Abstract
To understand how genotype influences fat patterning and obesity, we conducted an autosomal genome scan using male and female F2 hybrids between the C57BL/6ByJ and 129P3/J parental mouse strains. Mice were studied in middle-adulthood and were fed a low-energy, low-fat diet during their lifetime. We measured the weight of the retroperitoneal adipose depot (near the kidney) and the gonadal adipose depot (near the epididymis in males and ovaries in females). An important feature of the analysis was the comparison of linkage results for absolute adipose depot weight and depot weight adjusted for body size, i.e., relative weight. We detected 67 suggestive linkages for six phenotypes, which fell into one of three categories: those specific to absolute but not relative depot weight (Chr 5, 11, and 14), those specific to relative but not absolute depot weight (Chr 9, 15, and 16), and those involving both (Chr 2 and 7). Some quantitative trait loci (QTLs) affected one adipose depot more than another: Retroperitoneal depot weight was linked to Chr 8, 11, 12, and 17, but the linkage effects for the gonadal depot were stronger for Chr 5, 7, and 9. Several linkages were specific to sex; for instance, the absolute weight of gonadal fat was linked to Chromosome 7 in male (LOD = 3.4) but not female mice (LOD = 0.2). Refining obesity as a phenotype may uncover clues about gene function that will assist in positional cloning efforts.
Introduction
Although mice have served as models of human obesity since the 1920s (Danforth 1927), the placement of fat and the weight of depots have not received the same attention as has the morphology of other organs, e.g., bones or heart. Textbooks about the use of the mouse as a model organism contain anatomic descriptions of other organs, but there is little mention of specific adipose depots (Berry 1981; Gruneberg 1943; Iwaki et al. 2001). However, inbred strain surveys demonstrate that adipose depot weight can vary up to fourfold among normal mice (Festing 1979). Several lines of evidence suggest there are genetic influences on the distribution of fat among individual depots. For example, selective breeding of mice changes the weight of one adipose depot with little effect on overall body fatness (Allen and McCarthy 1980), and mice with genetically engineered alleles of specific genes have an unusual fat distribution pattern relative to control mice (Tsai et al. 2004).
Differences among individual fat depots are biologically and clinically significant. Certain genes are expressed in some adipose depots but not in others (Fukuhara et al. 2005; Gesta et al. 2006; Ramis et al. 2002), and these profiles are probably explained in part by the depot-specific functions of adipocytes or other cell types. For instance, adipose depots containing lymph nodes contribute fatty acids to fuel the neighboring immune cells when animals are fighting infection (Pond 2003). Likewise, some adipose depots provide a cushion for bones or organs and are resistant to depletion during starvation compared with other depots, which are mobilized for energy first when food is scarce (Pond 1998). In mice, the retroperitoneal adipose depot increases in response to an energy-dense diet less than does the gonadal depot (Bachmanov et al. 2001). Taken together, genetic and physiologic studies suggest that adipose depots differ in weight and function and that individual differences in adipose depot weight are at least partly due to genotype (Allen and McCarthy 1980; Eisen and Coffey 1990; Festing 1979; West et al. 1994).
The goal of this work was to understand the genetic architecture of individual adipose depots in the mouse and to identify the genomic regions that contribute to individual variation in these traits. To this end, we assessed the heritability and conducted a genome scan for different adipose depots (gonadal and retroperitoneal) in a cross between the C57BL/6ByJ and 129P3/J inbred strains, building on our previous work with these mice (Bachmanov et al. 2001; Reed et al. 2003). Mice were studied when they were about midway through their natural life and were fed a relatively low-fat, low-energy diet. Since adipose depot weight and location differ between male and female mice, we analyzed the data separately by sex, in addition to the combined sample. Adipose depot weight is related to body size, so we compared two sets of linkage results, absolute adipose depot weight and adipose depot weight adjusted for (or relative to) body size. Previous studies have shown that this approach distinguishes loci that affect fatness from those that affect overall size (Stylianou et al. 2006).
Method
Mice
C57BL/6ByJ (B6) and 129P3/J (129) inbred mice were obtained from the Jackson Laboratory (Bar Harbor, ME). The B6 × 129 F1 and F2 hybrids were bred at the Monell Chemical Senses Center. The mice were housed in a temperature-controlled vivarium at 23°C on a 12:12-h light:dark cycle and had free access to water and pelleted Teklad Rodent Diet 8604 (4.4% fat). All protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the Monell Chemical Senses Center. F2 pups were weaned at 21–30 days of age and reared in same-sex groups. A total of 457 F2 mice (228 female and 229 male) were bred from three types of reciprocal crosses: (B6♀ × 129♂) F1♀ × (B6♀ × 129♂) F1♂, (129♀ × B6♂) F1♀ × (129♀ × B6♂) F1♂, and (B6♀ × 129♂) F1♀ × (129♀ × B6♂) F1♂. Parental mice (ten of each strain and sex) were bred and tested simultaneously. As a part of another experiment, all mice were tested to determine their preferences for taste solutions. Results of the behavioral experiments are reported elsewhere (Bachmanov et al. 2002).
Adipose depot dissection
The retroperitoneal and gonadal depots were dissected and weighed in euthanized mice when they were 8–9 months old. The retroperitoneal depot included adipose tissue from the perirenal capsule as well as adipose tissue that attached to the dorsal body wall near the kidneys. The adrenal glands were removed from the tissue in cases where they were embedded in the adipose kidney capsule. The retroperitoneal fat also contains the renal artery and the ureter; no attempt was made to isolate and discard these structures. Gonadal fat in male mice was defined by the proximity to the epididymis and vesicular gland. For female mice, gonadal fat was defined as that which clung to the ovaries and uterus. Although the epididymal (male) and parametrial (female) depots are categorized with a single label (gonadal), these depots are different in structure and possibly differ in function. Adipose tissue associated with the omental membrane, ileum, jejunum, or duodenum was not removed. Variables measured were the weight of the right and left retroperitoneal and gonadal adipose depot (four depots total, weighed individually to the nearest 0.01 g), body weight (to the nearest 0.1 g), and body length (base of the lower incisors to anus, distance to the nearest 1 mm).
Phenotype analysis
Parental strain differences were evaluated by analysis of variance (ANOVA) using strain and sex as factors. In the F2 generation, sex differences were evaluated by t test, and the homogeneity of variances was assessed using the Levene test. Correlations among phenotypes in the F2 generation were assessed (Excel, Microsoft Corp., Redmond, WA) and tested to determine (1) whether the correlation coefficient was significantly different from zero and (2) differed between female and male mice, using previously described methods (Edwards 1973). We used a more stringent p-value criterion than usual (p < 0.01) to control for the effects of multiple testing.
The traits used in the linkage analysis were the weight of the left and right retroperitoneal depot combined, the weight of the left and right gonadal depot combined, and the sum of both depots. Adipose depot weight correlates with litter size, individual age, body weight, and body length. We have calculated two sets of fat-depot weight indexes: (1) absolute values unadjusted for body size and (2) relative values adjusted for body size (bs). The absolute values were calculated as residuals after linear regression analysis with age and litter size as covariates. The relative values were calculated as residuals after linear regression analysis with age, litter size, body weight, and body length as covariates. These residuals were calculated in males and females separately using the data from all F2 mice. To correct for sex differences, the residual values were standardized to a mean of 0 and a standard deviation of 1 within each sex and then were combined. The multiple regression method offers several advantages over ratio methods (e.g., fat weight/body weight) because it allows adjustment for more than one variable (e.g., age and litter size), is less likely to generate spurious correlations among traits, and leads to more accurate assessment of QTL effects (Lang et al. 2005). Linear regression analyses were conducted using Statistica (StatSoft, Tulsa, OK).
Heritability computation
To determine whether the weights of individual adipose depots were heritable in this experimental population and thus suitable for linkage analysis, we computed the degree of genetic determination (Falconer 1989), i.e., broad-sense heritability. This value was estimated based on variances in the parental strains and the F2 generation, and the data were adjusted as described above (absolute and relative to body size), using parental strain data, as well as data from the F2 generation. The environmental (nongenetic) variance was calculated as an average between the trait (total) variances for the two parental strains: VARE = ½(VARB6 + VAR129). The genetic variance was calculated as a difference between the phenotypic variance of the F2 generation and the environmental variance: VARG = VARF2 − VARE. The heritability estimate was calculated as a percentage of the genetic variance from the trait variance of F2: h2 = VARG/VARF2 × 100 (Wright 1968). Because it is not known whether adipose depot heritability in these strains differs by sex, and because one adipose depot is associated with the gonads and thus anatomic differences might be relevant to its weight, we considered male and female mice separately in the analysis. We computed heritability for the adipose depot traits and for the benchmarks of body weight and body length.
DNA extraction and genotyping
Genomic DNA was purified from mouse tails either by phenol/chloroform extraction and precipitation with ethanol (Hogan et al. 1986) or by a sodium hydroxide method (Truett et al. 2000). One hundred thirty-nine markers were selected to span the autosomes. The average distance between markers was 9.6 cM, with no gaps greater than 30 cM (Table 1). Micro-satellite markers were amplified by polymerase chain reaction (PCR) with primers purchased from Invitrogen (Carlsbad, CA) or Research Genetics (Huntsville, AL), with a protocol modified slightly from that of Dietrich et al. (1992). The denatured PCR products were separated by electrophoresis on a 6% polyacrylamide, 8.3 M urea sequencing gel, and the polymorphisms were visualized by autoradiography. Some genotyping was conducted by the Australian Genome Research Facility (Melbourne, Australia) using fluorescently labeled primers. For assessing single nucleotide polymorphisms (SNPs) (rs3705780 and rs3719256), we used fluorescently labeled primers and probes (Assay-by-Design, Applied Biosystems, Foster City, CA), as follows: Genomic DNA (5 ng/μl concentration, 4 μl/well) was transferred to a PCR 96-well optical reaction plate, and each well was supplemented with Taq-Man assay reagents and Universal PCR Master Mix™ to a final volume of 5–50 μl. The PCR product was heated to 50°C for 2 min and 95°C for 10 min, and then 40 amplification cycles were conducted at 95°C for 15 sec and 60°C for 60 sec. DNA amplification was performed in an ABI PRISM 7000 Sequence Detection System; the genotypes were determined by performing allelic discrimination. Genotyping was checked in some cases by DNA sequencing and in other cases by determining whether genotypes were compatible with the pre-existing haplotype. Suspicious genotypes, such as those that created double recombinants, were assayed again as needed.
Table 1.
Markers used in autosomal genome scan
| Marker | Pos. | Marker | Pos. | Marker | Pos. | Marker | Pos. |
|---|---|---|---|---|---|---|---|
| D1Mit294 | 8.3 | D5Mit1 | 5.0 | D10Mit87 | 16.0 | D15Mit13 | 6.7 |
| D1Mit430 | 10.0 | D5Mit128 | 24.0 | D10Mit194 | 29.0 | D15Mit130 | 14.5 |
| D1Mit123 | 21.0 | D5Mit6 | 50.0 | D10Mit186 | 40.0 | D15Mit85 | 15.4 |
| D1Mit19 | 36.9 | D5Mit24 | 60.0 | D10Mit10 | 51.0 | D15Mit29 | 42.8 |
| D1Mit48 | 54.0 | D5Mit214 | 70.0 | D10Mit162 | 59.0 | D15Mit96 | 48.9 |
| D1Mit139 | 65.0 | D5Mit374 | 79.0 | D10Mit180 | 64.0 | D15Mit193 | 57.9 |
| D1Mit102 | 73.0 | D5Mit286 | 86.0 | D10Mit205 | 69.0 | D15Mit35 | 61.7 |
| D1Mit14 | 81.6 | ||||||
| D1Mit15 | 87.9 | D6Mit86 | 0.5 | D11Mit77 | 2.0 | D16Mit55 | 3.4 |
| D1Mit37 | 101.0 | D6Mit77 | 15.8 | D11Mit21 | 20.0 | D16Mit3 | 21.0 |
| D1Mit17 | 106.3 | D6Mit188 | 32.5 | D11Mit23 | 28.1 | D16Mit4 | 27.3 |
| D6Mit177 | 38.5 | D11Mit4 | 37.0 | D16Mit47 | 43.0 | ||
| D2Mit1 | 1.0 | D6Mit36 | 46.0 | D11Mit41 | 49.0 | D16Mit6 | 63.2 |
| D2Mit151 | 15.0 | D6Mit55 | 49.7 | D11Mit199 | 62.0 | D16Mit71 | 70.7 |
| D2Mit7 | 28.0 | D6Mit201 | 74.1 | D11Mit184 | 78.0 | ||
| D2Mit61 | 34.0 | D17Mit46 | 11.7 | ||||
| D2Mit9 | 37.0 | D7Mit76 | 3.4 | D12Mit12 | 6.0 | D17Mit51 | 22.9 |
| D2Mit12 | 50.3 | rs37005780 | 13.1a | D12Mit46 | 16.0 | D17Mit6 | 31.0 |
| D2Mit224 | 74.0 | rs3719256 | 23.5a | D12Mit34 | 29.0 | D17Mit93 | 44.5 |
| D2Mit168 | 81.7 | D7Mit69 | 24.5 | D12Mit14 | 37.0 | D17Mit123 | 56.7 |
| Agouti (A) | 89.0 | Pink-eye (P) | 28.0 | D12Mit194 | 45.0 | ||
| D2Mit197 | 92.0 | Albino (Tyr) | 44.0 | D12Mit20 | 58.0 | D18Mit19 | 2.0 |
| D2Mit148 | 105.0 | D7Nds2 | 37.0 | D18Mit55 | 25.0 | ||
| D7Mit31 | 44.0 | D13Mit44 | 7.0 | D18Mit33 | 44.0 | ||
| D3Mit54 | 4.6 | D7Rp2 | 46.3 | D13Mit38 | 19.0 | D18Mit144 | 57.0 |
| D3Mit203 | 11.2 | D7Mit38 | 50.7 | D13Mit34 | 30.0 | ||
| D3Mit25 | 29.5 | D7Mit7 | 54.0 | D13Mit97 | 40.0 | D19Mit85 | 16.0 |
| D3Mit199 | 33.7 | D7Mit15 | 71.0 | D13Mit147 | 49.0 | D19Mit11 | 41.0 |
| D3Mit10 | 49.7 | D13Mit151 | 71.0 | D19Mit10 | 47.0 | ||
| D3Mit86 | 76.2 | D8Mit95 | 8.0 | D13Mit35 | 75.0 | D19Mit1 | 52.0 |
| D3Mit89 | 86.1 | D8Mit190 | 21.0 | D19Mit35 | 53.0 | ||
| D8Mit29 | 33.0 | D14Mit11 | 0.7 | ||||
| D4Mit264 | 1.9 | D8Mit41 | 41.0 | D14Mit52 | 11.5 | ||
| D4Mit4 | 12.1 | D8Mit271 | 57.0 | D14Mit55 | 10.5 | ||
| D4Mit7 | 35.5 | D8Mit56 | 73.0 | D14Mit82 | 19.5 | ||
| D4Mit58 | 48.5 | D14Mit37 | 27.5 | ||||
| D4Mit204 | 61.9 | D9Mit218 | 4.0 | D14Mit7 | 44.0 | ||
| D4Mit33 | 79.0 | D9Mit25 | 26.0 | D14Mit97 | 58.0 | ||
| D4Mit42 | 81.0 | D9Mit32 | 35.0 | ||||
| D4Ertd296e | 81.7 | D9Mit306 | 42.0 | ||||
| D4Mit256 | 82.7 | D9Mit182 | 55.0 | ||||
| D9Mit37 | 61.0 |
Pos. = position in centimorgans (cM). Map locations are based upon the cM position (starting at the centromere) from the Mouse Genome Database, accessed on 21 February 2006.
Single nucleotide polymorphism markers not integrated in the linkage map; the cM position is given for the nearest (in Mb) genetically mapped marker (UCSC Genome Bioinformatics, see Website References).
We also used the genotypes associated with coat and eye color as markers in the linkage analysis. In addition to the markers genotyped through PCR, several dominant coat and eye color markers were inferred from the appearance of the mice: agouti (A) on Chromosome 2 and tyrosinase (Tyr, formerly albino) and pink-eyed dilution (P) on Chromosome 7. The B6 mice have black eyes and fur determined by genotypes a/a, Tyr/Tyr, and P/P. The 129 mice have pink eyes and albino (genotype Aw/Aw, Tyrc/Tyrc, p/p) or cream (light chinchilla; genotype Aw/Aw, Tyrc-ch/Tyrc, p/p) fur (Roderick and Guidi 1989; Withham 1990). The F2 mice had several eye and coat color phenotypes. The F2 mice with pink eyes were albino (Tyrc/Tyrc), cream (Tyrc-ch/Tyrc), or light buff (Tyrc-ch/Tyrc-ch); agouti alleles could not be determined in these mice and were scored as unknown. The other variants were white-bellied agouti coat and black eyes (Aw/-, Tyr/-, P/-), black coat and black eyes (a/a, Tyr/-, P/-), yellow coat and pink eyes (Aw/-, Tyr/-, p/p), blue-gray coat and pink eyes (a/a, Tyr/-, p/p), chinchilla coat and black eyes (Aw/-, Tyrc-ch/Tyrc-ch, P/- and Aw/-, Tyrc-ch/Tyrc, P/-), and chocolate coat and black eyes (a/a, Tyrc-ch/Tyrc-ch, P/-) (Silvers 1979). For each coat and eye color marker, two genotypes could be distinguished: a homozygous genotype for a recessive allele (nonagouti for the B6 strain and albino and pink-eyed dilution for the 129 strain), and a heterozygous or homozygous genotype for a dominant allele. These alleles were coded by a trained observer, and the genotype was used in the linkage analysis.
Linkage analysis
Linkage maps were created using the computer program MAPMAKER/EXP. Trait analysis was undertaken using MAPMAKER/QTL (Lander et al. 1987) as follows: A genome scan was conducted on a randomly selected subset of mice (n = 164, 86 female and 78 male) using markers from all autosomes and the weight of the two adipose depots separately and the sum of two adipose depots (absolute and relative weight, for a total of six traits). In all linkage analyses, the traits were adjusted for age and litter size through multiple regression as described above, using all F2 mice that were bred and phenotyped. Thresholds for suggestive and significant linkage were used as described previously (Lander and Kruglyak 1995). To define a region of linkage, a confidence interval for each locus was computed. To minimize spurious results, we report the logarithm of the odds (LOD) score at individual markers. The percentage of variance accounted for is also reported at the marker nearest the maximal LOD score, and the direction of dominance is expressed relative to the behavior of the 129 allele; so, for instance, if either one or two copies of the 129 allele reduced the trait to the same extent, the mode of inheritance would be dominant. A separate category was used to identify which allele was associated with higher absolute trait values, referred to as the “plus” allele. In the case above, the 129 allele is dominant but the B6 allele is the “plus” allele. Where there was suggestive evidence for linkage, interactions among marker pairs were examined by two-way ANOVA, with marker genotypes as factors to estimate the contribution of epistasis. In a final and separate analysis, the percent of trait variance explained by all linked loci combined was determined using MAPMAKER/QTL under the unconstrained model.
Sex-dependent linkage analysis and epistasis
Male and female mice were included in the genome scan. Then the linkage analyses were conducted separately for males and females, using a suggestive or significant linkage in one sex and at least a 1-LOD difference, within the confidence interval, for the other sex as a criterion for sex-dependent linkage (Reed et al. 2003). Linkage results from the sex chromosomes (XY) will be the subject of a separate publication.
Results
Measurement of mice
A total of 457 F2 and 40 parental mice were bred, but several died before dissection. The final numbers of mice, strain, and sex-specific means and standard deviations are shown in Table 2. The parental strains differed by strain and sex for all the traits studied. For the F2 generation, the average weight for female mice was 29 g, and that for male mice was 39 g. Male F2 mice were longer than female mice by about 5 mm. The weight of the gonadal depot was, on average, about 1.26 g in F2 male mice, which was significantly higher than that in F2 female mice (0.82 g). Likewise, the retroperitoneal adipose depot was approximately twice the weight in male compared with female F2 mice. Among F2 mice, tests of homogeneity of variance revealed that female mice were significantly more variable than males in the weight of the retroperitoneal depot [F(1,441) = 5.6, p < 0.02], gonadal depot [F(1,441) = 7.1, p < 0.01], and both depots combined [F(1,441) = 8.6, p < 0.01], whereas male mice were more variable in body weight [F(1,441) = 7.85, p < 0.01].
Table 2.
Summary of traits (mean ± SD)
| Phenotype | Female | Male |
|---|---|---|
| Parental strain (C57BL/6ByJ) | n = 9 | n = 10 |
| Age at sacrifice (months) | 7.4 ± 0.9 | 7.9 ± 0.9 |
| Retroperitoneal adipose depot (g)a,b,a*b | 0.16 ± 0.05 | 0.46 ± 0.16 |
| Gonadal adipose depot (g)a,b,a*b | 0.50 ± 0.14 | 1.21 ± 0.38 |
| Sum of adipose depot (g)a,b,a*b | 0.66 ± 0.19 | 1.66 ± 0.54 |
| Body weight (g)a,b,a*b | 26.4 ± 1.6 | 33.7 ± 1.6 |
| Body length (cm)a,b,a*b | 8.4 ± 0.18 | 9.3 ± 0.18 |
| Parental strain (129P3/J) | n = 9 | n = 8 |
| Age at sacrifice (months) | 8.2 ± 0.70 | 8.2 ± 0.64 |
| Retroperitoneal adipose depot (g)a,b,a*b | 0.08 ± 0.06 | 0.23 ± 0.07 |
| Gonadal adipose depot (g)a,b,a*b | 0.43 ± 0.26 | 0.77 ± 0.20 |
| Sum of adipose depot (g)a,b,a*b | 0.51 ± .31 | 1.0 ± 0.27 |
| Body weight (g)a,b,a*b | 22.8 ± 2.2 | 27.4 ± 2.0 |
| Body length (cm)a,b,a*b | 8.6 ± 0.3 | 8.8 ± 0.3 |
| F2 generation | n = 217 | n = 225 |
| Age at sacrifice (months) | 8.9 ± 1.1 | 8.9 ± 1.2 |
| Retroperitoneal adipose depot (g) | 0.17 ± 0.22 | 0.33 ± 0.13* |
| Gonadal adipose depot (g) | 0.82 ± 0.83 | 1.26 ± 0.52* |
| Sum of adipose depot (g) | 0.99 ± 1.02 | 1.59 ± 0.63* |
| Body weight (g) | 29.1 ± 5.4 | 39.1 ± 6.2* |
| Body length (cm) | 8.6 ± 0.5 | 9.2 ± 0.6* |
For the parental strains, superscripts indicate significant effects of strain (a), sex (b), and strain-by-sex (a*b) interactions (p < 0.05; parental data only). For the F2 generation, * denotes significant differences between male and female mice (p < 0.05).
Adipose depot and body size are correlated
Measures of adipose depot weight, body length, and body weight were moderately or strongly correlated. These relationships were present for both female and male mice, but with differences in the strength of the relationship (Table 3). In both sexes, the correlation coefficients between body weight and body length were similar, near 0.50, and body length was also related to adipose depot weight, although to a lesser degree (r = ~ 0.30). However, body weight and adipose depot weight were more strongly correlated in female mice than in male mice (Table 3). We also found that the weights of each adipose depot were correlated with each other but not perfectly so (Fig. 1, Table 3). For both male and female mice, the gonadal adipose depot had a stronger correlation with body weight than did the retroperitoneal adipose depot (Table 3).
Table 3.
Correlations in F2 mice for adipose depot weights by sex
| Phenotype | RP | GON | RP & GON | Body weight | Body length |
|---|---|---|---|---|---|
| Retroperitoneal (RP) | 0.84 | 0.90 | 0.73 | 0.25 | |
| Gonadal (GON) | 0.74 | 0.99 | 0.85 | 0.33 | |
| RP & GON | 0.83 | 0.99 | 0.84 | 0.32 | |
| Body weight | 0.60 | 0.72 | 0.72 | 0.53 | |
| Body length | 0.33 | 0.38 | 0.39 | 0.54 |
RP & GON = combined weight of both adipose depots.
Male values are below the diagonal in boldface; female values are above the diagonal in italics. All correlations are significantly different from 0.0, p < 0.01. Correlations differing significantly between females and males, are underlined, p < 0.01.
Fig. 1.
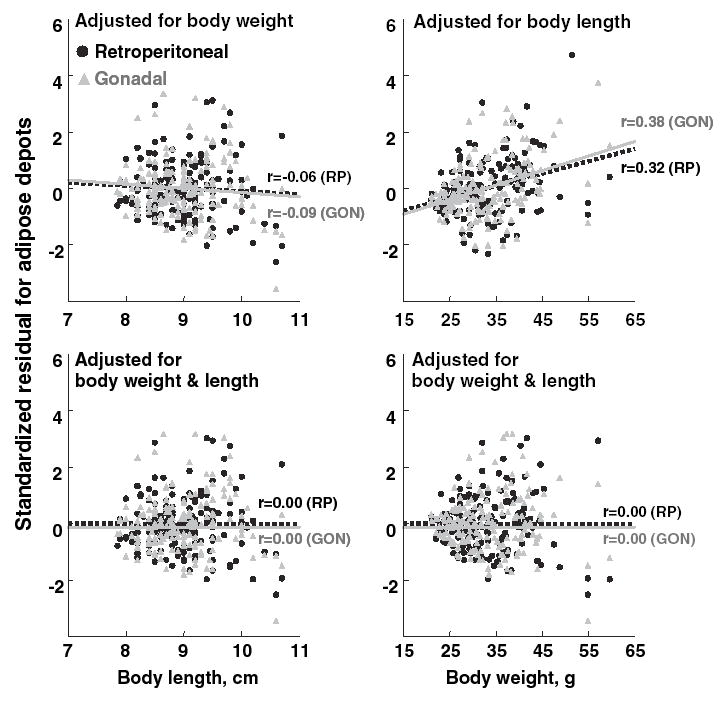
Adipose depot weight of F2, mice expressed as standardized residuals after multiple regression analysis using age and litter size as convariates, is plotted against body length or body weight. Linear regressions between body length or body weight and adipose depot weight are shown as lines, and individual data points are shown as black circles (retroperitoneal) or gray triangles (gonadal). Body weight positively correlates with both the weight of the retroperitoneal adipose depot (r value differs from 0.0; p = 1.9 × 10−7) and the weight of the gonadal depot (p = 1.3 × 10−5). Body length is related to the weight of the retroperitoneal depot but these relationships are not statistically significant (RP, p = 0.22; GON, p = 0.12). Correction for both body weight and body length removes all effects of body size on adipose depot weights (all final r values are 0.0).
Because the weight of adipose depots is related to both body weight and body length, we sought to obtain a relative measure of adipose depot weight that was independent of overall body size. We conducted three general multiple regression analyses to determine the optimal adjustment measures: We adjusted the retroperitoneal or gonadal adipose depot weight by body weight, by body length, and by both body length and body weight. The adjustment that showed the least dependence on body size included both body length and body weight as covariates (Fig. 1, lower left and right panels). Although body length did not contribute significantly to fat depot weight after regression for body weight, it is included in the adjustment procedure to remove its residual effects. Consequently, we used fat depot weight adjusted for both body weight and body length in subsequent analyses.
Heritability
Heritability estimates differed between male and female mice, between the two depots, and between unadjusted or adjusted depot weight (Table 4). There were several patterns: First, heritability was always higher in females regardless of the depot or whether the weight of the depot was relative or absolute. Another consistent pattern in the data was that heritability estimates declined, at least slightly, after adjustment for body size. The final pattern observed is that the relative weight of the gonadal depot was more heritable than the relative weight of the retroperitoneal depot for males, but the reverse was true for females. The most extreme example of sex-by-depot interactions occurred when male and female mice were compared for the absolute weight of the retroperitoneal depot: For female mice, it was nearly 95% determined by genotype, whereas in male mice, genotype accounted for less than 5% of the variation.
Table 4.
Heritable component of adipose depot weight (%)
| Phenotype | Female | Male |
|---|---|---|
| Retroperitoneal (bs) | 81.4 | 0.0 |
| Retroperitoneal | 94.4 | 4.7 |
| Gonadal (bs) | 60.2 | 45.5 |
| Gonadal | 93.2 | 79.2 |
| Retroperitoneal & gonadal (bs) | 61.1 | 25.5 |
| Retroperitoneal & gonadal | 93.4 | 41.0 |
| Body weight | 87.5 | 91.4 |
| Body length | 77.2 | 82.4 |
See text for computation of heritability. bs = phenotype adjusted for body size, see Method section.
Values are percent of trait variance accounted for by genetic effects.
Linkages
The results of the genome scan for loci associated with the weight of the retroperitoneal and gonadal depots are shown in Figure 2 and summarized in Table 5. Twelve of 19 autosomes harbored at least one suggestive locus for adipose depot weight, confirming that these traits are polygenic. Three types of results were found: (1) suggestive evidence only for absolute adipose depot weight (linkages to Chr 5, 11, 14, and 17); (2) suggestive linkage only for relative adipose depot weight (Chr 8, 9, 15, and 16); and (3) suggestive linkages for both (Chr 2, 7, and 12). For 8 of the 12 linkages, the B6 allele was increasing adipose depot weight, which recapitulates the direction of the parental differences (B6 mice have larger depots than 129 mice). For Chr 5, 8, and 9, there was the opposite effect, with the 129 allele increasing adipose depot weight. In some cases, the linkage was most pronounced for the retroperitoneal depot (e.g., the QTL on Chr 11 or 17), and in other cases, for the gonadal depot (e.g., the QTL on Chr 7 or 9). For each trait, the percentage of variance explained by the combined influence of all suggestive loci is shown in Table 6. No significant pairwise interactions between markers were detected using two-way ANOVA (p > 0.05).
Fig. 2.
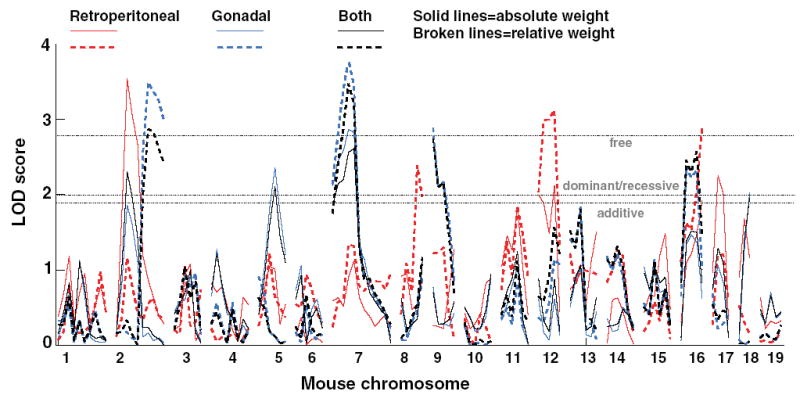
Genome scan results for the retroperitoneal adipose depot (red lines), the gonadal adipose depot (blue lines), and the sum of both depots (black lines), expressed as both absolute (solid lines) and relative to body size (dashed lines). The horizontal lines indicate the suggestive thresholds for additive (1.9), dominant and recessive (2.3), and free (2.8) models. Data are plotted for the free model and therefore some linkages do not meet the criterion for suggestive linkage for a free model but do meet the threshold for a constrained model. See Table 5 and text for other details.
Table 5.
Summary of genome scan results
| Chr | Phenotype | Peak (CI) (cM) | Sex dependent? | Sex | LOD score | Mode | Plus allele | % Variance |
|---|---|---|---|---|---|---|---|---|
| 2 | RP | 38 (21–56) | Yes | Both | 3.5* | free | B6 | 9.5 |
| 2 | RP | 30 (27–34) | Yes | Male | 3.8* | free | B6 | 20.3 |
| 2 | RP | NA | Yes | Female | 0.7 | free | NA | NA |
| 2 | GON (bs) | 91 (56–110) | No | Both | 3.5* | free | 129 | 9.5 |
| 2 | GON | 30 (27–36) | Yes | Both | 1.9* | add | B6 | 5.1 |
| 2 | GON | 30 (25–35) | Yes | Male | 1.9* | add | B6 | 10.8 |
| 2 | GON | NA | Yes | Female | 0.3 | add | NA | NA |
| 2 | RP & GON (bs) | 91 (54–110) | Yes | Both | 2.9* | add | 129 | 7.9 |
| 2 | RP & GON (bs) | 81 (71–86) | Yes | Female | 2.6* | dom | 129 | 14.4 |
| 2 | RP & GON (bs) | NA | Yes | Male | 0.9 | free | NA | NA |
| 2 | RP & GON | 38 (5–59) | Yes | Both | 2.3* | add | B6 | 6.3 |
| 2 | RP& GON | 30 (27–34) | Yes | Males | 2.4* | add | B6 | 20.3 |
| 2 | RP & GON | NA | Yes | Female | 0.4 | free | NA | NA |
| 5 | GON | 75 (0–145) | No | Both | 2.2* | dom | 129 | 6.1 |
| 5 | RP & GON | 72 (58–77) | No | Both | 2.0* | dom | 129 | 5.7 |
| 7 | GON (bs) | 32 (0–42) | Yes | Both | 3.8* | free | B6 | 10.2 |
| 7 | GON (bs) | 32 (18–34) | Yes | Male | 3.2* | free | B6 | 17.2 |
| 7 | GON (bs) | NA | Yes | Female | 0.9 | free | NA | NA |
| 7 | GON | 25 (0–51) | Yes | Both | 2.9* | free | B6 | 7.7 |
| 7 | GON | 33 (32–34) | Yes | Male | 3.4* | free | B6 | 18.0 |
| 7 | GON | NA | Yes | Female | 0.2 | free | NA | NA |
| 7 | RP & GON (bs) | 32 (0–44) | Yes | Both | 3.5* | free | B6 | 9.3 |
| 7 | RP & GON (bs) | 32 (18–36) | Yes | Male | 2.6* | add | B6 | 14.4 |
| 7 | RP & GON (bs) | NA | Yes | Female | 0.9 | free | NA | NA |
| 7 | RP & GON | 45 (0–54) | Yes | Both | 2.6* | add | B6 | 7.2 |
| 7 | RP & GON | 34 (32–36) | Yes | Males | 2.9* | add | B6 | 17.2 |
| 7 | RP & GON | NA | Yes | Female | 0.2 | free | NA | NA |
| 8 | RP (bs) | 42 (39–52) | No | Both | 2.3* | rec | 129 | 9.5 |
| 9 | GON (bs) | 18 (0–76) | No | Both | 2.9* | rec | 129 | 8.1 |
| 9 | RP & GON (bs) | 18 (0–74) | No | Both | 2.7* | rec | 129 | 7.8 |
| 10 | RP (bs) | NA | Yes | Both | 0.4 | dom | NA | NA |
| 10 | RP (bs) | 23 (15–29) | Yes | Female | 2.1* | dom | B6 | 11.3 |
| 10 | RP (bs) | NA | Yes | Male | 0.6 | dom | NA | NA |
| 11 | RP | NA | Yes | Both | 1.7 | dom | NA | NA |
| 11 | RP | 39 (31–44) | Yes | Male | 2.0* | dom | B6 | 11.3 |
| 11 | RP | NA | Yes | Female | 0.1 | free | NA | NA |
| 12 | RP (bs) | 24 (0–70) | Yes | Both | 3.1* | rec | B6 | 8.8 |
| 12 | RP (bs) | 20 (12–30) | Yes | Males | 2.7* | rec | B6 | 19.9 |
| 12 | RP (bs) | NA | Yes | Female | 0.5 | free | NA | NA |
| 12 | RP | NA | No | Both | 2.0 | free | NA | NA |
| 12 | RP | 16 (0–30) | No | Male | 3.3* | free | B6 | 14.0 |
| 12 | RP | NA | No | Female | 2.5 | free | NA | NA |
| 14 | RP | NA | Yes | Both | 0.1 | dom | NA | NA |
| 14 | RP | 27 (19–31) | Yes | Female | 2.3* | dom | B6 | 15.5 |
| 14 | RP | NA | Yes | Male | 0.4 | free | NA | NA |
| 14 | GON | NA | Yes | Both | 0.5 | add | NA | NA |
| 14 | GON | 28 (21–37) | Yes | Female | 2.2* | add | B6 | 11.9 |
| 14 | GON | NA | Yes | Male | 0.0 | free | NA | NA |
| 14 | RP & GON | NA | Yes | Both | 0.4 | dom | NA | NA |
| 14 | RP & GON | 28 (19–33) | Yes | Female | 2.4* | dom | B6 | 13.5 |
| 14 | RP & GON | NA | Yes | Male | 0.0 | free | NA | NA |
| 15 | GON (bs) | NA | Yes | Both | 1.1 | add | NA | NA |
| 15 | GON (bs) | 27 (13–35) | Yes | Male | 2.1* | add | B6 | 11.5 |
| 15 | GON (bs) | NA | Yes | Female | 0.0 | free | NA | NA |
| 15 | RP & GON (bs) | NA | Yes | Both | 1.1 | add | NA | NA |
| 15 | RP & GON (bs) | 27 (10–37) | Yes | Male | 2.1* | add | B6 | 11.5 |
| 15 | RP & GON (bs) | NA | Yes | Female | 0.0 | free | NA | NA |
| 16 | RP (bs) | 65 (5–72) | Yes | Both | 2.8* | add | B6 | 8.6 |
| 16 | RP (bs) | 64 (56–65) | Yes | Male | 2.9* | free | B6 | 27.3 |
| 16 | RP (bs) | NA | Yes | Female | 0.5 | free | NA | NA |
| 16 | GON (bs) | 32 (15–42) | Yes | Both | 2.3* | dom | B6 | 7.7 |
| 16 | GON (bs) | 50 (15–67) | Yes | Male | 2.4* | dom | B6 | 26.3 |
| 16 | GON (bs) | NA | Yes | Female | 0.4 | free | NA | NA |
| 16 | RP & GON (bs) | 32 (27–42) | Yes | Both | 2.5* | dom | B6 | 9.4 |
| 16 | RP & GON (bs) | 50 (41–60) | Yes | Male | 2.9* | free | B6 | 31.4 |
| 16 | RP & GON (bs) | NA | Yes | Female | 0.4 | free | NA | NA |
| 17 | RP | 27 (22–31) | No | Both | 2.1* | dom | B6 | 5.6 |
CI = confidence interval; 1-LOD drop from peak; Chr = chromosome; RP = retroperitoneal adipose depot; GON = gonadal adipose depot; bs indicates that the trait was adjusted for body size; cM refers to the location of the peak linkage based on the experimentally derived map, anchoring the first marker of each chromosome to the cM position found in the Mouse Genome Database. Each peak cM position is associated with a confidence interval. To evaluate sex-dependent linkages, the maximum LOD score for both sexes is provided for comparison. See text for other criteria for sex-dependent locus. N/A = not applicable.
Suggestive linkage (LOD threshold = 1.9 for additive, 2.3 for dominant/recessive, and 2.8 for unconstrained models). LOD scores are for the most likely mode of inheritance as computed by MAPMAKER/QTL: free; recessive (rec); dominant (dom); additive (add). The “plus” allele is defined as the strain-specific allele associated with higher values of the trait. The % variance is the amount of trait variance accounted for by the QTL as estimated by MAPMAKER/QTL. Male and female-dependent results are not reported unless they meet the criterion for sex-dependent linkage. See text for other details.
Table 6.
Percentage of variance accounted for by multiple loci
| Phenotype | Female | Male | Both |
|---|---|---|---|
| Retroperitoneal (bs) | 11.8 | 46.1 | 28.5 |
| Retroperitoneal | 28.3 | 58.8 | 43.9 |
| Gonadal (bs) | 37.9 | 44.0 | 33.5 |
| Gonadal | 18.2 | 26.7 | 14.4 |
| Retroperitoneal and gonadal (bs) | 43.6 | 42.8 | 32.2 |
| Retroperitoneal and gonadal | 14.6 | 27.5 | 17.8 |
Estimate of the percentage of total trait variance accounted for when suggestive linkages are considered together; see text for other details.
Sex-specific linkages to adipose depot weights
Because there were large differences between males and females in heritability of the weight of the retroperitoneal depot and, to a lesser extent, the gonadal depot, we determined whether linkage relationships might be sex-dependent. Of the 12 chromosomes with evidence for linkage, eight contained loci that were sex-dependent (Chr 2, 7, 10, 11, 12, 14, 15, and 16); of these chromosomes, two harbored only female-dependent loci (Chr 10 and 14) and five chromosomes harbored only male-dependent loci (Chr 7, 11, 12, 15, and 16). Chr 2 had two linkages, one male-dependent and the other female-dependent, which differed in location. The male-dependent linkage was more centrally located and the female-dependent linkage was more distally located. Data presented in Table 5 and Figure 3 summarize these sex-dependent effects. Overall, there were three times the number of male-dependent linkages (n = 15) than female-dependent linkages (n = 5). Male-dependent linkages were evenly split between unadjusted and adjusted trait values, whereas female-dependent linkages were found almost exclusively for adjusted adipose depot weights. For male-only and female-only groups, the percent of variance explained when all suggestive loci were considered simultaneously is shown in Table 6.
Fig. 3.
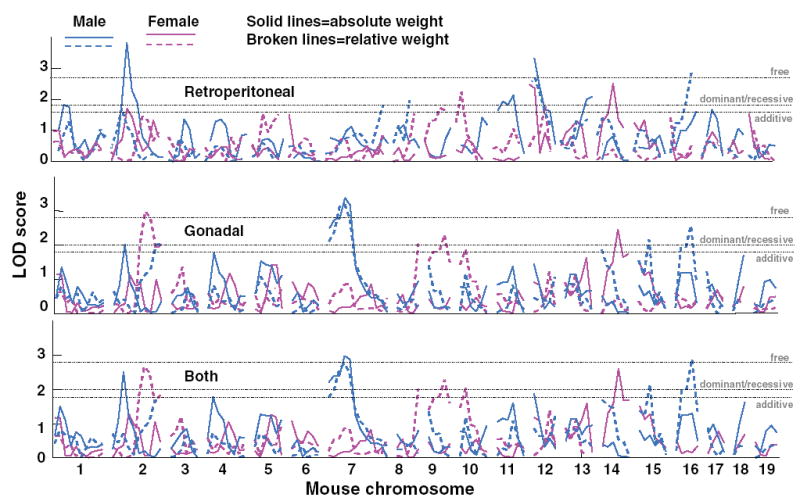
Genome scan results for the retroperitoneal adipose depot (upper panel), gonadal adipose depot (middle panel), and sum of both the retroperitoneal and gonadal adipose depots (lower panel), either absolute (solid lines) or relative to body size (dashed lines), for females (pink lines) and males (blue lines). The horizontal dotted line indicates the threshold for suggestive linkage for different models. See Table 5 and text for other details.
Discussion
The results of the genome scan indicated that the weight of adipose depots, like that of other organs, was under polygenic control, and the linkage relationships were dependent upon sex and adipose depot. In this study, the weight of the adipose depots was more heritable in female than in male mice. This sex difference in heritability, as well as linkage relationships by adipose depot, was found in other crosses of mice (Cheverud et al. 2004). Despite a very low heritability estimate for the retroperitoneal adipose depot in males, we detected several male-dependent linkages for this depot. This low heritability is probably due to high variation in retroperitoneal fat weight in B6 males, which must have inflated nongenetic variance and thus decreased the heritability estimate. Other investigators have also observed a variable fat gain by male B6 mice in response to overfeeding (Burcelin et al. 2002; Koza et al. 2006; Schadt et al. 2003). This variable expressivity of fatness present in the inbred B6 background in males appears to be reduced in a mixed genetic background of F2 mice.
The current study differs from a previous analysis of this genetic cross (Reed et al. 2003) because it uses several different analytical approaches. First, we analyzed retroperitoneal and gonadal fat-depot weights separately, as well as the weights combined. Second, we used both absolute and relative depot weights as variables. Third, the traits were adjusted using multiple regression within each sex rather than using sex as a covariate when the group was analyzed as a whole. Adjusting within each sex is advantageous because there are sex-specific relationships between body length, body weight, and adipose depot weight. These new analytical approaches allowed us to detect several new linkages, e.g., to Chromosome 7. One drawback to the sex-dependent analysis presented here and in other studies of this type is that it results in a reduction in sample size and statistical power, leading to type II error. Thus, there may be more sex-dependent linkages than were detected here because of a relatively small sample size in male-only and female-only groups.
When linkages for absolute or relative adipose depot weights are compared, they fell into three categories: (1) for absolute depot weight only, (2) for relative depot weight only, and (3) for both relative and absolute weights. In the first case, the relevant locus contributed to adiposity because the mouse was larger (and therefore had more grams of fat) or smaller (and therefore had less grams of fat). In the second case, effects of genes influencing adipose depot weight do not depend on body size. In the third case, genes contribute to both body size and relative adipose depot weight, e.g., leading to larger mice that were relatively fatter. Some of the variability in linkage results from different investigators might be due to differences in how they express adipose depot (or other organ) weights and also may be due to variation in body size in the experimental population. In groups where all mice have similar body size, there is less reason to be concerned about the distinction between relative and absolute adipose depot weights. However, in cases where mouse body size differs, the relative size of the fat depots needs to be considered.
The outcome of all studies of this kind depends on the choice of parental strains, which must be considered when comparing the present results with those from other studies. The most common strain of B6 mice (C57BL/6J) was one of the first strains to be used for obesity-related research and is a standard strain choice for investigators (Collins et al. 2004), but we used a slightly different substrain of B6. However, these two B6 substrains are almost identical and probably interchangeable in obesity QTL studies (Bailey 1978; Petkov et al. 2004; Smith et al. 2000). In contrast, the 129 strains (e.g., 129P3/J, 129X1/SvJ, 129S1/SvImJ) are not as interchangeable because they are more genetically distinct (Simpson et al. 1997). The results of a pilot study in our laboratory suggest that mice from different 129 substrains differ in body size and adipose depot weight. Other investigators report 129 substrain differences in spontaneous physical activity (Mouzeyan et al. 2000). The results of previous obesity linkage studies differ depending on the 129 substrain used (Almind and Kahn 2004; Ishimori et al. 2004), and we suspect that the differences among results may be due to the relatively distant genetic relationships among these substrains.
Although the large number of suggestive linkages found here precludes a description of each one, several results are worth highlighting. The linkages with the highest LOD score were for the relative and absolute weights of the gonadal and retroperitoneal depots on Chromosome 2. This region is the most widely reported obesity-related QTL, with 24 positive reports of linkage found through query of the Mouse Genome Database (MGD; see Website References). The large number of linkage reports to Chromosome 2 could be the result of multiple genes that influence body size and fatness, a hypothesis supported by studies using congenic lines of mice (Diament et al. 2004; Jerez-Timaure et al. 2004) and the observation (here) that there appear to be two distinct linkages, one for the retroperitoneal adipose depot near the middle of the chromosome, and the other for the gonadal depot, located more distally. The chromosome with the next largest number of obesity-related QTL reports was Chromosome 7 (16 results from the MGD) which was also detected in this cross. A candidate gene (Atp10c) has been found (Dhar et al. 2000) that maps near the peak LOD score reported here. As was the case with Chromosome 2, however, there may be multiple linked loci on chromosome 7 (Diament and Warden 2004). Both of these linkages (Chr 2 and 7) are near coat color markers, agouti (A), in the case of Chromosome 2, and pink-eye dilution (P) and albino (Tyr) in the case of Chromosome 7, however suggesting that these genes are candidates for adipose depot weight is premature. Another area of linkage was on Chromosome 9. There have been 13 QTLs linked to obesity for this chromosome, and it is the focus of the fine-mapping work reported in the companion article in this issue (McDaniel et al. 2006). Chromosome 9 was chosen for further study because of the strength of the linkage results and because of the lack of prior fine-mapping and positional cloning attempts. Pursuing the gene(s) on Chromosome 9 that contribute to differences among strains in obesity is therefore likely to uncover novel results. Other linkages found in this report are novel, especially a linkage for the size of the retroperitoneal depot to Chromosome 12, which is an example of a linkage that would have been missed if depots were pooled rather than analyzed individually.
Mouse models of human obesity vary in what aspects they are intended to represent. Most studies focus on younger mice to reduce the time invested in each experiment and the costs of housing the mice. This study is unusual because these mice are older, almost middle-aged by human standards. Because they were fed a standard rodent chow (containing 4.4% fat) throughout life, those mice that become the fattest could therefore be considered cases of spontaneous rather than diet-induced obesity. Therefore, this current model represents middle-age obesity not exacerbated by a high-calorie, palatable, or high-fat diet. These mouse strains could also be used to study dietary obesity because the parental strains differ markedly in adiposity when offered a high-fat supplement to the diet (Bachmanov et al. 2001), and they differ in the sensory and metabolic responses to both high-sugar and high-fat foods and drinks (Bachmanov et al. 1996, 2001; Lewis et al. 2005, 2006; Sclafani and Glendinning 2005). Identifying genes and alleles that contribute to mouse strain differences in adipose depot weight, specifically through the use of consomic and congenic mice as a part of a positional cloning strategy, will be of future benefit in the study of human obesity, which shares similar features.
Website References
http://www.genome.ucsc.edu/: University of California at Santa Cruz Genome (UCSC) Bioinformatics
http://www.informatics.jax.org/: Mouse Genome Database
Acknowledgments
Grants from the National Institutes of Health funded this research (R01DK 058797 to DRR, R01AA011028 and R01DC00882 to AAB, and R01DK046791 and R01AA12715 to MGT). The authors acknowledge Dr. Kelly Ewen-White and Paige Stevenson at the Genotyping Section at the Australian Genome Research Facility for part of the genotyping. Maria Theodorides provided excellent technical assistance. Early data collection for this experiment was conducted in the laboratory of R. Arlen Price, and his support is gratefully acknowledged. Patricia Watson provided helpful editorial advice. Discussions with Hong Ji, Mark I. Friedman, and Caroline M. Pond enhanced the quality of this work.
References
- 1.Allen P, McCarthy JC. The effects of selection for high and low body weight on the proportion and distribution of fat in mice. Anim Prod. 1980;31:1–11. [Google Scholar]
- 2.Almind K, Kahn CR. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes. 2004;53:3274–3285. doi: 10.2337/diabetes.53.12.3274. [DOI] [PubMed] [Google Scholar]
- 3.Bachmanov AA, Reed DR, Tordoff MG, Price RA, Beauchamp GK. Intake of ethanol, sodium chloride, sucrose, citric acid, and quinine hydrochloride solutions by mice: a genetic analysis. Behav Genet. 1996;26:563–573. doi: 10.1007/BF02361229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmanov AA, Reed DR, Tordoff MG, Price RA, Beauchamp GK. Nutrient preference and diet-induced adiposity in C57BL/6ByJ and 129P3/J mice. Physiol Behav. 2001;72:603–613. doi: 10.1016/s0031-9384(01)00412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachmanov AA, Reed DR, Li X, Li S, Beauchamp GK, et al. Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ x 129P3/J F2 intercross. Genome Res. 2002;12:1257–1268. doi: 10.1101/gr.129702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey DW. Sources of subline divergence and their relative importance for sublines of six major inbred strains of mice. In: Morse HC, editor. Origins of inbred mice. New York: Academic Press; 1978. [Google Scholar]
- 7.Berry RJ. Biology of the House Mouse. In: Berry RJ, editor. Symposium Proceedings: Zoological Society of Lodon. Vol. 47. London: Zoological Society of London; 1981. [Google Scholar]
- 8.Burcelin R, Crivelli V, Dacosta A, Roy-Tirelli A, Thorens B. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am J Physiol Endocrinol Metab. 2002;282:E834–E842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 9.Cheverud JM, Ehrich TH, Kenney JP, Pletscher LS, Semenkovich CF. Genetic evidence for discordance between obesity- and diabetes-related traits in the LGXSM recombinant inbred mouse strains. Diabetes. 2004;53:2700–2708. doi: 10.2337/diabetes.53.10.2700. [DOI] [PubMed] [Google Scholar]
- 10.Collins S, Martin TL, Surwit RS, Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiol Behav. 2004;81:243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Danforth CH. Hereditary adiposity in mice. J Hered. 1927;18:153–162. [Google Scholar]
- 12.Dhar M, Webb LS, Smith L, Hauser L, Johnson D, et al. A novel ATPase on mouse chromosome 7 is a candidate gene for increased body fat. Physiol Genomics. 2000;4:93–100. doi: 10.1152/physiolgenomics.2000.4.1.93. [DOI] [PubMed] [Google Scholar]
- 13.Diament AL, Warden CH. Multiple linked mouse chromosome 7 loci influence body fat mass. Int J Obes Relat Metab Disord. 2004;28:199–210. doi: 10.1038/sj.ijo.0802516. [DOI] [PubMed] [Google Scholar]
- 14.Diament AL, Farahani P, Chiu S, Fisler J, Warden CH. A novel mouse Chromosome 2 congenic strain with obesity phenotypes. Mamm Genome. 2004;15:452–459. doi: 10.1007/s00335-004-2352-x. [DOI] [PubMed] [Google Scholar]
- 15.Dietrich W, Katz H, Lincoln SE, Shin H-S, Friedman J, et al. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992;131:423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards AL. Statistical Methods. New York: Holt: Rinehart and Winston; 1973. [Google Scholar]
- 17.Eisen EJ, Coffey MT. Correlated responses in body composition based on selection for different indicator traits in mice. J Anim Sci. 1990;68:3557–3562. doi: 10.2527/1990.68113557x. [DOI] [PubMed] [Google Scholar]
- 18.Falconer D. Introduction to Quantitative Genetics. New York: Wiley & Sons; 1989. [Google Scholar]
- 19.Festing MFW. The inheritance of obesity in animal models of obesity. In: Festing MFW, editor. Animal models of obesity. Vol. 1. New York: Oxford University Press; 1979. pp. 15–37. [Google Scholar]
- 20.Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, et al. Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science. 2005;307:426–430. doi: 10.1126/science.1097243. [DOI] [PubMed] [Google Scholar]
- 21.Gesta S, Bluher M, Yamamoto Y, Norris AW, Berndt J, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci USA. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruneberg H. The genetics of the mouse. Cambridge: Cambridge University Press; 1943. [Google Scholar]
- 23.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 24.Ishimori N, Li R, Kelmenson PM, Korstanje R, Walsh KA, et al. Quantitative trait loci that determine plasma lipids and obesity in C57BL/6J and 129S1/SvImJ inbred mice. J Lipid Res. 2004;45:1624–1632. doi: 10.1194/jlr.M400098-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Iwaki T, Yamashita H, Hayakawa T. A color atlas of sectional anatomy of the mouse. Tokyo: Adthree Publishing Co., Ltd; 2001. [Google Scholar]
- 26.Jerez-Timaure NC, Kearney F, Simpson EB, Eisen EJ, Pomp D. Characterization of QTL with major effects on fatness and growth on mouse chromosome 2. Obes Res. 2004;12:1408–1420. doi: 10.1038/oby.2004.177. [DOI] [PubMed] [Google Scholar]
- 27.Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2:e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 29.Lander E, Green P, Abrahamson J, Barlow A, Daley M, et al. MAPMAKER: An interactive complex package for constructing primary linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 30.Lang DH, Sharkey NA, Mack HA, Vogler GP, Vandenbergh DJ, et al. Quantitative trait loci analysis of structural and material skeletal phenotypes in C57BL/6J and DBA/2 second-generation and recombinant inbred mice. J Bone Miner Res. 2005;20:88–99. doi: 10.1359/JBMR.041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis SR, Ahmed S, Dym C, Khaimova E, Kest B, et al. Inbred mouse strain survey of sucrose intake. Physiol Behav. 2005;85:546–556. doi: 10.1016/j.physbeh.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Lewis SR, Ahmed S, Khaimova E, Israel Y, Singh A, et al. Genetic variance contributes to ingestive processes: A survey of 2-deoxy-d-glucose-induced feeding in eleven inbred mouse strains. Physiol Behav. 2006;87:595–601. doi: 10.1016/j.physbeh.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 33.McDaniel AH, Tordoff MG, Bachmanov AA, Reed DR. A locus on mouse chromosome 9 (Adip5) affects the relative size of the gonadal but not retroperitoneal adipose depot. Mamm Genome. 2006;17:xxx–xxx. doi: 10.1007/s00335-006-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouzeyan A, Choi J, Allayee H, Wang X, Sinsheimer J, et al. A locus conferring resistance to diet-induced hypercholesterolemia and atherosclerosis on mouse chromosome 2. J Lipid Res. 2000;41:573–582. [PubMed] [Google Scholar]
- 35.Petkov PM, Cassell MA, Sargent EE, Donnelly CJ, Robinson P, et al. Development of a SNP genotyping panel for genetic monitoring of the laboratory mouse. Genomics. 2004;83:902–911. doi: 10.1016/j.ygeno.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Pond CM. The fats of life. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 37.Pond CM. Paracrine interactions of mammalian adipose tissue. J Exp Zool A Comp Exp Biol. 2003;295:99–110. doi: 10.1002/jez.a.10215. [DOI] [PubMed] [Google Scholar]
- 38.Ramis JM, Franssen-van Hal NL, Kramer E, Llado I, Bouillaud F, et al. Carboxypeptidase E and thrombospondin-1 are differently expressed in subcutaneous and visceral fat of obese subjects. Cell Mol Life Sci. 2002;59:1960–1971. doi: 10.1007/PL00012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed DR, Li X, McDaniel AH, Lu K, Li S, et al. Loci on chromosomes 2, 4, 9 and 16 for body weight, body length and adiposity identified in a genome scan of an F2 intercross between the 129P3/J and C57BL/6ByJ mouse strains. Mamm Genome. 2003;14:302–313. doi: 10.1007/s00335-002-2170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roderick TH, Guidi JN. Strain distribution of polymorphic variants. In: Lyon MF, Searle AG, editors. Genetic variants and strains of the laboratory mouse. New York: Oxford University Press; 1989. pp. 663–772. [Google Scholar]
- 41.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 42.Sclafani A, Glendinning JI. Sugar and fat conditioned flavor preferences in C57BL/6J and 129 mice: oral and postoral interactions. Am J Physiol Regul Integr Comp Physiol. 2005;289:R712–R720. doi: 10.1152/ajpregu.00176.2005. [DOI] [PubMed] [Google Scholar]
- 43.Silvers WK. The coat colors of mice, a model for mammalian gene action and interaction. New York: Springer-Verlag; 1979. [Google Scholar]
- 44.Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, et al. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- 45.Smith BK, Andrews PK, West DB. Macronutrient diet selection in thirteen mouse strains. Am J Physiol Regul Integr Comp Physiol. 2000;278:R797–R805. doi: 10.1152/ajpregu.2000.278.4.R797. [DOI] [PubMed] [Google Scholar]
- 46.Stylianou IM, Korstanje R, Li R, Sheehan S, Paigen B, Churchill GA. Quantitative trait locus analysis for obesity reveals multiple networks of interacting loci. Mamm Genome. 2006;17:22–36. doi: 10.1007/s00335-005-0091-2. [DOI] [PubMed] [Google Scholar]
- 47.Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, et al. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- 48.Tsai YS, Kim HJ, Takahashi N, Kim HS, Hagaman JR, et al. Hypertension and abnormal fat distribution but not insulin resistance in mice with P465L PPARgamma. J Clin Invest. 2004;114:240–249. doi: 10.1172/JCI20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.West DB, Goudey-Lefevre J, York B, Truett GE. Dietary obesity linked to genetic loci on chromosome 9 and 15 in a polygenic mouse model. J Clin Invest. 1994;94:1410–1416. doi: 10.1172/JCI117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Withham B. Coat colors of sublines of 129 mice. 1990. JAX Notes 170, No 441. [Google Scholar]
- 51.Wright S. Evolution and the Genetics of Populations. Vol. 1. Chicago: University of Chicago Press; 1968. [Google Scholar]


