Abstract
A set of 1438 human exons was subjected to nested PCR. The initial success rate using a standard PCR protocol required for ligation-independent cloning was 83.4%. Logistic regression analysis was conducted on 27 primer- and template-related characteristics, of which most could be ignored apart from those related to the GC content of the template. Overall GC content of the template was a good predictor for PCR success; however, specificity and sensitivity values for predicted outcome were improved to 84.3 and 94.8%, respectively, when regionalized GC content was employed. This represented a significant improvement in predictability with respect to GC content alone (P < 0.001; χ2) and is expected to increase in relative sensitivity as template size increases. Regionalized GC was calculated with respect to a threshold of 61% GC content and a sliding window of 21 bp across the target sequence. Fine-tuning of PCR conditions is not practicable for all target sequences whenever a large number of genes of different lengths and GC content are to be amplified in parallel, particularly if total open reading frame or domain coverage is essential for recombinant protein synthesis. Thus, the present method is proposed as a means of grouping subsets of genes possessing potentially difficult target sequences so that PCR conditions can be optimized separately in order to obtain improved outcomes.
INTRODUCTION
The recent mapping of the human, mouse, fly, yeast and other genomes has paved the way to an era of massive intra- and inter-genomic comparisons. In parallel, biomedical research laboratories, biotechnology and pharmaceutical companies have developed high-throughput methods for genomic and proteomic applications (1). Many of these methods depend upon amplification of nucleic acids by PCR (2–5).
PCR requires a DNA template and a pair of primers flanking the target DNA. An important parameter to be considered when selecting PCR primers is the ability of the primers to form a stable duplex exclusively with the specific site on the target DNA. The use of the nearest-neighbor thermodynamic parameters for computing DNA or RNA duplex stability has been shown to produce reliable predictions (6–9). These methods calculate the melting temperature (Tm) of the primers, which is correlated with the GC/AT ratio of the primers. Typically, primers should have a GC/AT ratio similar to or higher than that of the amplified template (10). Other considerations that increase the specificity of PCR include: (i) avoidance of complementarity at the 3′ termini of the primers, as this promotes the formation of primer dimer artifacts; and (ii) avoidance of stable self-complementary hairpin loops that increase primer stability (10).
The DNA template used for PCR is often overlooked when compared with the effort put into primer design. The most commonly used parameters that relate to the DNA template are the PCR product size and the Tm of the product (10–12). However, it is known that DNA templates with a very high or very low GC/AT ratio can be difficult to amplify (13–15).
PCR has become a well-understood in vitro process (16). Many tools exist that help to achieve a high yield of PCR products, such as primer design software (12,17), optimization kits and well-characterized protocols (18,19). However, these tools are often designed for a small number of reactions, or indeed a specific gene whereby the temperature and/or ion concentrations are varied to achieve maximal recovery of desired product (18). This is not feasible when hundreds of genes are to be amplified in parallel.
Several recent studies have evaluated the success of primer extension for genotyping (2,20) and for generation of gene sequence tags (21). Vieux et al. (2) reported a 96% success rate in PCR using a very strict primer selection strategy combined with stringent PCR conditions for analysis of single nucleotide polymorphisms. These applications have the luxury of scanning long nucleotide sequences until the optimal primers are found. However, amplifying a particular DNA sequence of interest does not usually allow a stringent primer selection strategy, especially if the target sequence is a few hundred base pairs in length or contains the whole open reading frame (ORF) or specific portions of it for recombinant protein synthesis (22–25). The latter are thought to become increasingly important in a proteomics context.
Here we report on the amplification of 1438 human exons and efforts to establish a suitable predictor of PCR outcome.
MATERIALS AND METHODS
Selection of exons
We randomly selected 1438 human ORFs from disease-related genes available in publicly accessible clone libraries in late 2001 and retrieved their DNA coding sequence from GenBank (http://www.ncbi.nlm.nih.gov). Coding sequences were compared with the human genome (Genbank build 25) using BLAST (26), and the exons were extracted and set in-frame. For ORFs containing multiple exons, the first was discarded to reduce the likelihood of a signal protein, and from the remaining exons the longest was chosen.
Primer design strategy
We selected by default the first and last 21 nucleotides of each target sequence as the primers and modified each primer only if more than four Gs or four Cs were present in the last five nucleotides of the 3′ end, or if more than three consecutive Ts were present at the 3′ end. In such cases, up to five nucleotides were removed from the 3′ end, allowing a minimum primer length of 16 nucleotides. This study was conducted with a view to subsequent cloning in the Gateway™ system (Invitrogen). Therefore, two long adaptors, named attB1 and attB2, had to be attached to both sides of the PCR product in a two-step procedure. First, an oligonucleotide of 14 bases was attached to the 5′ end of the forward primer (AAAAAGCAGGCTTG) and an oligonucleotide of 13 bases was attached to the 5′ end of the reverse primer (AGAAAGCTGGGTA). Secondly, two universal primers were employed that bound to the adaptors from the first PCR. The forward universal primer GGGGACAAGTTTGTACAAAAAAGCAGGCTTG and the reverse universal primer GGGGACCACTTTGTACAAGAAAGCTGGGTA were used to complete the attB1 and attB2 site. All primers were synthesized by Sigma Genosys.
PCR
Genomic DNA was isolated from purified human white blood cells using a Genomic tip™ 500/g from Qiagen. A two-step PCR was performed in 96-well plates with a GeneAmp PCR system 9700 from Applied Biosystems. The standard PCR conditions were: 0.1 µg of template DNA, 0.05 µl of TaKaRa Ex Taq, 1 µl of 10× Ex Taq buffer (2 mM Mg2+), 0.8 µl of dNTP mixture (2.5 mM each) and 0.5 µM of each primer in a 10 µl reaction mixture. In all PCR cycles, denaturation lasted 30 s at 94°C and polymerization 2 min at 72°C. The annealing step was for 30 s at varying temperatures, namely 58°C in the first PCR and 45°C for five cycles followed by 65°C for 25 cycles in the second PCR. PCR products were visualized with 0.5 µl/ml ethidium bromide on a 1.2% agarose gel. Images were taken using GeneGenious from Syngene® and analyzed with the bundled GeneTools software.
Logistic regression
A stepwise backward likelihood ratio (LR) logistic regres sion was performed with SPSS version 10. Entry and removal P-values were set to <0.05. The receiver operating characteristic (ROC) curve was used as a measure of model performance. It was employed graphically to represent the trade-off between false-positive and false-negative rates for every possible cut off. The false-positive rate was plotted on the x-axis and the true positive rate (1 – the false-negative rate) on the y-axis. The area under the curve was of primary interest as it measured the correlation between the category predicted by the test and the true category into which the case falls (27,28).
Informatics
Software for sequence analysis of primers and DNA template was written in Python (http://www.python.org), and all data and results were stored in a FileMaker database (http://www.filemaker.com). SPSS software version 10 was used for data analysis and statistical modeling. The parameters employed for the study of primers and DNA template are summarized in Table 1. In all statistical tests, the primers were labeled 1 and 2 according to their GC content. Primer 1 is the primer with the higher GC content of the two primers and not necessarily the forward primer.
Table 1. Description of parameters used for analyzing PCR primers and template.
| Parameter name | Description | Reference |
|---|---|---|
| Tmprimer | Melting temperature of the primers | (7,10) |
| IntStab3 | Internal stability of the primer at the 3′ end | (7,10) |
| IntStab5 | Internal stability of the primer at the 5′ end | (7,10) |
| IntStabD | IntStab5 – IntStab3 | |
| GCprimer | Primer (G + C)/primer length | |
| GC_DIFF | GCprimer/GCtemplate | |
| SigHits | Number of significant hits when comparing the primer sequence with the human genome using BLAST A blast-hit was considered significant when 10 identical nucleotides occurred at the 3′ end | |
| Bend | Bending value at the 3′ end of the primer | EMBOSS, banana (31) |
| Curve | Curvature value at the 3′ end of the primer | EMBOSS, banana (31) |
| SelfAny | The maximum local alignment score when testing a primer for annealing with itself or with the other primer Computed by Primer3. | (12) |
| SelfEnd | The maximum 3′-anchored global alignment score when testing a primer for annealing with itself or with the other primer. Computed by Primer3 | (12) |
| TmProduct | Melting temperature of the PCR template | (32) |
| GCtemplate | Template (G + C)/template length | |
| TmDiff | |Tmprimer1 – Tmprimer2| | |
| TaOpt | Optimal temperature for PCR | (17) |
| Dimer | Highest possible duplex stability between both primers, calculation based on free energy values. | (7) |
| MaxCurve | Highest DNA curvature in the PCR template. | EMBOSS, banana (31) |
| AUCTm | Area under the Tm curve and above 75°C of the PCR template | |
| AUCGC | Area under the GC curve and above 65% of the PCR template | |
| ratioGC | Number of GC windows with values above 65% divided by the length of the PCR template | |
| ratioTm | Number of Tm windows with values above 75°C divided by the length of the PCR template | |
| NormAUCGC | RatioGC × AUCGC | |
| NormAUCTm | RatioTm × AUCTm | |
| MinDist | Shortest distance from either ends of the PCR template and the first high GC region | |
| MINGC | Low value of the GC content of the first and last 60 nucleotides of the PCR template. | |
| MAXGC | High value of the GC content of the first and last 60 nucleotides of the PCR template. | |
| SIZE | PCR product length |
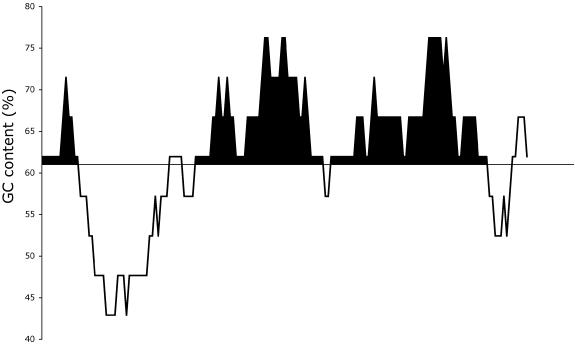
Regionalized GC content within template DNA was calculated using a sliding window of 21 nucleotides, shifted one nucleotide at a time. The results were plotted and the area under the GC curve (AUCGC) above a 61% threshold was calculated using the trapezoid method (Fig. 1). A high GC content region was considered significant if it was >61% for 10 consecutive windows. Similarly, regionalized Tm and the area under the Tm curve (AUCTm) above a threshold of 74°C were calculated. The thresholds for both the GC curve and the Tm curve were chosen initially as 65% and 75°C so as to reflect population extremes. Subsequently, these threshold values were made more precise with respect to their ability to discriminate between ‘good’ and ‘failed’ groups for all integer values between 50 and 70% and between 65 and 85°C, respectively, while employing the LR logistic regression. Table 1 summarizes the methods and parameters employed for statistical analysis, while associated software is available from: http://wwwcmc.pharm.uu.nl/moret/pub/benita.
Figure 1.
A slide window of GC content plotted against the DNA sequence of an example template. The black regions represent areas above a 61% threshold for GC content and measured across 31 bp, i.e. ×10 sliding windows of 21 bp.
RESULTS
Primers and template properties
Out of 2876 primers, 2501 were not altered, while 1–4 nucleotides were removed from the 3′ end of the remaining 375. Properties employed for primer evaluation are shown in Table 2. A wide range of values was allowed for each primer property, yet average values of Tm, GC content and internal stability were well within the recommended range, as defined by Rychlik (10,29) and McPherson and Møller (18). However, when combined parameters are examined, for example, Tm and 3′ end internal stability together, results are less clear-cut. In the latter example, only 60% of the primers were within the recommended range and only 37% of the primer pairs were both within the range. Primers had 4.8 significant hits, on average, when compared with the human genome using BLAST. A search of the entire human genome for potential PCR products that required the primers to be on opposite strands and not more than 2000 nucleotides apart predicted that only one PCR product could be formed by in silico predictions. In silico, all pairs of primers generated a single target PCR product. A combined analysis of both primers and the PCR template was performed to evaluate the success of the reaction as shown in Table 3. The observed range was very wide for most of the parameters. Analysis of DNA curvature was included to identify DNA structural oddities that might have affected the ability of Taq polymerase to duplicate the template.
Table 2. Properties of the 2876 primers employed.
| Tm (°C) | GC content (%) | GC Diff | IntStab3 (kcal/mol) | IntStab5 (kcal/mol) | IntStabD (kcal/mol) | SelfAny (kcal/mol) | SelfEnd (kcal/mol) | |
|---|---|---|---|---|---|---|---|---|
| Average | 60.6 ± 7.12 | 49.0 ± 11.3 | 0.95 ± 0.18 | 7.6 ± 1.2 | 7.83 ± 1.35 | 0.23 ± 1.73 | 5.1 ± 1.9 | 2.6 ± 2.2 |
| Range | 36.1–86.8 | 14.0–86.0 | 0.34–1.73 | 5–13.1 | 4.8–13 | –5.17–6.3 | 0–14 | 0–12 |
| Recommended | 55–70 | 30–70 | >1 | <9 | NA | >0 | <8 | <3 |
Table 3. Template and primer properties.
| Ta Opt (°C) | Tm Diff (°C) | Lowest GC Diff | ΔG dimer (kcal/mol) | Tm product (°C) | GC content (%) | DNA max curvature (°) | |
|---|---|---|---|---|---|---|---|
| Average | 56.5 ± 4.8 | 7.3 ± 5.7 | 0.8 ± 0.2 | 4.5 ± 7.0 | 78.2 ± 3.9 | 51.9 | 32.4 |
| Range | 32.8–69.6 | 0.01–32.8 | 0–1.4 | 0–30.5 | 68.2–89.6 | 31.2–77.1 | 9.7–120.4 |
| Recommended | 58 | ≤5 | ≥1 | ≤12 | None | 30–70 | None |
PCR
A PCR product with an expected size >350 bp was considered ‘good’ if the observed band was 10% longer or shorter than the expected size. A maximal deviation of 15% was allowed for smaller products, due to the inherent insensitivity of on-gel mobility measurements. All bands <120 bp were discarded and interpreted as representing primer dimers or PCR artifacts. A band of the expected size was observed for 1226 (83.4%) sequences. The other 212 (14.7%) failed in duplicate experiments to produce the correct band size, 69 (32.5%) had no product at all and 44 (20.7%) were associated with a product of incorrect size. Of all the ‘good’ products, 858 (70%) had one clear band and the other 305 (25%), 54 (4.4%) and eight (0.75%) had two, three and four bands, respectively.
Numerical analysis
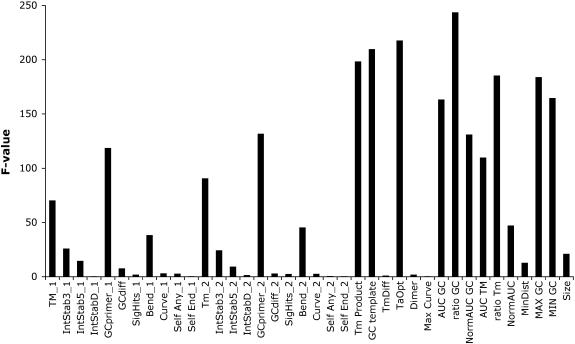
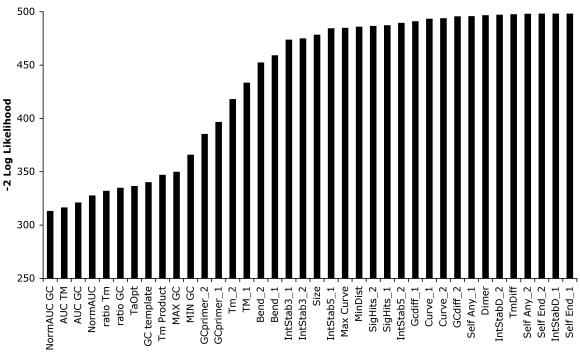
Two data sets were created for the analysis: data set A contained 212 sequences that failed to PCR twice and data set B contained 318 sequences, which twice produced a clear visible band. We avoided including too many samples in data set B since it could bias the statistical analysis. Seventy percent of the data in each set was used for statistical analysis as selected by a random function, while the remainder was used as a test set for the prediction model. The mean values of groups A and B for the parameters described in Table 1 were compared using a one-way ANOVA. The ANOVA results showed that Tm and GC content are the most significant parameters at the primer level, and parameters that are correlated with total GC content of the template are the most significant at the template level (Fig. 2). All the primer and template parameters were used in a stepwise backward LR logarithmic regression. Although total GC content, GC ratio, TmProduct and TaOpt are the most significant parameters in the ANOVA test, NormAUCGC and NormAUCTm were shown to be much better predictors in the logistic regression. A logistic regression for each variable was performed separately, and the goodness of fit was assessed by –2 log likelihood. The strongest single predictor of success and failure of PCR using the logistic regression model was NormAUCGC (Fig. 3). The best logistic regression model contained both the primer with lower GC content (GCprimer2) and the NormAUCGC. Wald χ2 values for GCprimer2, NormAUCGC and the constant were 19.2, 40.1 and 38.2, respectively, each with similar degrees of freedom. Thus, a good level of confidence was obtained for the expected PCR success, as shown by the following equation:
Figure 2.
F-values for a one-way ANOVA computed from a learning set of 370 DNA target templates. Large F-values represent a significant difference between the failed and successful PCR. P = 0.05 for F = 3.87 and P = 0.001 for F = 11.
Figure 3.
Likelihood values –2 log obtained by logistic regression for each variable separately. These values are commonly used to indicate the goodness of fit. The lower the value, the better the model.
where P is the probability of a successful PCR and GCprimer2 and NormAUCGC are the parameters described in Table 1. The area under the ROC curve was 0.87, and the Nagelkerke R2 was 0.58. Both are high and suggest the model’s performance is good. A PCR is predicted successful for P ≥ 0.5. Using this equation on our test set (n = 160, i.e. 30% of data set A + B), 86.3% of PCRs were predicted correctly. The sensitivity of the model is the probability of correctly predicting a positive example, while the specificity is the probability that a positive example is correct (30). The specificity and sensitivity values of the test set were 84.3 and 94.8%, respectively; and 94.9 and 85.2%, respectively for the 1438 PCRs examined in this study.
The logistic regression equation was able to predict that, for a given value of NormAUCGC, a reduction of the GC content of the primer should increase the probability of PCR success. This was due to the significantly lower GCprimer2 of group B when compared with that of group A, at NormAUCGC ≤ 340. The mean values of GCprimer2 for NormAUCGC ≤ 340 for groups A and B were 48 ± 10% and 44 ± 9% (P < 0.05), respectively, corresponding to a 2°C difference in mean Tmprimer2 values. Nevertheless, for NormAUCGC > 340, the probability of PCR success was significantly reduced even for low GCprimer2 values, since 67.2% of the sequences in group A possessed a NormAUCGC > 340 compared with only 5.2% for group B. Therefore, PCR success was also predicted based on NormAUCGC alone with an upper threshold of 340. The complete set of 1438 sequences was divided into three groups, sequences with NormAUCGC ≤ 340; NormAUCGC between 340 and 750 (95th percentile of successful PCR); and NormAUCGC > 750. In the first category (n = 1143), the success rate was 93.8%, while in the second (n = 139) and third (n = 156) it was 71.2 and 35.2%, respectively. Thus, the high predictive value of the index, NormAUCGC ≤ 340, was clearly demonstrated.
DISCUSSION
For more than a decade, primer design has evolved into an efficient and mature science. Although the primer sequence can be modified by biologists, the target DNA cannot. Therefore, little attention is usually paid to the analysis of the PCR template prior to experimental procedures, i.e. apart from its relevance to primer design. Faced with the challenge of large-scale PCR, protocol optimization becomes increasingly important, yet is increasingly problematic due to the variation in template sequence and length. As a result, overall PCR success rates can be compromised. In this study, we found regionalized GC content to be a good predictor of PCR success across multiple templates. Indeed, any parameter able to be correlated with the GC content of the PCR template, such as TmProduct and TaOpt, was statistically significant when PCR success and failure were compared. However, NormAUCGC was seen to be a better predictor for PCR success (P < 0.001; χ2) for the total data set of 1438 PCRs. NormAUCGC was much more sensitive to fluctuations in GC content than other methods that simply relied on averaging overall GC. The performance of this predictor is expected to improve as template size increases due to the greater likelihood of problematic regions occurring within a given template.
The evidence presented here would suggest that the primer was most often not the cause of PCR failure, but rather the template, i.e. because all primers met similar stringency demands. In all cases, the average values of 63 and 52% for GCprimer1 and GCprimer2 of failed PCRs were acceptable, even if the most stringent primer design criteria were employed (2). Furthermore, Rychlik et al. (11) showed that primer design was significant for a low number of PCR cycles, while this diminished after 25 cycles. When employing nested primer PCR of 30 cycles for each reaction, as here for ligation-independent cloning experiments, strong amplification can be expected to depend less upon stringent primer design due to the addition of 14 and 13 bases to the 5′ ends of the specific forward and reverse primers, respectively, and the associated increase in affinity of primers for template. Thus, provided obvious homologies and self-annealing attributes of primers are minimized, then most 20mer strings will be associated with sufficient target specificity. As a consequence, our results would suggest that more effort should be put towards analysis of the PCR template. Stringent primer design might result in high amounts of very pure PCR product, but it comes at the expense of sequence coverage. The latter is most important when entire ORFs or domains are being targeted and template coverage is essential for recombinant protein synthesis.
For templates possessing a NormAUCGC > 340, it is predicted that the success of PCR will be more dependent on a suitable protocol than on primer selection of primers. Therefore, when faced with the task of large-scale PCR, we recommend dividing the samples into three groups and subsequently optimizing the PCR for successful outcomes in each of the following categories: sequences with NormAUCGC ≤ 340; NormAUCGC between 340 and 750; and NormAUCGC ≥ 750. The first group is likely to be successfully amplified using standard PCR protocols, and as a result primer stringency may be relaxed without deleterious effects, thereby allowing maximal target sequence coverage. The second and third groups should each be optimized in turn, with increasing attention being given to protocol and primer design.
In summary, the NormAUCGC of a PCR template was found to represent a more sensitive predictor of PCR outcome than parameters previously described, while its predictive value as an improvement on GC content alone is likely to increase concomitantly with template size. Although the learning set examined during this study was derived from nested primer PCR, the index, NormAUCGC, is expected to maintain its relevance for standard PCR experiments, as most primer-related failures probably occur during initial cycles only.
Acknowledgments
ACKNOWLEDGEMENTS
Rogier Donders, Ed Moret and David Gestel are acknowledged for their valuable input. We wish to thank Glaucus Proteomics B.V. for financial and logistic support of this study.
REFERENCES
- 1.Chapman T. (2003) Lab automation and robotics: automation on the move. Nature, 421, 661–666. [DOI] [PubMed] [Google Scholar]
- 2.Vieux E.F., Kwok,P.Y. and Miller,R.D. (2002) Primer design for PCR and sequencing in high-throughput analysis of SNPs. Biotechniques, Suppl. 28–30, 32. [PubMed] [Google Scholar]
- 3.Zhang Y., He,Y. and Yeung,E.S. (2001) High-throughput polymerase chain reaction analysis of clinical samples by capillary electrophoresis with UV detection. Electrophoresis, 22, 2296–2302. [DOI] [PubMed] [Google Scholar]
- 4.Markoulatos P., Siafakas,N. and Moncany,M. (2002) Multiplex polymerase chain reaction: a practical approach. J. Clin. Lab. Anal., 16, 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Myakishev M.V., Khripin,Y., Hu,S. and Hamer,D.H. (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res., 11, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslauer K.J., Frank,R., Blocker,H. and Marky,L.A. (1986) Predicting DNA duplex stability from the base sequence. Proc. Natl Acad. Sci. USA, 83, 3746–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freier S.M., Kierzek,R., Jaeger,J.A., Sugimoto,N., Caruthers,M.H., Neilson,T. and Turner,D.H. (1986) Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA, 83, 9373–9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu D.Y., Ugozzoli,L., Pal,B.K., Qian,J. and Wallace,R.B. (1991) The effect of temperature and oligonucleotide primer length on the specificity and efficiency of amplification by the polymerase chain reaction. DNA Cell Biol., 10, 233–238. [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto N., Nakano,S., Yoneyama,M. and Honda,K. (1996) Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res., 24, 4501–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rychlik W. (1993) Selection of primers for polymerase chain reaction. In White,B.A. (ed.), PCR Protocols: Current Methods and Applications. Humana Press, Totowa, NJ, Vol. 15, pp. 31–40. [DOI] [PubMed] [Google Scholar]
- 11.Rychlik W., Spencer,W.J. and Rhoads,R.E. (1990) Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res., 18, 6409–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozen S. and Skaletsky,H. (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol., 132, 365–386. [DOI] [PubMed] [Google Scholar]
- 13.Chenchik A., Diachenko,L., Moqadam,F., Tarabykin,V., Lukyanov,S. and Siebert,P.D. (1996) Full-length cDNA cloning and determination of mRNA 5′ and 3′ ends by amplification of adaptor-ligated cDNA. Biotechniques, 21, 526–534. [DOI] [PubMed] [Google Scholar]
- 14.Baskaran N., Kandpal,R.P., Bhargava,A.K., Glynn,M.W., Bale,A. and Weissman,S.M. (1996) Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res., 6, 633–638. [DOI] [PubMed] [Google Scholar]
- 15.Varadaraj K. and Skinner,D.M. (1994) Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene, 140, 1–5. [DOI] [PubMed] [Google Scholar]
- 16.Mullis K.B., Ferre,F. and Gibbs,R.A. (eds) (1994) The Polymerase Chain Reaction. Birkhauser, Boston, MA. [Google Scholar]
- 17.Rychlik W. and Rhoads,R.E. (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res., 17, 8543–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPherson M.J. and Møller,S.G. (2000) PCR. BIOS Scientific Publishers, Oxford, UK. [Google Scholar]
- 19.White B.A. (ed.) (1993) PCR Protocols: Current Methods and Applications. Humana Press, Totowa, NJ. [Google Scholar]
- 20.Yuryev A., Huang,J., Pohl,M., Patch,R., Watson,F., Bell,P., Donaldson,M., Phillips,M.S. and Boyce-Jacino,M.T. (2002) Predicting the success of primer extension genotyping assays using statistical modeling. Nucleic Acids Res., 30, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varotto C., Richly,E., Salamini,F. and Leister,D. (2001) GST-PRIME: a genome-wide primer design software for the generation of gene sequence tags. Nucleic Acids Res., 29, 4373–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braun P., Hu,Y., Shen,B., Halleck,A., Koundinya,M., Harlow,E. and LaBaer,J. (2002) Proteome-scale purification of human proteins from bacteria. Proc. Natl Acad. Sci. USA, 99, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christendat D., Yee,A., Dharamsi,A., Kluger,Y., Gerstein,M., Arrowsmith,C.H. and Edwards,A.M. (2000) Structural proteomics: prospects for high throughput sample preparation. Prog. Biophys. Mol. Biol., 73, 339–345. [DOI] [PubMed] [Google Scholar]
- 24.Albala J.S., Franke,K., McConnell,I.R., Pak,K.L., Folta,P.A., Rubinfeld,B., Davies,A.H., Lennon,G.G. and Clark,R. (2000) From genes to proteins: high-throughput expression and purification of the human proteome. J. Cell. Biochem., 80, 187–191. [DOI] [PubMed] [Google Scholar]
- 25.Hammarstrom M., Hellgren,N., van Den Berg,S., Berglund,H. and Hard,T. (2002) Rapid screening for improved solubility of small human proteins produced as fusion proteins in Escherichia coli. Protein Sci., 11, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centor R.M. and Schwartz,J.S. (1985) An evaluation of methods for estimating the area under the receiver operating characteristic (ROC) curve. Med. Decis. Making, 5, 149–156. [DOI] [PubMed] [Google Scholar]
- 28.Beck J.R. and Shultz,E.K. (1986) The use of relative operating characteristic (ROC) curves in test performance evaluation. Arch. Pathol. Lab. Med., 110, 13–20. [PubMed] [Google Scholar]
- 29.Rychlik W. (1995) Selection of primers for polymerase chain reaction. Mol. Biotechnol., 3, 129–134. [DOI] [PubMed] [Google Scholar]
- 30.Baldi P. and Brunak,S. (2001) Bioinformatics: The Machine Learning Approach, 2nd Edn. MIT Press, London, UK. [Google Scholar]
- 31.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 32.Baldino F. Jr, Chesselet,M.F. and Lewis,M.E. (1989) High-resolution in situ hybridization histochemistry. Methods Enzymol., 168, 761–777. [DOI] [PubMed] [Google Scholar]