Abstract
The study of pathogenic mitochondrial DNA mutations has, in most cases, relied on the production of transmitochondrial cybrids. Although the procedure to produce such cybrids is well established, it is laborious and cumbersome. Moreover, the mechanical enucleation procedure is inefficient and different techniques have to be used depending on the adherence properties of the cell. To circumvent these difficulties, we developed a chemical enucleation method that can have wide applicability for the production of transmitochondrial cybrids. The method is based on the use of actinomycin D to render the nuclear genome transcription/replication inactive and unable to recover after treatment. Such treated cells are fused to cells devoid of mitochondrial DNA and selected for the presence of a functional oxidative phosphorylation system. Our results showed that 95% of the clones recovered by this procedure are true transmitochondrial cybrids. This method greatly facilitates the production of transmitochondrial cybrids, thereby increasing the number of mtDNA mutations and the recipient cell types that can be studied by this system.
INTRODUCTION
The development of animal cells devoid of mitochondrial DNA (mtDNA), termed ρ° cells (1), has spearheaded the study of mammalian mtDNA mutations associated with human diseases. King and Attardi (2) showed that ρ° cells could be repopulated with exogenous mitochondria from mechanically enucleated cells. Because ρ° cells are auxotrophic for uridine and pyruvate (2), a selection system can be applied to obtain cells harboring exogenous mtDNA after fusion of ρ° cells with cytoplasts (mtDNA donor) isolated by mechanical enucleation after disruption of the cytoskeleton by cytochalasin B treatment (2,3). In addition, the ρ° cells require a recessive nuclear marker, in most cases a thymidine kinase mutant, to avoid the isolation of hybrids or mtDNA donor cells that fail to enucleate during centrifugation. Although extremely useful, this system is cumbersome, and some cell lines do not enucleate well under standard conditions of mechanical enucleation (3). Moreover, as mentioned above, a recessive nuclear marker in the ρ° cells is required to eliminate the non-enucleated mtDNA donor cells and hybrids.
We developed an efficient and simple method to chemically enucleate cells that can subsequently be used as mtDNA donors. The method takes advantage of actinomycin D, an antibiotic (produced by Streptomyces antibioticus) that intercalates into DNA duplexes, thereby interfering with the action of enzymes engaged in replication and transcription. Actinomycin D consists of a phenoxazone ring system, to which two cyclic pentapeptides are attached. The aromatic ring system is able to intercalate into DNA, preferably at GC steps (4,5). Actinomycin D has been used extensively as an inhibitor of transcription and replication (6) as well as an anti-tumor drug in the clinic (7).
MATERIALS AND METHODS
The cell line TEX is a human skin fibroblast modified by infection with: (i) a retrovirus containing a hTERT (human telomerase) and a puromycin-resistance gene; (ii) a retrovirus containing the adenovirus E6/E7 and a neomycin-resistance gene. The ρ° cell line is an osteosarcoma 143B derivative which had its mtDNA depleted by chronic treatment with ethidium bromide (3).
Cells were cultured in DMEM high glucose supplemented with 50 µg/ml uridine and 1 mM pyruvate (complete medium) unless noted otherwise. Actinomycin D was purchased from Sigma (cat A-9415). Cell fusions were performed with 45% polyethylene glycol (MW 1450; Sigma cat P-5402) for 60 s as described (2,3). Briefly, 0.3 × 106 cells, plated on a 35 mm dish were treated with (0.5–4) µg/ml actinomycin D for different times (Table 1) and washed with complete medium for 10 min to remove the drug. One million ρ° cells were overlaid on the dish and after 3 h (time to attach) were washed three times with DMEM without serum. The medium was then completely removed and cells subjected to fusion with PEG. After fusion, cells were washed three times with DMEM supplemented with 10% DMSO and once with DMEM. Three milliliters of complete medium was finally placed on the dish for the subsequent 48 h. The cells were then trypsinized and plated onto ten 100 mm dishes.
Table 1. Optimization of actinomycin D chemical enucleation for the production of transmitochondrial cybrids.
| Actinomycin D (µg/ml) | Time (h) | Viabilitya |
|---|---|---|
| 1.0 | 1 | +++ |
| 1.0 | 2 | +++ |
| 1.0 | 4 | +++ |
| 1.5 | 1 | ++ |
| 1.5 | 2 | ++ |
| 1.5 | 4 | + |
| 4.0 | 4 | +/– |
| 0.5 | 15 | – |
aViability was assessed by the relative number of cells that recovered from the treatment and regained proliferative capacity; 10–30% death (+++), 30–60% death (++), 60–90% death (+), 90–95% death (+/–), 100% death (–). Clones obtained by fusing the TEXPN actinomycin D-treated cells (0.5 µg/ml, 15 h) with the osteosarcoma ρ° cell were grown in the presence of G418 or puromycin. Only 1 out of 20 clones was able to grow in both G418 and puromycin.
Nuclear markers were obtained from Research Genetics/Invitrogen (Carlsbad). The following loci with polymorphic tetranucleotide repeats were used: D3S2427, D9S934, D14S587, D17S1290 and D21S1270. Mitochondrial DNA was analyzed by Southern blot using a probe corresponding to mtDNA positions 4120–4530 (8). Chromosomal haplotypes were analyzed by PCR of the polymorphic nucleotide repeats as suggested by the manufacturer (Research Genetics/Invitrogen). Mitochondrial DNA was analyzed by Southern blot as described (9).
Cell respiration was determined in a Hansatech oxygen measurement chamber in 600 µl of medium as described (10).
RESULTS AND DISCUSSION
To assess the efficiency of the chemical enucleation method, we used a human fibroblast line (TEXPN) containing two nuclear markers, namely puromycin- and neomycin-resistance (Fig. 1). This cell line was treated with different concentrations of actinomycin D for various times (Table 1). It was important to find conditions that would render the cells unviable after the treatment was halted. Higher concentrations (1–4 µg/ml) for short times (1–4 h) were not able to kill all the cells. We found that a treatment of this immortalized fibroblast line with 0.5 µg/ml for 15 h was the best compromise for causing irreversible cell death, but still obtaining viable cytoplasts that could be used for fusion with ρ° cell.
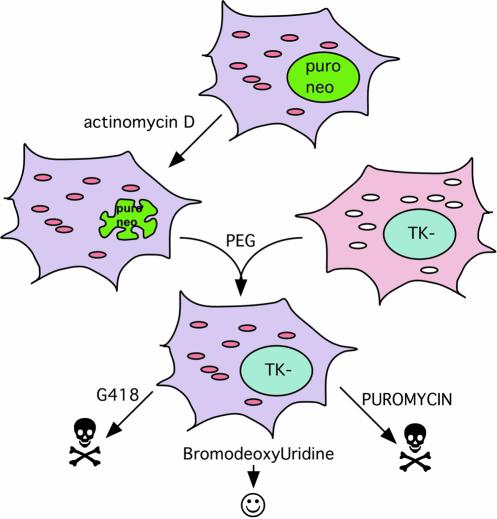
Figure 1.
Chemical enucleation procedure to transfer mtDNA to ρ° cells. The mtDNA donor cell is treated with actinomycin D at conditions that preclude recovery of treated cells. This cell with a degenerating nucleus is a suitable mtDNA donor and can be fused to ρ° cells with PEG as described in Materials and Methods. In our experiment, transmitochondrial cybrid clones were obtained without a nuclear selection (they were isolated in media lacking uridine to inhibit growth of ρ° cells) and later checked for resistance to G418, puromycin and bromodeoxyuridine (BrdU). Most of the cybrids were sensitive to the drug-resistance present in the actinomycin-treated cells and resistant to BrdU.
Approximately 0.3 × 106 TEXPN cells, treated with 0.5 µg/ml of actinomycin D, were fused to 1 × 106 osteosarcoma ρ° cells and plated at low density as described in Materials and Methods. After approximately 15 days of no uridine selection (to eliminate unfused ρ° cells), clones were isolated by the cloning ring method (3). Analysis of 20 independent clones showed that all but one clone (clone 15) died in the presence of G418 (a neomycin analogue) or puromycin (Fig. 1 and Table 1). All but clone 15 were also resistant to bromodeoxyuridine. Clone 15 was resistant to puromycin and G418 and was sensitive to bromodeoxyuridine, suggesting that it had retained chromosomes from the actinomycin-treated cells. Therefore, it is evident that a minority of clones (approximately 5%) can contain chromosomes that were not replication-arrested or damaged (to the point of no repair) by actinomycin D. We confirmed this phenomenon by culturing a pool of approximately 100–200 unselected clones in G418. Despite a massive initial death, some cells survived the G418 treatment. These drug-treated and untreated pools, as well as the original unselected clones were analyzed for five different nuclear markers, located in five different chromosomes. This analysis showed that the vast majority (19 out of 20 clones) had chromosomes originated exclusively from the osteosarcoma nuclear donor (Fig. 2). As expected, the pool of clones also had only the osteosarcoma nuclear markers, but after selection in G418 + puromycin, both the osteosarcoma and the fibroblast nuclear markers became apparent (Fig. 2). It appears that it is an all or none phenomenon for rare clones that maintain the actinomycin-treated chromosomes. We have confirmed this observation in subsequent experiments with different cell lines (not shown). Because we did not observe surviving cells after this treatment, it is likely that chromosomes from damaged cells were able to recover after fusion with a healthy cell.
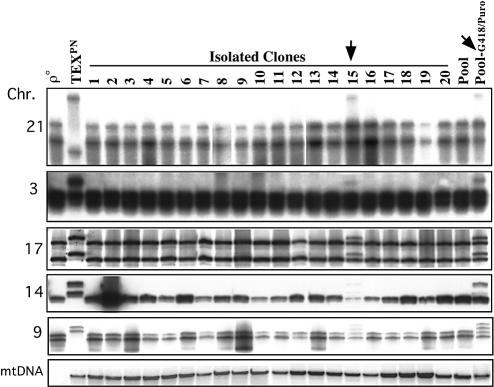
Figure 2.
Genetic make-up of transmitochondrial cybrids produced by chemical enucleation. Twenty individual transmitochondrial cybrid clones were analyzed for the presence of chromosomes from the mtDNA donor cell. Polymorphic tetranucleotide repeat markers located in chromosomes 3, 9, 14, 17 and 21 (see Materials and Methods) were used to distinguish the nuclear background (TEX versus 143B). Nineteen of 20 clones had exclusively 143B-derived chromosomes. Only clone 15, and a pool of clones selected for G418 resistance had TEX-derived chromosomes. The presence of mtDNA was also determined by Southern blot.
MtDNA analyses showed that the cybrids were repopulated with mtDNA from the actinomycin D-treated cells (Fig. 2, lower panel). To assess for the recovery of mitochondrial oxidative phosphorylation, representative clones were analyzed for respiratory function. Figure 3 shows that the different cell lines regained respiratory function and therefore can be considered bona fide transmitochondrial cybrids.
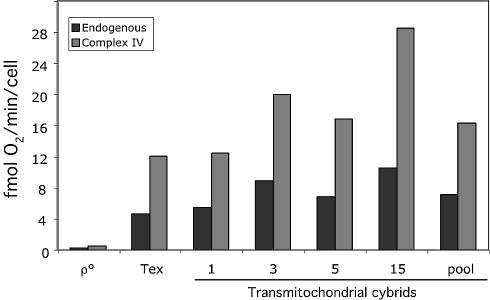
Figure 3.
Cell respiration in transmitochondrial cybrids. Approximately 106 cells (in 600 µl of no glucose DMEM) were injected in a Hansatech oxygen measurement chamber and oxygen consumption recorded over time. Antimycin A was added to the chamber to inhibit complex I- and II-driven respiration and Ascorbate/TMPD was added to initiate complex IV-driven respiration. The latter was inhibited by KCN. The oxygen consumption rates (endogenous minus antimycin and Asc/TMPD minus KCN) were plotted for representative clones and parental cell lines. Respiration was restored in transmitochondrial cybrids.
To evaluate the efficiency of rescue of actinomycin D-treated cells by nuclear donors other than the human osteosarcoma, we treated the human fibroblast (TEX) with 0.5 µg/ml actinomycin D for 15 h and fused it to chimpanzee or gorilla fibroblasts that were depleted of mtDNA. Three nuclear polymorphic markers located in three different chromosomes were used to assess the nuclear identity of the clones. In the TEX-chimpanzee fusion, 11 out of 14 (79%) of the clones analyzed contained exclusively chimpanzee chromosome alleles. Among TEX-gorilla fusion products, 14 of 14 (100%) had only gorilla alleles. As expected the mtDNA origin was human in both cases. These results were very similar to the ones obtained using the human osteosarcoma cell line as the nuclear donor, indicating that actinomycin D-treated cells are not committed to apoptosis, and can be rescued by fusion to different nuclei backgrounds.
Although we have not treated a primary cell line (i.e. without overexpression of telomerase) with actinomycin D, we suspect that the procedure would be equally efficient. Cells overexpressing telomerase have stable chromosomes and although they have been shown to have increased resistance to certain apoptotic insults, they were not more resistant to DNA replication inhibitors (11).
It is unclear why actinomycin D does not block mtDNA replication, or damage mtDNA as severely as it does to the nuclear DNA. It may be related to the different packing and replication modes between these genomes. Actinomycin D is a strong transcriptional inhibitor, and the human mtDNA depends on an RNA primer for replication (12). It is possible that even if arrested, enough mtDNA molecules remain intact and are able to repopulate fusion products. Although actinomycin D has been shown to affect mitochondrial transcription (13,14), mtDNA replication may be less sensitive than that of the nuclear DNA. In support of this concept, actinomycin D was found to increase mitotic recombination in Saccharomyces cerevisiae but not in Escherichia coli (15).
Although this method is very efficient, we make two important recommendations. First, individual clones should be isolated after the fusion with PEG. The easiest procedure to ensure cell clones are not hybrids is by testing two nuclear markers. Our results demonstrate that partial chromosome transfer is unlikely, and any two polymorphic marker (see Materials and Methods) should indicate the cybrid nature of the cell clone. Moreover, although a minority of cells retains the nuclear genome from the mitochondrial donor, such hybrid clones may have a faster population doubling and steadily increase their presence in the pool. Second, the actinomycin D treatment conditions should be optimized for different cell lines. The mildest treatment that results in complete cell death after drug removal should be selected. The experiments described here provide starting points for this cell-specific optimization.
CONCLUSIONS
This simple method has a large number of applications and advantages for cell biology investigators. (i) It allows enucleation of essentially any cell type, as long as the treatment conditions are optimized for that specific system. This means that the treatment is severe enough to not allow cells to recover after the drug is removed, but mild enough to preserve mtDNA integrity and cytoplast viability. (ii) It avoids mechanical enucleation, a method that is inefficient, cumbersome, laborious and time consuming. (iii) In contrast to mechanical enucleation, the ρ° cells do not need a recessive nuclear marker to block the growth of non-enucleated mtDNA donor cells. This is a major advantage of the chemical enucleation method because it allows any cell type, which is devoid of mtDNA, to function as mtDNA acceptor for the production of transmitochondrial cybrids. In combination with rhodamine 6G treatment, which can eliminate mitochondria from a nuclear donor (16), transmitochondrial cybrids could be produced exclusively by chemical ablation of nuclear and mitochondrial compartments. (iv) This method should also be adaptable to other cytoplast or nuclear transfer experiments, including embryo cloning.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by grants from the National Institutes of Health.
REFERENCES
- 1.Desjardins P., de Muys,J.M. and Morais,R. (1986) An established avian fibroblast cell line without mitochondrial DNA. Somat. Cell Mol. Genet., 12, 133–139. [DOI] [PubMed] [Google Scholar]
- 2.King M.P. and Attardi,G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 3.Moraes C.T., Dey,R. and Barrientos,A. (2001) Transmitochondrial technology in animal cells. Methods Cell Biol., 65, 397–412. [DOI] [PubMed] [Google Scholar]
- 4.Krugh T.R., Mooberry,E.S. and Chiao,Y.C. (1977) Proton magnetic resonance studies of actinomycin D complexes with mixtures of nucleotides as models for the binding of the drug to DNA. Biochemistry, 16, 740–747. [DOI] [PubMed] [Google Scholar]
- 5.Chiao Y.C. and Krugh,T.R. (1977) Actinomycin D complexes with oligonucleotides as models for the binding of the drug to DNA. Paramagnetic induced relaxation experiments on drug-nucleic acid complexes. Biochemistry, 16, 747–755. [DOI] [PubMed] [Google Scholar]
- 6.Waring M.J. and Bailly,C. (1994) DNA recognition by intercalators and hybrid molecules. J. Mol. Recognit., 7, 109–122. [DOI] [PubMed] [Google Scholar]
- 7.Lurain J.R. (2002) Advances in management of high-risk gestational trophoblastic tumors. J. Reprod. Med., 47, 451–459. [PubMed] [Google Scholar]
- 8.Anderson S., Bankier,A.T., Barrell,B.G., de Bruijn,M.H., Coulson,A.R., Drouin,J., Eperon,I.C., Nierlich,D.P., Roe,B.A., Sanger,F. et al. (1981) Sequence and organization of the human mitochondrial genome. Nature, 290, 457–465. [DOI] [PubMed] [Google Scholar]
- 9.Moraes C.T., DiMauro,S., Zeviani,M., Lombes,A., Shanske,S., Miranda,A.F., Nakase,H., Bonilla,E., Werneck,L.C., Servidei,S. et al. (1989) Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns-Sayre syndrome. N. Engl. J. Med., 320, 1293–1299. [DOI] [PubMed] [Google Scholar]
- 10.Barrientos A. (2002) In vivo and in organello assessment of OXPHOS activities. Methods, 26, 307–316. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama M., Yamada,O., Kanda,N., Akita,S., Kawano,T., Ohno,T., Mizoguchi,H., Eto,Y., Anderson,K.C. and Yamada,H. (2002) Telomerase overexpression in K562 leukemia cells protects against apoptosis by serum deprivation and double-stranded DNA break inducing agents, but not against DNA synthesis inhibitors. Cancer Lett., 178, 187–197. [DOI] [PubMed] [Google Scholar]
- 12.Clayton D.A. (1982) Replication of animal mitochondrial DNA. Cell, 28, 693–705. [DOI] [PubMed] [Google Scholar]
- 13.Connor M.K., Takahashi,M. and Hood,D.A. (1996) Tissue-specific stability of nuclear- and mitochondrially encoded mRNAs. Arch. Biochem. Biophys., 333, 103–108. [DOI] [PubMed] [Google Scholar]
- 14.Gaines G. and Attardi,G. (1984) Intercalating drugs and low temperatures inhibit synthesis and processing of ribosomal RNA in isolated human mitochondria. J. Mol. Biol., 172, 451–466. [DOI] [PubMed] [Google Scholar]
- 15.Nestmann E.R., Nasim,A., Haynes,R.H. and Kowbel,D.J. (1981) Genetic activity of actinomycin D in Saccharomyces cerevisiae but not in Escherichia coli.Mutat Res., 89, 229–236. [PubMed] [Google Scholar]
- 16.Trounce I. and Wallace,D.C. (1996) Production of transmitochondrial mouse cell lines by cybrid rescue of rhodamine-6G pre-treated L-cells. Somat. Cell Mol. Genet., 22, 81–85. [DOI] [PubMed] [Google Scholar]