of the replicates and the amount of adjustment of each data point is computed by substituting the spot intensity into the regression function. Therefore, the dye bias correction performed by iAN is:
Mjk ← Mjk – f(Ajk), A2
where f(.) is the regression function which has been parameterised by the pooled data.
The pooled data set contains both measurements from forward labelling slides and reverse labelling slides and they are matched into pairs, hence the influence of m(Aj) (in forward labelled measurements) and –m(Aj) (in reverse labelled measurements) cancels out. Therefore, the fitting of the iAN technique only picks up the systematic error items ek(Ajk) and c and not the terms m(Aj) and ck. Another basic point of the iAN method is the commonly used assumption that dye bias is spot intensity-dependent. We further assume that the dye bias function for a pool of replicated data sets and for a single data set will behave similarly. Based on these assumptions, the main reason for a given spot being measured with a different dye bias contribution is that the spot intensity will differ between replicates. The influence of the difference between dye bias functions (for pooled replicates and for each of the replicates) will be much weaker. This point has been confirmed by analysis of the yeast data described in the main text. The variation (measured by variance) of dye bias caused by spot intensity differences is about 100 times as large as that caused by differences due to changes in the form of dye bias function between replicates. Therefore, iAN can be expected to perform well.
Self-normalisation (SN) can only be applied to data from a dye-flipped pair of slides as otherwise the operation will remove any genuine expression ratio. We assume that the number of replicates is an even number n = 2s. Let slide k and k + s be a dye-flip pair. A single measurement is obtained from a pair of replicates:
Mjk ← (Mjk – Mj,k + s)/2 for k = 1, 2, …, s. A3
Regional normalisation (RN) is applied to each block of a replicate. The position-dependent error is identified by a 2D regression that takes the spot position as the independent variable and M as the dependent variable. Let fk(Pj) represent the fit for feature j on slide k from the regional fitting. The correction is:
Mjk ← Mjk – fk(Pj) for k = 1, 2, …, s. A4
Feature normalisation (FN) is identical to SN except that it is carried out after AN and RN (or after iAN and RN). The transformation is identical to equation A3 except that Mjk in the right hand side has been transferred by the other normalisations before this transformation.
In the following sections we consider the process and performance of different normalisation approaches in removing the experimental error contained in microarray data.
Self-self replicates without dye-flip
For this situation, the statistical model is shown by equation 2 in the main text; iAN, SN and FN are not usable, and so we use AN+RN (adaptive normalisation plus regional normalisation) to normalise the data.
In the AN process, the fit is fk(.) = c + ck + ek(Ajk). Therefore, after transformation A1, the measured expression ratio becomes:
Mjk = e(Fj) + ek(Pj) + εjk. A5
In the RN process, the fit is fk(Pj) = ek(Pj) and, after transformation A4, we get:
Mjk = e(Fj) + εjk. A6
Finally, it is sensible to take the mean of the n replicates as the final result. Let m̄j represent the mean of the log ratio for a gene on spot j. After normalisation, m̄j can be represented by equation 3 in the main text.
Self-self replicates with dye-flip
For this situation, we can take the n replicated measurement as s pairs (n = 2s). The model is then given by equation 4 in the main text. The two normalisation approaches AN+RN+FN and SN+RN are considered below.
AN+RN+FN. The outcome of AN and RN can still be represented by equation A6. Moreover, because dye-flip replicates are available, we can apply FN to the outcome in equation A6, and using the transformation in equation A3 we get:
Mjk = [e(Fj) – e(Fj) + εjk – εj,k + s]/2 = (εjk – εj,k + s)/2 A7
Finally, we take the mean of the s outcomes as the normalised result shown by equation 5 in the main text.
SN+RN. The SN process performs the transform A3, after which we have:
Mjk = (ck – ck + s)/2 + [ek(Ajk) – ek + s(Aj,k + s) + ek(Pj) – ek + s(Pj) + εjk – εj,k + s]/2 A8
after the FN transformation in A4 and, taking the mean of s sets of results as the normalised result, the final result is shown by equation 6 in the main text.
Non-self-self slides without dye-flip
We split mj into an intensity-independent term mj* and an intensity-dependent term m(Aj) so that mj= mj* + m(Aj). The statistical model is then shown by equation 7 in the main text. We consider the AN+RN approach. For AN the fit fk(.) will pick up the terms m(Aj), c, ck and ek(Ajk) and the expression ratio after transformation A1 becomes:
Mjk = mj* + e(Fj) + ek(Pj) + εjk. A9
For the RN process, the fit fk(.) will pick up the term ek(Pj) and after the correction based on equation A4 the outcome can be shown to be:
Mjk = mj* + e(Fj) + εjk. A10
Finally, we make use of the relationship mj* = mj – m(Aj) and the mean of the n replicated measurements is represented by equation 8 in the main text.
Non-self-self slides with dye-flip
The model of the data set is then given by equation 9 in the main text. We consider the three different normalisation approaches AN+RN+FN, SN+RN and iAN+RN+FN.
AN+RN+FN. It is easy to see that the outcome of AN is:
Mjk = mj* + e(Fj) + ek(Pj) + εjk A11
Mj,k + s = –mj* + e(Fj) + ek + s(Pj) + εj,k + s
RN will pick up ek(Pj) and the transformation A4 will remove it, so we obtain:
Mjk = mj* + e(Fj) + εjk A12
Mj,k + s = –mj* + e(Fj) + εj,k + s.
Finally, applying FN to each pair of slides and computing the mean of the s measurements we obtain equation 10 in the main text.
iAN+RN+FN. As before, iAN removes the error terms ek(Ajk) and c, but leaves m(Aj) and ck untouched, so that:
Mjk = mj + ck + e(Fj) + ek(Pj) + εjk A13
Mj,k + s = –mj + ck + s + e(Fj) + ek + s (Pj) + εj,k + s
RN removes the terms ck and ek(Pj) and we find:
Mjk = mj + e(Fj) + εjk A14
Mj,k + s = –mj + e(Fj) + εj,k + s
Finally, use of the FN process removes the term e(Fj), and taking the mean of the outcomes we obtain equation 11 in the main text.
SN+RN. The result is similar to the self-self case and the result is given by equation 12 in the main text.
REFERENCES
- 1.Chen Y., Dougherty,E.R. and Bittner,M.L. (1997) Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Optics, 24, 364–374. [DOI] [PubMed] [Google Scholar]
- 2.Hill A.A., Brown,E.L., Whitley,Z.M., Tucker-Kellogg,G., Hunter,P.C. and Slonim,K.D. (2001) Evaluation of normalization procedures for oligonucleotide array data based on spiked cRNA controls. Genome Biol., 2, research0055.1–research0055.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y.H., Dudoit,S., Luu,P., Lin,D.M., Peng,V., Ngai,J. and Speed,T.P. (2002) Normalisation for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res., 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr M.K., Martin,M. and Churchill,G. (2000) Analysis of variance for gene expression microarray data. J. Comput. Biol., 7, 819–837. [DOI] [PubMed] [Google Scholar]
- 5.Zien A., Aigner,T., Zimmer,R. and Lengauer,T. (2001) Centralization: a new method for the normalization of gene expression data. Bioinformatica, 1, 1–9. [DOI] [PubMed] [Google Scholar]
- 6.Yang Y.H., Dudoit,S., Luu,P. and Speed,T.P. (2001) Normalization for cDNA microarray. In Bittner,M.L., Chen,Y., Dorsel,A.N. and Dougherty,E.R. (eds), Microarrays: Optical Technologies and Informatics. SPIE, Society for Optical Engineering, San Jose, CA. [Google Scholar]
- 7.Suzuki T., Higgins,P.J. and Crawford,D.R. (2000) Control selection for RNA quantitation. Biotechniques, 29, 332–337. [DOI] [PubMed] [Google Scholar]
- 8.Goldsworthy S.M., Goldsworthy,T.L., Sprankle,C.S. and Butterworth,B.E. (1993) Variation in expression of genes used for normalization of Northern blots after induction of cell proliferation. Cell Prolif., 26, 511–518. [DOI] [PubMed] [Google Scholar]
- 9.Tseng G.C., Oh,M.-K., Rohlin,L., Liao,J.C. and Wong,W.H. (2001) Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res., 29, 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleveland W.S. and Delvin,S.J. (1988) Locally weighted regression: an approach to regression analysis by local fitting. J. Am. Statist. Assoc., 83, 596–610. [Google Scholar]
- 11.Rocke D.M. and Durbin,B. (2001) A model for measurement error for gene expression arrays. J. Comput. Biol., 8, 557–569. [DOI] [PubMed] [Google Scholar]
- 12.Brown A.J.P., Planta,R.J., Restuhadi,F., Bailey,D.A., Butler,P.R., Cadahia,J.L., Cerdan,M.E., De Jonge,M., Gardner,D.C.J., Gent,M.E., Hayes,A., Kolen,C.P.A.M., Lombardia,L.J., Murad,A.M.A., Oliver,R.A., Sefton,M., Thevelein,J., Tournu,H., Van Delft,Y.J., Verbart,D.J., Winderickz,J. and Oliver,S.G. (2001) Transcript analysis of 1003 novel yeast genes using high throughput northern hybridizations. EMBO J., 20, 3177–3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayes A., Zhang,N., Wu,J., Butler,P.R., Hauser,N.C., Hoheisel,J.D., Lim,F.L., Sharrocks,A.D. and Oliver,S.G. (2002) Hybridization array technology coupled with chemostat: tools to interrogate gene expression in S. cerevisiae. Methods, 26, 281–290. [DOI] [PubMed] [Google Scholar]