Abstract
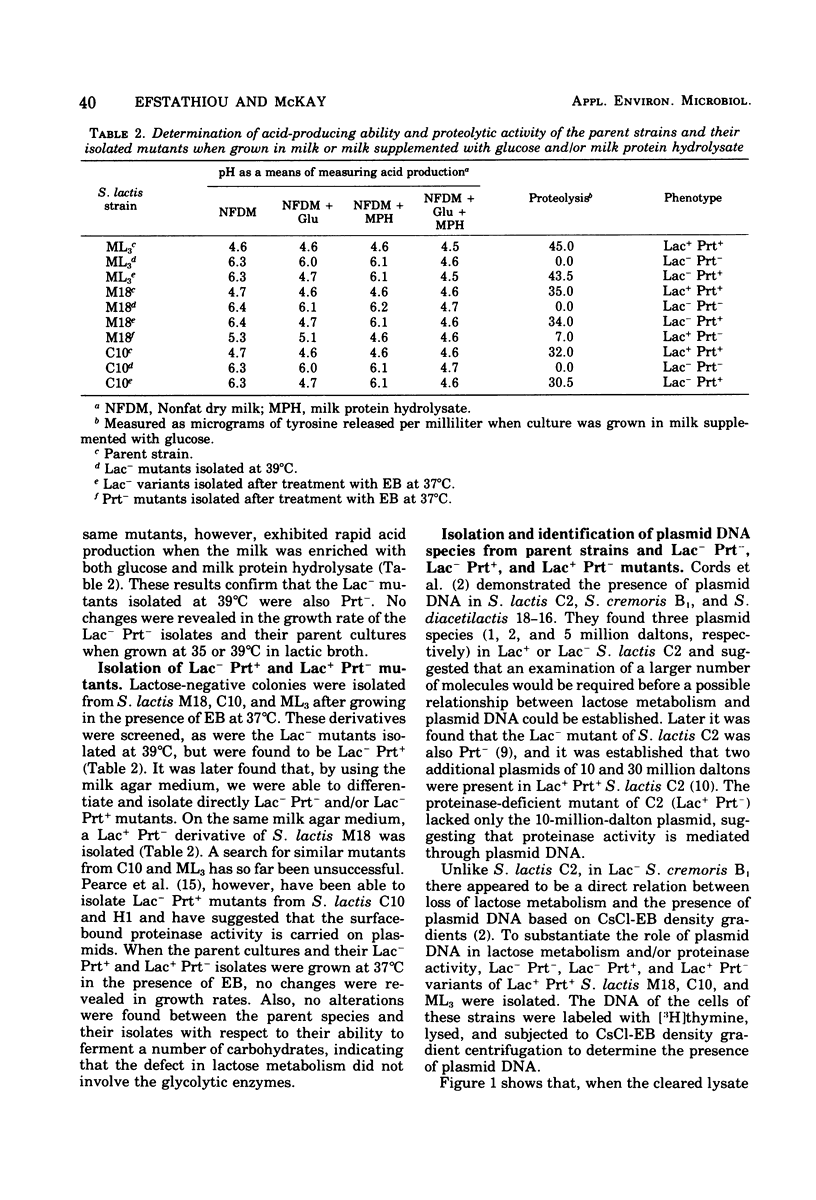
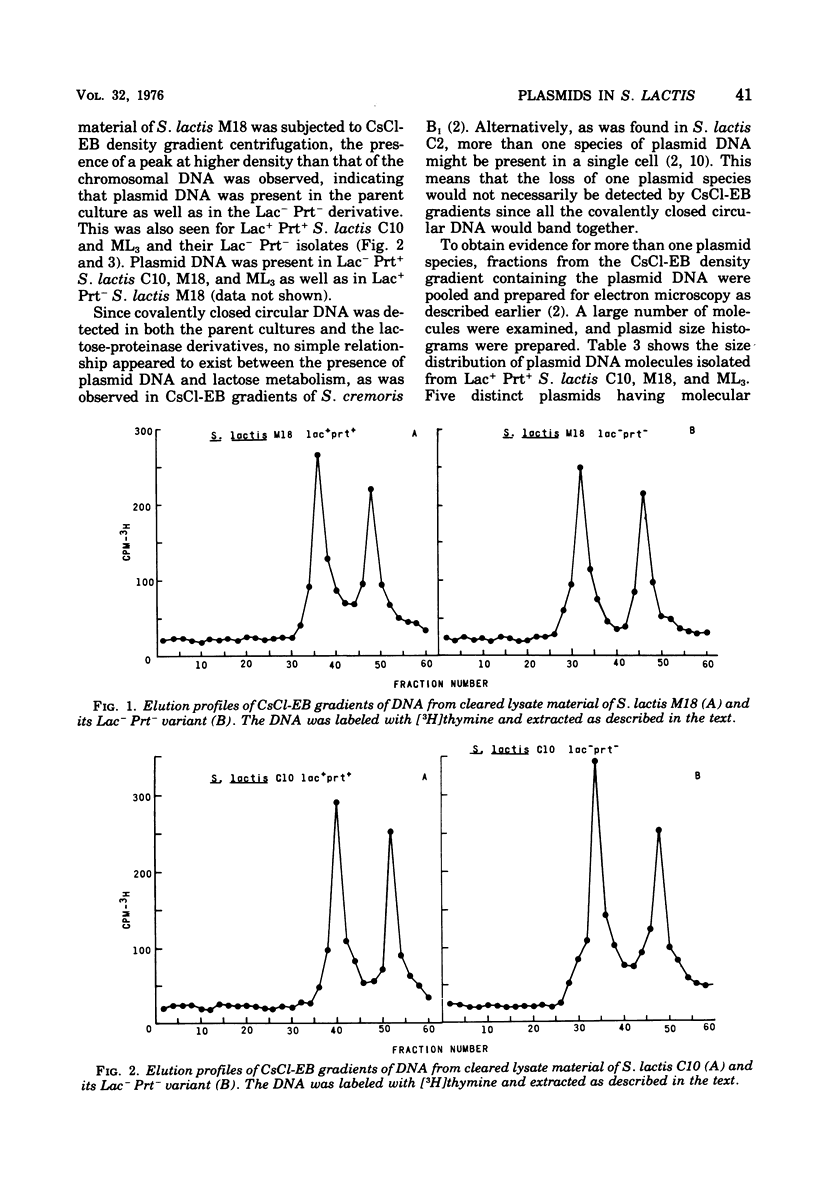
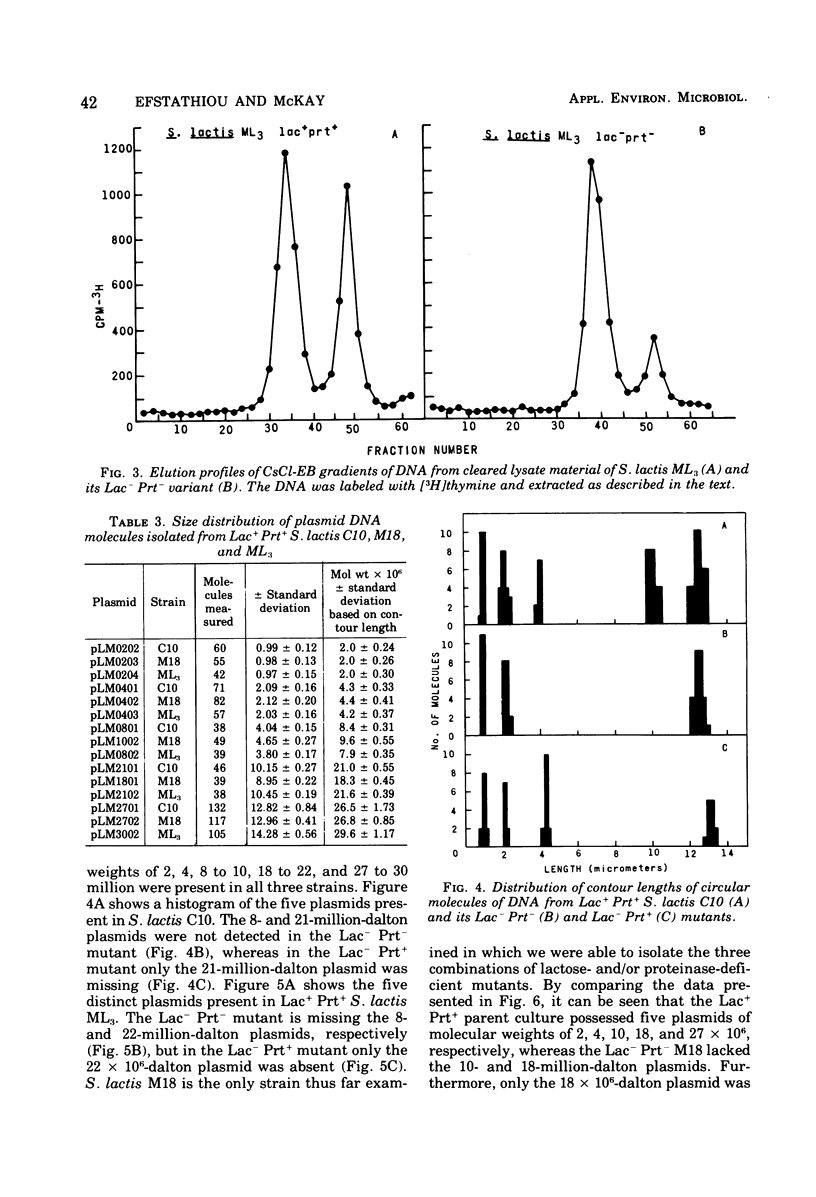
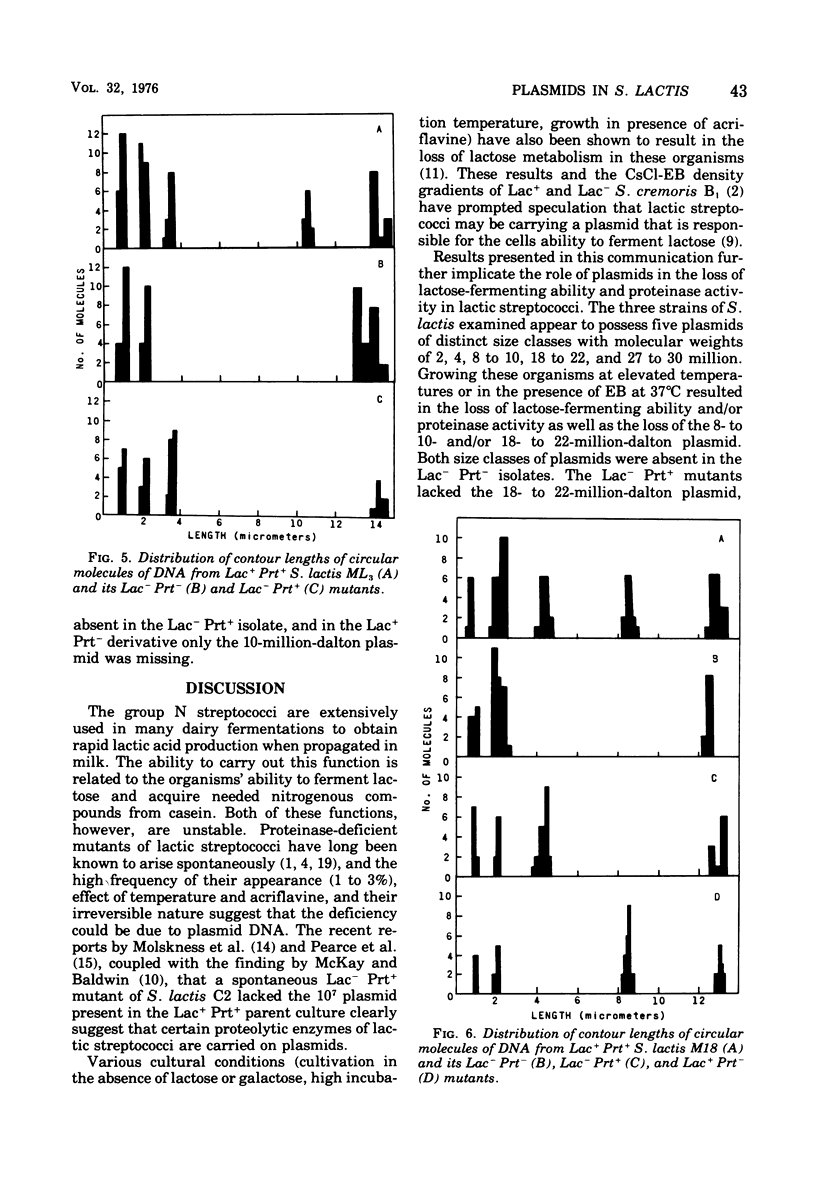
Populations of lactose positive (Lac+) and proteinase positive (Prt+) cells from Streptococcus lactis M18, C10, and ML3 grown at 39 degrees C gave rise to increasing proportions of Lac- Prt- clones. The deficiencies did not appear until after a number of generations at the elevated temperature, and the rate depended on the strain.Lac- Prt+ and Lac+ Prt- mutants were isolated after treatment with ethidium bromide. Plasmid deoxyribonucleic acid was isolated by cesium chloride-ethidium bromide equilibrium density gradient centrifugation from the parent cultures as well as from their Lac- Prt-, Lac- Prt+, and Lac+ Prt- mutants. Five distinct plasmid sizes of approximate molecular weights of 2,4, 8, 21, and 27 million were found in S. lactis C10, whereas the Lac- Prt- derivative lacked the 8- and 21-million-dalton plasmids, but the 8-million-dalton plasmid was present in the Lac-Att mutant. In S. lactis m18 five plasmids possessing molecular weights of about 2, 4, 10, 18 and 27 million were observed. The 10- and 18-million-dalton plasmids were not detected in the Lac- Prt- mutants, whereas the Lac- Prt+ derivative lacked only the 18-million-dalton plasmid and the Lac+ Prt- mutant lacked only the 10-million-dalton plasmid. In S. lactis ML3 five distinct plasmids, with approximate molecular weights of 2, 4, 8, 22, and 30 million, were present. The 8- and 22-million-dalton plasmids were not detected in the Lac- Prt- derivative, but the 8-million-dalton plasmid was present in the Lac- Prt+ mutant. The evidence suggests that lactose-fermenting ability and proteinase activity in these organisms are mediated through two distinct plasmids having molecular weights of 8 x 10(6) to 10 x 10(6) for proteinase activity and 18 x 10(6) to 22 x 10(6) for lactose metabolism.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CITTI J. E., SANDINE W. E., ELLIKER P. R. COMPARISON OF SLOW AND FAST ACID-PRODUCING STREPTOCOCCUS LACTIS. J Dairy Sci. 1965 Jan;48:14–18. doi: 10.3168/jds.s0022-0302(65)88152-8. [DOI] [PubMed] [Google Scholar]
- Cords B. R., McKay L. L., Guerry P. Extrachromosomal elements in group N streptococci. J Bacteriol. 1974 Mar;117(3):1149–1152. doi: 10.1128/jb.117.3.1149-1152.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury D. T., Helinski D. R. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli. I. Isolation and specificity of host and plasmid mutations. Genetics. 1973 May;74(1):17–31. doi: 10.1093/genetics/74.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang D. Molecular weights of coliphages and coliphage DNA. 3. Contour length and molecular weight of DNA from bacteriophages T4, T5 and T7, and from bovine papilloma virus. J Mol Biol. 1970 Dec 28;54(3):557–565. doi: 10.1016/0022-2836(70)90126-9. [DOI] [PubMed] [Google Scholar]
- MAY J. W., HOUGHTON R. H., PERRET C. J. THE EFFECT OF GROWTH AT ELEVATED TEMPERATURES ON SOME HERITABLE PROPERTIES OF STAPHYLOCOCCUS AUREUS. J Gen Microbiol. 1964 Nov;37:157–169. doi: 10.1099/00221287-37-2-157. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Plasmid distribution and evidence for a proteinase plasmid in Streptococcus lactis C2-1. Appl Microbiol. 1975 Apr;29(4):546–548. doi: 10.1128/am.29.4.546-548.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Simultaneous loss of proteinase- and lactose-utilizing enzyme activities in Streptococcus lactis and reversal of loss by transduction. Appl Microbiol. 1974 Sep;28(3):342–346. doi: 10.1128/am.28.3.342-346.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Cords B. R., Baldwin K. A. Transduction of lactose metabolism in Streptococcus lactis C2. J Bacteriol. 1973 Sep;115(3):810–815. doi: 10.1128/jb.115.3.810-815.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molskness T. A., Sandine W. E., Brown L. R. Characterization of lac+ transductants of Streptococcus lactis. Appl Microbiol. 1974 Nov;28(5):753–758. doi: 10.1128/am.28.5.753-758.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L. E., Skipper N. A., Jarvis B. D. Proteinase activity in slow lactic acid-producing variants of Streptococcus lactis. Appl Microbiol. 1974 May;27(5):933–937. doi: 10.1128/am.27.5.933-937.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff D. C., Cowman R. A., Speck M. L. Isolation and partial characterization of a particulate proteinase from a slow acid producing mutant of Streptococcus lactis. J Dairy Sci. 1971 Sep;54(9):1253–1258. doi: 10.3168/jds.S0022-0302(71)86016-2. [DOI] [PubMed] [Google Scholar]



