Abstract
Background
Invitrogen Gateway technology exploits the integrase/att site-specific recombination system for directional cloning of PCR products and the subsequent subcloning into destination vectors. One or three DNA segments can be cloned using Gateway or MultiSite Gateway respectively. A vast number of single-site Gateway destination vectors have been created while MultiSite Gateway is limited to few destination vectors and therefore to few applications. The aim of this work was to make the MultiSite Gateway technology available for multiple biological purposes.
Results
We created a construct, pDONR-R4-R3, to easily convert any available Gateway destination vector to a MultiSite Gateway vector in a single recombination reaction. In addition, we designed pDONR-R4-R3 so that DNA fragments already cloned upstream or downstream of the Gateway cassette in the original destination vectors can still be utilized for promoter-gene or translational fusions after the conversion.
Conclusion
Our tool makes MultiSite Gateway a more widely accessible technology and expands its applications by exploiting all the features of the Gateway vectors already available.
Background
Recombinase-based cloning technologies are becoming increasingly popular because of their easy use and high efficiency. These tools exploit bacterial or viral site-specific recombinases like the bacteriophage P1 Cre, the Saccharomyces cerevisiae FLP or the bacteriophage lambda integrase [1-3]. These enzymes catalyze a reciprocal double-stranded DNA exchange between two specific DNA sites [4].
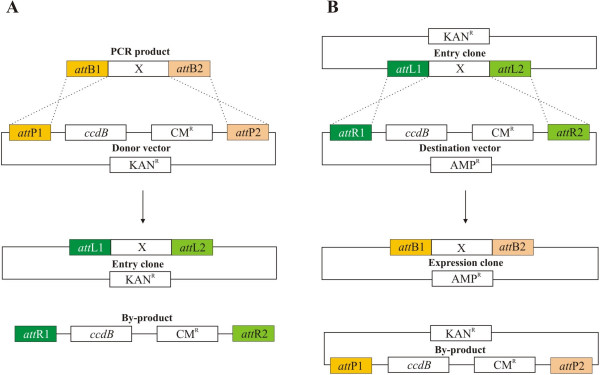
Gateway (Invitrogen) is one of the most popular recombination cloning technologies [3]. It is based on the Escherichia coli bacteriophage lambda integrase/att system [5]. Two sets of reactions are employed in this technology: LR and BP recombinations (Fig 1). The BP reaction is catalyzed by the BP Clonase enzyme mix (Invitrogen), which recombines attB sites with attP sites. The LR Clonase mix (Invitrogen) is responsible for recombination of attL sites with attR sites. Any DNA fragment of interest can be PCR amplified with primers containing attB sites and cloned into a donor vector carrying attP sites in the presence of the BP Clonase mix. The recombination of attB with attP sites leads to the formation of attL and attR sites. The reaction creates an entry clone bearing the insert of interest flanked by attL sites and a byproduct flanked by attR sites (Fig 1A). The insert can then be mobilized into any destination vector having attR sites via an LR reaction (Fig 1B). The resulting construct can be easily selected using a combination of a positive (antibiotic resistance) and a negative (the cytotoxic ccdB gene) selection which does not allow the growth of E. coli transformed with either the initial vectors or the by-product [3,6]. The vast number of Gateway destination vectors designed for many different biological analyses and the large collections of entry clones carrying entire genome open reading frames make this technology highly attractive.
Figure 1.
Gateway BP and LR reactions. (A) BP recombination of a PCR product "X" flanked by attB sites with a Gateway donor vector [3]. (B) LR recombination of an entry clone bearing a DNA fragment "X" with a Gateway destination vector [3].
A variant of Gateway, named MultiSite Gateway (Invitrogen), has been developed to facilitate the cloning of multiple DNA fragments [7]. By using four different attB sites (attB1, attB2, attB3, and attB4), three PCR products flanked by specific attB sites can be cloned into three donor vectors bearing the corresponding attP sites (Fig 2A). The LR Clonase Plus mix (Invitrogen) specifically recombines all attL sites with their corresponding attR sites. A single LR Plus reaction with the three entry clones and the pDEST-R4-R3 destination vector (Invitrogen) creates a construct with the three inserts in a defined and oriented order (Fig 2B). Any Gateway entry clone can be mobilized into a MultiSite Gateway destination vector. Compared to Gateway, MultiSite Gateway is limited by the availability of a small number of destination vectors which prevents this powerful technology from being used for multiple biological purposes.
Figure 2.
MultiSite Gateway. (A) BP recombinations of three PCR products "A", "B" and "C" flanked by attB sites with three Gateway donor vectors [7]. (B) LR plus recombination of three entry clones bearing the DNA fragments "A", "B" and "C" with the MultiSite Gateway destination vector pDEST-R4-R3 (Invitrogen) [7].
Here we present a method to convert any Gateway vector to a MultiSite Gateway vector via a single recombination event.
Results and discussion
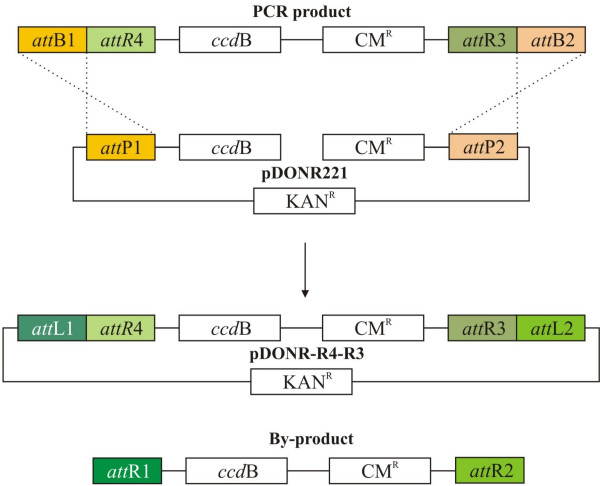
We decided to take advantage of the large number of Gateway single-site destination vectors and developed a new method to easily convert them to MultiSite Gateway. The functional part of MultiSite Gateway destination vectors is the attR4-attR3 cassette which can recombine with three Gateway donor vectors carrying the appropriate attL sites (Fig 2B) [7]. We constructed a vector, pDONR-R4-R3, to mobilize the attR4-attR3 cassette into any Gateway Destination vector. To create pDONR-R4-R3, we cloned a Gateway cassette inside another Gateway cassette by choosing a combination of att sites that cannot recombine with one another. We exploited the ability of the BP Clonase enzyme mix to recombine attB sites with attP sites while leaving attR sites untouched [3]. The attR4-attR3 cassette was amplified using attB primers and cloned into pDONR221 (Invitrogen), a vector carrying an attP1-attP2 cassette. The BP recombination created the pDONR-R4-R3 vector that contains the attR4-attR3 cassette flanked by attL1-attL2 sites (Fig 3).
Figure 3.
pDONR-R4-R3. BP recombination of the attR4-attR3 Gateway cassette flanked by attB1 and attB2 sites with a linear Gateway donor vector pDONR221 (Invitrogen) which leads to the creation of the vector pDONR-R4-R3 and a cytotoxic by-product.
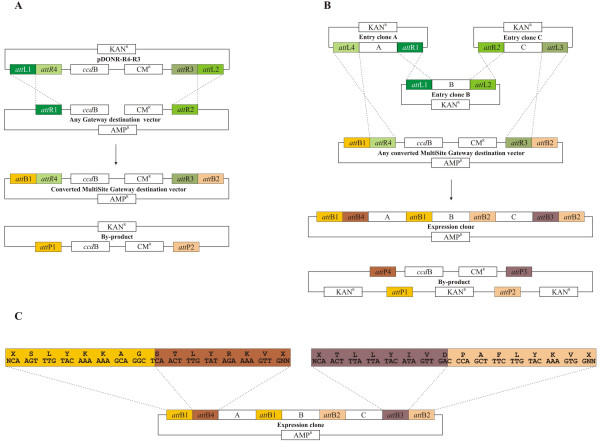
In order to use pDONR-R4-R3 to deliver the attR4-attR3 cassette into any Gateway destination vector in a single LR reaction, we took advantage of the fact that attR4 and attR3 sites cannot recombine with the flanking attL1 and attL2 sites (Fig 2B) [7]. To test it, we performed LR reactions with pDONR-R4-R3 and three different Gateway vectors: pDEST32 (Invitrogen), pDEST22 (Invitrogen) and pMDC163 [8]. These specific destination vectors were chosen to demonstrate the feasibility of the experiment. The linearization of pDEST32 and pDEST22 inside the Gateway cassette and their antibiotic resistance allowed us to specifically select for and recover the entry clones pDEST32-R4-R3 and pDEST22-R4-R3 bearing the attR4-attR3 cassette flanked by attB1-attB2 sites (Fig 4A). Some destination vectors, like pMDC163, carry the Kanamycin resistance gene in discordance with the Invitrogen recommendations. In this case, the linearization of the entry clone efficiently works as a negative selector against the non-recombined entry clone. The linearization of pDONR-R4-R3 in the vector backbone and pMDC163 inside the Gateway cassette allowed us to specifically recover pMDC163-R4-R3 (data not shown). The large number of Gateway vectors already available for various biological experiments can be easily converted to MultiSite Gateway vectors after an LR recombination with pDONR-R4-R3.
Figure 4.
Conversion of any Gateway destination vector to a MultiSite Gateway destination vector. (A) LR recombination of the pDONR-R4-R3 vector with a linear Gateway destination vector. We tested this reaction using pDEST32 (Invitrogen) and pDEST22 (Invitrogen) destination vectors. (B) LR Plus recombination of three entry clones bearing the DNA fragments "A", "B" and "C" with a converted MultiSite Gateway destination vector. We tested this reaction by recombining three entry clones carrying a 3434 bp, 3116 bp and 561 bp DNA fragment with pDEST32-R4-R3 and pDEST22-R4-R3. (C) DNA and amino acid sequence of the external borders of the expression clone.
Last, we performed an LR Plus reaction with pDEST22-R4-R3 and pDEST32-R4-R3 to test the functionality of the attR4-attR3 cassette flanked by attB1-attB2 sites (Fig 4B). We used three entry clones bearing a 3434 bp, 3116 bp and 561 bp DNA fragment cloned into pDONR-P4-P1R (Invitrogen), pDONR221 and pDONR-P2R-P3 (Invitrogen) respectively [7]. Two LR Plus reactions were performed with the entry clones and either pDEST22-R4-R3 or pDEST32-R4-R3. Both reactions led to the creation of pDEST22 and pDEST32 expression clones containing the inserts in the expected order (Fig 4B). Correct functioning of the technology was further tested by cloning a different set of three entry clones into pDEST22-R4-R3 (data not shown). When the converted MultiSite Gateway destination vector bears a kanamycin resistance gene, like pMDC163-R4-R3, the entry clones need to be linearized in order to specifically recover the expression clone.
Figure 4C shows the external attB1-attB4 and attB3-attB2 sites of a converted MultiSite Gateway expression clone which maintain the Gateway frame and lack start and stop codons to allow promoter-gene or translational fusions. Thus, converted MultiSite Gateway vectors can take advantage of all reporter genes, tags or promoter sequences already present in the original Gateway destination vectors.
Conclusion
We have developed a new method to convert virtually all Gateway vectors to MultiSite Gateway. A careful combination of compatible att sites allowed us to use the Gateway recombination technology to deliver a MultiSite Gateway cassette. This strategy opens a new way of thinking about recombination sites as cloning and clonable elements at the same time and could apply to other recombination cassettes. Our method makes the Invitrogen technology available to many biological systems for translational fusion of different proteins or tags, expression of genes or reporter genes under the control of specific promoters and 3'-UTRs, and combinatorial analysis of promoters, genes or other DNA fragments. Our improvement also expands the potential use of this recombination system by utilizing all the features of the large number of Gateway vectors already created.
Methods
Construction of pDONR-R4-R3
We PCR amplified the attR4-attR3 cassette from the pDEST-R4-R3 vector (Invitrogen) using the primers attB1-attR4-F (5'GGGGACAAGTTTGTACAAAAAAGCAGGCTCAACTTTGTATAGAAAAGTTGAAC3') and attB2-attR3-R (5'GGGGACCACTTTGTACAAGAAAGCTGGGTCAACTATGTATAATAAAGTTGAAC3'). We digested the pDONR221 vector (Invitrogen) with EcoRI and used it in a BP recombination reaction (Invitrogen) according to the manufacturer with the attR4-attR3 cassette PCR fragment. We used the reaction to transform DB3.1 E. coli cells (Invitrogen) and selected them for kanamycin resistance. We named the resultant vector pDONR-R4-R3.
Construction of pDEST22-R4-R3, pDEST32-R4-R3 and pMDC163-R4-R3
We digested the pDEST22 (Invitrogen), pDEST32 (Invitrogen) and pMDC163 [8] vectors with EcoRI, XbaI and NcoI respectively. We conducted two independent LR reactions (Invitrogen) according to the manufacturer combining pDONR-R4-R3 with digested pDEST22 and pDEST32. An additional LR reaction was performed combining EcoNI linearized pDONR-R4-R3 with digested pMDC163. We used the three reactions to transform DB3.1 E. coli cells (Invitrogen) and selected them for ampicillin (pDEST22/pDONR-R4-R3 reaction), gentamicin (pDEST32/pDONR-R4-R3 reaction) or kanamycin (pMDC163/pDONR-R4-R3 reaction) resistance. We named the resultant vectors pDEST22-R4-R3, pDEST32-R4-R3 and pMDC163-R4-R3 respectively.
Construction of pDONR-P4-P1R-pPNY, pDONR221-PNY and pDONR-P2R-P3-YC
We PCR amplified 3434 bp upstream the start codon of the Arabidopsis thaliana PENNYWISE (PNY) gene [9] using pny-attB4F (GGGGACAACTTTGTATAGAAAAGTTGTTGGCACGATTCTGAAACACG) and pny-attB1R (GGGGACTGCTTTTTTGTACAAACTTGGGGAAAGGATGATGTCGATGAG) primers, the PNY gene (3116 bp) using pny-attB1F (GGGACAAGTTTGTACAAAAAAGCAGGCTTGATGGCTGATGCATACGAGCC) and pny-attB2R (GGGGACCACTTTGTACAAGAAAGCTGGGTAACCTACAAAATCATGTAGAA) primers and a 561 bp fragment of the pUC-SPYCE vector [10] using attB2-SPYCE-F (GGGGACAGCTTTCTTGTACAAAGTGGGGATGTACCCATACGATGTTCCA) and attB3-SPY-R (GGGGACAACTTTGTATAATAAAGTTGGAATTCCCGATCTAGTAACATAGATG) primers. The PNY promoter region, the PNY gene and pUC-SPYCE PCR fragments were cloned into the pDONR-P4-P1R (Invitrogen), pDONR221 and pDONR-P2R-P3 (Invitrogen) vectors respectively via three independent BP reactions according to the manufacturer. We used the reactions to transform TOP10 E. coli cells (Invitrogen) and selected them for kanamycin resistance. We named the resultant vectors pDONR-P4-P1R-pPNY, pDONR221-PNY and pDONR-P2R-P3-YC respectively.
Construction of pDEST22-pPNY-PNY-YC and pDEST32-pPNY-PNY-YC
We performed an LR Plus reaction (Invitrogen) according to the manufacturer combining pDONR-P4-P1R-pPNY, pDONR221-PNY, pDONR-P2R-P3-YC, and pDEST22-R4-R3. We used the reaction to transform TOP10 E. coli cells and selected for ampicillin resistance. We named the resultant vectors pDEST22-pPNY-PNY-YC. We performed another LR Plus reaction according to the manufacturer combining pDONR-P4-P1R-pPNY, pDONR221-PNY, pDONR-P2R-P3-YC, and pDEST32-R4-R3. We used the reaction to transform TOP10 E. coli cells and selected for gentamicin resistance. We named the resultant vectors pDEST32-pPNY-PNY-YC.
List of abbreviations
AMPr, ampicillin resistance.
CMR, chloramphenicol resistance.
KANR, kanamycin resistance.
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
EM conceived and designed the study, performed the experiments and drafted the manuscript. LB created the entry clones, performed one LR plus reaction and sequenced the final expression clones. SH helped draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Hector Candela, Elisa Fiume, Jennifer Fletcher, Elise Kikis, China Lunde, and Peter Quail for critically reading the article. EM and LB were supported by NRI 04-03387.
Contributor Information
Enrico Magnani, Email: emagnani@berkeley.edu.
Linnea Bartling, Email: lkbartling@gmail.com.
Sarah Hake, Email: maizesh@nature.berkeley.edu.
References
- Liu Q, Li MZ, Leibham D, Cortez D, Elledge SJ. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/S0960-9822(07)00560-X. [DOI] [PubMed] [Google Scholar]
- Sadowski PD. The Flp double cross system a simple efficient procedure for cloning DNA fragments. BMC Biotechnol. 2003;3:9. doi: 10.1186/1472-6750-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark WM, Boocock MR, Sherratt DJ. Catalysis by site-specific recombinases. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226:735–745. doi: 10.1016/0022-2836(92)90629-X. [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;14:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HM, Hake S. The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell. 2003;15:1717–1727. doi: 10.1105/tpc.012856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, Batistic O, Weckermann K, Nake C, Blazevic D, Grefen C, Schumacher K, Oecking C, Harter K, Kudla J. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]