Abstract
The gene encoding mt-tRNALeu(UUR), MT-TL1, is a hotspot for pathogenic mtDNA mutations. Amongst the first to be described was the 3302A>G transition which resulted in a substantial accumulation in patient muscle of RNA19, an unprocessed RNA intermediate including mt-16S rRNA, mt-tRNALeu(UUR) and MTND1. We have now been able to further assess the molecular aetiology associated with 3302A>G in transmitochondrial cybrids. Increased steady-state levels of RNA19 was confirmed, although not to the levels previously reported in muscle. This data was consistent with an increase in RNA19 stability. The mutation resulted in decreased mt-tRNALeu(UUR) levels, but its stability was unchanged, consistent with a defect in RNA19 processing responsible for low tRNA levels. A partial defect in aminoacylation was also identified, potentially caused by an alteration in tRNA structure. These deficiencies lead to a severe defect in respiration in the transmitochondrial cybrids, consistent with the profound mitochondrial disorder originally associated with this mutation.
INTRODUCTION
Mutations of mitochondrial DNA (mtDNA) are responsible for a broad range of diseases usually affecting multiple sites, particularly the central nervous system, muscle and retina. In some cases disease is limited to one tissue, even a single cell type (1). Although we continue to find new mutations in mtDNA, our understanding of how mitochondrial tRNA mutations cause their cellular defect has not grown to the same extent.
In the present work, we further studied the 3302A>G mutation of the MT-TL1 gene that affects the 3′ terminal part of the encoded mt-tRNALeu(UUR) (2). This mutation is rare (2,3) and causes a severe myopathy with respiratory insufficiency. The rarity of this mutation and severity of its phenotype are in keeping with the fact that this nucleotide is highly conserved among mammals (4). Our previous studies showed that the 3302A>G mutation probably interferes with processing of an RNA precursor, RNA 19, that contains 16S rRNA, mt-tRNALeu(UUR) and MTND1, and also showed that different precursor processing pathways are present in skeletal muscle and fibroblasts (2). Cytoplasts generated from fibroblasts of the patient were fused to 143B osteosarcoma rho0 cells and the cybrid clones were used in the present study to analyse in detail the molecular consequences of the mutation.
MATERIALS AND METHODS
Cell culture, generation of cybrid clones and measurement of proliferation rate
Cybrids carrying the 3302A>G mutation were generated by fusion of enucleated fibroblasts from the index patient with 143B rho0 cells (5). Cells were grown in DMEM, 5% fetal calf serum (FCS), 110 μg/ml sodium pyruvate, 50 μg/ml uridine and 1% penicillin/streptomycin at 37°C and 5% CO2. Using the cylinder method 60 individual cell clones were isolated and initial analysis for level of heteroplasmy showed that all clones analysed carried ∼85% mutant load, the same level that was present in the patient's fibroblasts. To generate clones with higher levels of heteroplasmy, the bulk cybrid population was cultivated in the presence of 100 ng/ml ethidium bromide (EtBr) for 2 weeks. Subsequently, EtBr was removed and cells were allowed to increase their mtDNA content for 2 weeks before being cloned once more. Again, individual clones were generated and analysed for their mutant content.
The following control cell lines cultivated in the same medium were included in further experiments: the parental 143B cells, 143B rho0 cells repopulated with wild-type (wt)-mtDNA from fibroblasts of an unrelated individual (control 1), 143B rho0 cells repopulated with wt-mtDNA from the patient's healthy maternal aunt (control 2), 143B rho0 cells repopulated with 100% 3243A>G mtDNA (A3243G 100%), 143B rho0 cells repopulated with 100% wt-mtDNA obtained during fusion experiments aimed at generating a cybrid clone containing the 3243A>G MELAS mutation (A3243G wt). The A3243G clones were kindly given to us by Dr Eric Shoubridge, the fibroblast fusions were carried out by Dr Karl Morton.
To analyse cell proliferation, cells were seeded at a density of ∼5000 cells per cm2, cultivated with daily changes of growth medium, harvested by trypsinization at selected time points and counted using a CASY1 cell counter (Schärfe System GmbH, Reutlingen, Germany). This device also measures cell size distribution by pulse area analysis, making it possible to discriminate living from dead cells.
To analyse the degradation rate of mitochondrial transcripts, 2 × 105 cells were pre-seeded in 60 mm dishes. The medium was changed after 24 h and cybrids were cultivated in EtBr (50 ng/ml) for up to 48 h before extraction of RNA.
Fibroblasts of healthy control individuals were cultivated in DMEM supplemented with 10% FCS, 1× non-essential amino acids, 2 mM l-glutamine and 1% penicillin/streptomycin at 37°C and 5% CO2. Primary human myoblast cultures were established from muscle samples obtained from healthy individuals undergoing orthopaedic hip surgery using standard techniques (6), which had been approved by the local ethics committee of the Medical Faculty of the University of Köln. Myoblasts were cultivated in Ham's F10 medium supplemented with 20% FCS, 2% chicken embryo extract, 2 mM l-glutamine and 1% penicillin/streptomycin at 37°C and 5% CO2. Myoblasts were fused to multi-nucleated myotubes by changing to DMEM supplemented with 4% horse serum, 100 μg/ml human transferrin, 10 μg/ml human insulin, 4 mM l-glutamine and 1% penicillin/streptomycin. Myotubes were identified by the presence of a minimum number of three nuclei within each fused cell when inspected under the microscope.
Analysis of heteroplasmy levels in different cybrid clones
The level of mutation load was determined using a radioactive PCR/RFLP (restriction fragment length polymorphism) method. Total DNA was extracted from individual cell populations as described previously (2). For the 3302A>G mutation a 725 bp fragment encompassing the mutation site was amplified with the forward primer mt-3232-s: 5′-TAAGATGGCAGAGCCCG-3′ and the reverse primer mt-3957-as: 5′-TCGGGCGTATTCGATGTT-3′. Samples were subjected to 30 cycles of amplification [3 min at 94°C; 45 s at 94°C, 1.5 min at 57°C, 1.5 min at 72°C (30×), final extension at 72°C for 7 min]. After renewed addition of 30 pmol of each primer, 5 μCi [α-32P]dCTP (5000 Ci/mmol; Hartmann Analytics, Braunschweig, Germany) and 1 U Taq polymerase (Promega, Heidelberg, Germany), the PCRs were subjected to an additional cycle of amplification. Labelled amplicons were precipitated and equal amounts (2000 counts) digested with 10 U MseI (New England Biolabs, Frankfurt am Main, Germany). Restriction fragments were separated on 3.3% non-denaturing polyacrylamide gels, dried on a support and analysed with ImageQuant software following exposure to a PhosphorImager (Molecular Dynamics, Krefeld, Germany). Three MseI sites are present in the wt product generating fragments of 32, 35, 172 and 486 bp. The 3302A>G mutation destroys an MseI site and results in the three remaining fragments of 32, 172 and 521 bp.
An analogous method was employed to analyse the A3243G wt or mutant cybrids using the following primers: mt-2928-s 5′-CCTAGGGATAA GCGCA-3′ and mt-3305-as: 5′-TAATACGACTCACTATATTGTTAAGAAGAGGAATTG-3′ and HaeIII digestion as described previously (7). Resolution of digestion products was carried out on 8% non-denaturing polyacrylamide gels.
Biochemical characterization of cybrid clones
Cells were grown until they were almost confluent, and the medium was changed the day before the measurements. Cells were harvested by trypsinization, centrifuged and washed twice in phosphate-buffered saline (PBS). The final cell pellet contained 106–107 cells and was resuspended at a density of 104–105 cells/μl in PBS. The cellular density of the suspension was evaluated by automated counting (CASY, Schärfe Systems, Reutlingen, Germany) while the protein content was determined by Bradford assay (Bio-Rad, München, Germany). This cell suspension was then used in part for oxygen consumption assays and was in part frozen for further spectrophotometric studies. Oxygen consumption studies were done using two Clark chambers (Hansatech, King's Lynn, UK), recording the intact cell respiration rate and then, after permeabilization with digitonine, the rates for pyruvate (+malate), malate (+glutamate), succinate and glycerol-3-phosphate as substrates, as described in detail previously (8).
Spectrophotometric measurements were performed using a Beckmann DU-600 spectrophotometer (Beckmann Instruments, Fullerton, CA, USA) for determination of the enzymatic activities of succinate–cytochrome c reductase (SCCR, CII+III), decylubiquinol–cytochrome c reductase (QCCR, C III), cytochrome c oxidase (COX, C IV), as well as citrate synthase (CS) and lactate dehydrogenase (LDH) as reference enzymes (8).
Analysis of mitochondrial transcripts, tRNA levels and properties by northern blotting
Total RNA was isolated from frozen skeletal muscle tissue of the patient and of muscle samples obtained from healthy individuals, ground to a fine powder in liquid nitrogen, and from cultured cells (cybrids, fibroblasts, myoblasts, myotubes; ∼2 × 106 cells) using Trizol reagent (Invitrogen, Karlsruhe, Germany). For the analysis of mRNA and precursor RNAs, 5 μg of total RNA was separated through 1.2% agarose gels containing 6.5% formaldehyde, 1× MOPS buffer (40 mM morpholinosulfonic acid, 10 mM sodium acetate and 1 mM EDTA, pH 7.0) and 1× MOPS as a running buffer. Separated samples were capillary blotted on to GeneScreen-plus membrane (Perkin Elmer, Zaventem, The Netherlands) in 10× SSPE and immobilized by UV fixation. For the northern-blot analysis of polyadenylated RNA species poly(A) RNA was isolated from 100 μg of total RNA using the Oligotex mRNA Mini Kit (Qiagen, Hilden, Germany). The total yield from the extraction procedure was subjected to northern-blot analysis as described above.
For quantification of tRNAs and determination of RNA length, samples of 0.25–2 μg of total RNA were denatured (90°C for 5 min) and separated on 13% denaturing polyacrylamide gels containing 8 M urea, using 1× TBE as a running buffer (termed neutral to distinguish from acidic gels). Separated samples were electroblotted onto GeneScreen-plus membrane in 0.25× TBE and immobilized by UV fixation. For the analysis of tRNA structure, unheated samples of 0.5 μg total RNA were subjected to electrophoresis through a 8% non-denaturing polyacrylamide gel using 1× TBE as gel and running buffer. Blotting and immobilization was performed as before.
To investigate aminoacylation of tRNAs, we extracted total RNA from cultured cells under acidic conditions as described previously (9). RNA isolated using Trizol reagent was kept on ice during the procedure and the final RNA pellet was resuspended in 0.3 M sodium acetate (pH 4.5) and 1 mM EDTA. Total acidic RNA (10 μg) was electrophoresed through a 6.5% acidic, denaturing polyacrylamide gel (20 × 43 cm2, 1 mm thickness) containing 8 M urea and 0.1 M sodium acetate/0.1 M acetic acid, pH 5.0, as running buffer at 4°C and 30 mA over 48 h. Final running distance of xylenecyanol was 23 cm. Deacylation of individual RNA samples was carried out in 0.3 M Tris–HCl, pH 9.0, at 75°C for 15 min. Separated samples were electroblotted and immobilized onto GeneScreen-plus membranes as described above.
Templates for in vitro transcription were amplified by PCR from a synthetic single stranded 144mer DNA encoding a hammerhead ribozyme downstream of a T7-promotor and followed by the sequence of wt human mt-tRNALeu(UUR) or the 3302A>G mutation (10). Upon transcription with T7-RNA polymerase, the hammerhead ribozyme cleaves at its designated target site to release the tRNA sequence with a 5′-hydroxyl end (11). After purification over a 12% 8 M urea PAGE, transcripts were passively eluted by agitation in 0.5 M NH4OAc. 5′-Labelling with ATP and T4-PNK and subsequent purification by PAGE were performed as described previously (12).
Regions of mtDNA encompassing various mRNA and tRNA genes amplified by PCR were used as probes for the northern blots. As template for the PCRs, DNA derived from 143B control cells was used. The mRNAs probes were amplified using the following forward and reverse primers.
For ND1: mt-3384-s: 5′-AATTCTAGGCTATATACAAC-3′ and mt-7395-as: 5′-ATCCATATAGTCACTCCA-3′, which was digested to give specific probes for ND1, ND2 and COI; for COI: mt-5907-s: 5′-TAAGGGAGGGTAGACACG-3′ and mt-6299-as: 5′-TCGCCGACCGTTGACTAT-3′. For 28S rRNA a 497 bp fragment was amplified using the forward primer 5′-AAGATGGTGAACTATGCCTG-3′and reverse primer 5′-GCAGGTGAGTTGTTACACAC-3′. Transfer RNA probes were generated as follows: for tRNALeu(UUR): mt-3232-s: 5′-TAA GAT GGC AGA GCC CG-3′ and mt-3305-as: 5′-TAA TAC GAC TCA CTA TAT TGT TAA GAA GAG GAA TTG-3′; for tRNAVal: mt-1602-s: 5′-CAGAGTGTAGCTTAACACAAA-3′ and mt-1670-as: 5′-TCAGAGCGGTCAAGTTAA-3′; for tRNALys: mt-8259-s: 5′-TTACCCTATAGCACCCCCT-3′ and mt-8390-as: 5′-ATACGGTAGTATTTAGTTGGG-3′.
For the normalization of tRNAs to a non-mitochondrial small RNA species blots were hybridized to a 5S rRNA probe. The oligonucleotide 5′-GGGTGGTATGGCCGTAGAC-3′ (13) was end labelled using [γ-32P]ATP (5000 Ci/mmol, Hartmann Analytics, Braunschweig, Germany) and T4-PNK. Purified PCR products (QIAquick PCR purification columns, Qiagen, Hilden, Germany) were radiolabeled with [α-32P]dCTP (5000 Ci/mmol; Hartmann Analytics, Braunschweig, Germany) by the random-primer method, and unincoorporated nucleotides were removed from labelling reactions by gel filtration through ChromaSpin columns (Clontech, Heidelberg, Germany).
Hybridization was carried out at 65°C (exceptions: 50°C: ND1, 55°C: tRNALeu(UUR)) overnight in 10 ml of a solution of 1% SDS, 10% dextran sulphate and 1 M NaCl containing 2 × 106 c.p.m. radiolabelled probe plus 4 mg sonicated salmon sperm DNA. After hybridization, two 10-min washes were performed at 45°C–65°C with 2× SSC, 0.1% SDS, followed by a 3–10-min wash at 45°C–65°C with 0.1× SSC, 0.1% SDS. Blots were subjected to PhosphorImager analysis and/or autoradiography.
RESULTS
Genotype, proliferation and mitochondrial activity in cybrid clones
After treatment with and recovery from EtBr, last hot cycle PCR analysis showed that 3 of 35 clones harboured the 3302A>G mutation at levels close to homoplasmy. One clone carried the mutation at a much lower level (77%, Supplementary Figure 1). All other clones were 85–95% mutant. A clone containing the 3243A>G MELAS mutation at 100% (3243 100%) was included in further experiments to allow comparison of two mutations affecting the same tRNA. Proliferation rates for the clones containing high levels of the mutated mtDNA showed no significant difference when compared with the wild-type control strains (data not shown).
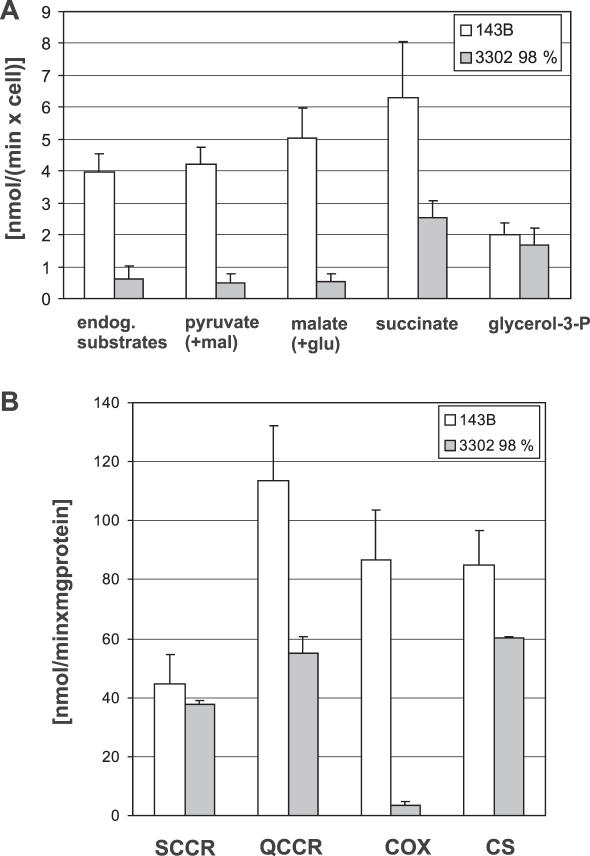
Two cell lines were used to characterize extensively the biochemical defect, parental 143B cells and clone 3 containing 98% of the 3302A>G mutation (Figure 1). In intact cells, 3302A>G cells showed a 6-fold reduction of respiration, from ∼4 to 0.6 fmol O2/cell × min, compared with parental 143B cells. Oxidation of pyruvate, malate and succinate, measured following permeabilization were severely reduced (to ∼10, 10 and 40% of mean control values, respectively), while oxidation of glycerol-3-phosphate was similar to the control (Figure 1A). Spectrophotometric assays of the activities of Complexes II+III (SCCR), Complex III (QCCR) and Complex IV (COX) of the mitochondrial respiratory chain revealed marked deficiency of Complex (C) III and C IV, while C II+III remained unchanged (Figure 1B). The clone containing the mutation at 77% showed no respiration defect.
Figure 1.
Respiratory chain phenotype of cybrid cell lines. (A) Polarography. Graphs show mean values and standard deviations for oxygen consumption of 143B control cells (white bars, n = 10) and of cybrids containing the 3302A>G mutation at 98% (grey bars, n = 3) in intact cells and permeabilized cells respiring on the indicated substrates. (mal = malate; glu = glutamate). (B) Respiratory chain complex activities. Graphs show activities for SCCR—Complex II+III, QCCR—Complex III, COX—Complex IV and CS as mitochondrial matrix marker. Graphs are mean values and standard deviations for 143B control cells (white bars, n = 5) and cybrids containing the 3302A>G mutation (grey bars, n = 3).
In conclusion, the 3302A>G mutation causes a dramatic defect in respiratory chain function in 143B osteosarcoma cybrid cells when present at high levels, as it did in skeletal muscle of the patient (14).
Levels of mt-tRNALeu(UUR) in cybrid clones and patients muscle
Densitometric analysis of a PAGE-northern blot showed that tRNALeu(UUR) was reduced in clones bearing the 3302A>G mutation (Figure 2A). This was more obvious in cells with near homoplasmic levels of the mutation, where the amount was <60% of controls, than in the clone with 77% heteroplasmy. The level of mt-tRNALeu(UUR) was even lower in the 3243 100% clone (Figure 2), confirming earlier studies with this mutation (15,16). In our initial analysis of the patient's muscle (2), the extraction procedure used (17) did not allow recovery of small RNAs, thus no information was available concerning tRNA levels in vivo. We now used a different method to extract total RNA from the remainder of the skeletal muscle biopsy that had been stored in liquid N2 for almost 15 years. Analysis showed that the tRNAs were still intact and yielded sharp bands (Supplementary Figure 2, upper panel). The level of mt-tRNALeu(UUR) is also reduced in the muscle of the patient when compared with healthy controls (Supplementary Figure 2, lower panel). This is seen when normalized to nuclear encoded 5S rRNA (Supplementary Figure 2, black bars), but becomes especially evident when normalized to two other randomly selected mitochondrial tRNAs, mt-tRNALys or mt-tRNAVal, encoded on the same DNA strand (Supplementary Figure 2, white and grey bars). As is often seen for randomly selected human individuals, the variability between the two controls is high, but still allows the conclusion that the 3302A>G mutation leads to decreased levels of mt-tRNALeu(UUR) also in the patient's muscle, while interestingly a general increase of other mitochondrial tRNAs is observed.
Figure 2.
Determination of steady-state levels of tRNALeu(UUR) in cybrid clones. (A) Levels of tRNAs analysed by high-resolution PAGE followed by blot hybridization in two control cell lines and the cybrid clone containing 77% of the 3302A>G mutation and 98% of the 3302A>G mutation, as well as a clone containing the 3243A>G mutation (3243 100%). RNA was loaded in two concentrations in order to ensure linearity of the densitometric signal. (B) The densitometric values for 1 μg of 143B RNA were arbitrarily set to 1.0 in order to get an estimate of the variability of the measurements. Upper panel: levels of mt-tRNALeu(UUR) (black bars) and tRNAVal (grey bars) were normalized to nuclear encoded 5S rRNA; lower panel: Levels of mt-tRNALeu(UUR) were normalized to tRNAVal.
These findings suggest that either the rate of synthesis or the stability of mt-tRNALeu(UUR) is impaired by the 3302A>G mutation.
Accumulation of the unprocessed precursor RNA19 and stability of mt-tRNALeu(UUR)
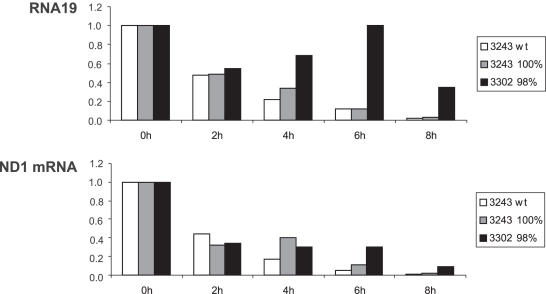
The most profound finding in our initial description of the patient was that the mutation at nt 3302 leads to a dramatic, up to 1000-fold, accumulation of RNA19 in muscle (2). This precursor transcript consists of the mt-tRNALeu(UUR) flanked by 16S rRNA and ND1 mRNA (18). Northern-blot analysis of cybrid cell RNA consecutively hybridized with mt-tRNALeu(UUR) and ND1 mRNA probes (Figure 3) showed that increased amounts of RNA19 were also present in the cybrid cells containing the 3302A>G mutation at 77 and 98%, but at much lower levels than in the patient's muscle. Accumulation of RNA19 was also found in the clone bearing the 3243A>G mutation at homoplasmic levels, confirming earlier reports (18–21), however the ratio RNA19/ND1 was clearly higher in the presence of the 3302A>G mutation.
Figure 3.
Analysis of mitochondrial transcript processing in cybrid clones, human skeletal muscle and primary human myoblasts, myotubes and fibroblasts. Levels of the precursor transcript RNA19 and its processing intermediates as well as ND1 and COX I mRNA analysed by agarose gel electrophoresis followed by blot hybridization. Samples were from two healthy human skeletal muscles (lanes 1 and 3), primary human myoblasts from these biopsies (lanes 4 and 5), primary human myotubes differentiated in vitro from such myoblasts (lanes 6 and 7), primary human fibroblasts (lanes 8 and 9), control cell lines (143B, 3243 wt, lanes 10 and 12) and from the cybrid clone containing 77% of the 3302A>G mutation (lane 13), 98% of the 3302A>G mutation (lane 14), as well as from the clone containing the A3243G mutation (3243 100%, lane 11). RNA from 143B rho0 cells was also loaded (lane 15). 5 μg of total RNA was loaded in each lane. Probes used for hybridization are indicated on the right, RNA species identified on the left.
In our earlier study, we also showed that RNA19 appears to be cleaved by different pathways in skeletal muscle compared with proliferating fibroblasts (2). This would lead to different intermediates and we postulated that this might be the underlying mechanism explaining the striking tissue selectivity of this mutation. When hybridized with a mt-tRNALeu(UUR) probe (Figure 3), northern-blot analysis of skeletal muscle RNA confirmed that RNA19 is predominantly cleaved to a 1.07 kb intermediate, probably consisting of mt-tRNALeu(UUR) + ND1 mRNA. In all other samples, including human primary myoblasts, human myotubes and human primary fibroblasts, in addition to the 143B clones, this RNA species was virtually absent and instead a 1.6 kb intermediate was present. Similar to RNA19, this intermediate, comprising tRNALeu(UUR) + 16S rRNA was especially prominent in the clones containing mt-tRNALeu(UUR) mutations. The resolution of the gel was insufficient to differentiate between the mature ND1 mRNA and the muscle specific precursor tRNALeu(UUR) + ND1 mRNA because of the small difference in length, thus the ND1 probe only shows one band in all samples. Levels of the mRNA for MTCOI were similar in all cell lines, while the ND1 mRNA was slightly increased in the clones containing high levels of the 3302A>G and 3243A>G mutations. Mitochondrial transcripts were absent in 143B rho0 cells (right lane), as expected.
In conclusion, RNA19 accumulates in 143B cybrid cells containing the 3302A>G mutation, however to a lesser degree than in the patients muscle. The skeletal muscle specific processing pattern for this transcript could only be observed in vivo in mature muscle, and could not be recapitulated in cultured cells, not even in differentiated primary human myotubes.
We postulated earlier that accumulation of RNA19 could result in impaired release and synthesis of mature mt-tRNALeu(UUR) (2) and might thereby explain decreased steady-state levels of this tRNA. To study this question, mtDNA transcription was blocked by EtBr and RNA decay analysed over an 8 hr time period. In three independent experiments, we found RNA19 to be more stable in the clones containing the 3302A>G mutation compared with either a control line or cells containing the 3243A>G mutation. One experiment is shown in Figure 4; this demonstrates that RNA19 does not display an exponential decay in 3302 cybrid cells, but appears to be stabilized after EtBr treatment, leading to increased steady-state levels between 4 and 6 h. The half-life of the mature ND1 mRNA was also found to be increased in the clone containing the 3302A>G mutation, whereas half-life of COX I mRNA was similar (data not shown). Thus, our results suggest that the 3302A>G mutation does indeed cause a processing defect, leading to an increased stability and, consequently, increased steady-state levels of RNA19.
Figure 4.
Determination of the half-life of RNA19 and ND1 mRNA in cybrid clones treated with EtBr for up to 8 h. Decay of mitochondrial transcripts was analysed by agarose gel electrophoresis followed by blot hybridization to a probe for ND1 mRNA in isogenic control cells containing 100% wt-mtDNA (3243 wt), in the cybrid clone containing the 3302A>G mutation (3302 98%), as well as in the clone containing the 3243A>G mutation (3243 100%).
A decreased rate of RNA19 processing due to mutations of mt-tRNALeu(UUR) may cause a protein synthesis defect, not only due to lowered availability of the tRNA (loss of function), but also due to the accumulation of the precursor itself [gain-of-function, (22)]. RNA19 could adversely affect translation in two ways: If the 16S rRNA component is incorporated into the ribosome this may cause stalling of a resulting abnormal ribosome (18). Alternatively, polyadenylated RNA19 may potentially be loaded onto mitoribosomes. Although little is known concerning the structural requirements for mRNA binding in mammalian mitochondrial ribosomes, polyadenylation seems to be important. We therefore analysed poly(A+)-RNA prepared from cybrid cell RNA by northern blotting and found that RNA19 is indeed present in higher amounts in the poly(A+)-fraction from cells containing the 3302A>G mutation than in cells containing the 3243A>G mutation or in control cells, especially when compared with polyadenylated ND1 mRNA (Figure 5).
Figure 5.
Analysis of polyadenylation of RNA19. Polyadenylation of RNA19 was analysed by agarose gel electrophoresis of total RNA (5 μg) and poly(A+)-RNA (the total yield from 100 μg of total RNA) in 143B control cells, in the cybrid clone containing the 3243A>G mutation, as well as in the clone containing 98% of the 3302A>G mutation. Probes used for hybridization are indicated on the right, RNA species identified on the left.
In addition to sequestration in RNA19, reduced levels of mt-tRNALeu(UUR) may be caused by an increased rate of degradation due to destabilization of the tRNA structure by the mutation. We measured degradation rate by analysing the decay of tRNAs following inhibition of mtDNA transcription by low concentrations of EtBr (Figure 6A). Inhibition was maintained for 48 h and two independent experiments showed identical results. Decay of mt-tRNALeu(UUR) was compared with that of two other tRNAs encoded on the same strand (tRNAVal and tRNALys). There were no obvious differences in tRNA decay in clones containing high levels of the 3302A>G or 3243A>G mutation nor between degradation rate for mt-tRNALeu(UUR), tRNAVal and tRNALys situated upstream and downstream of the mutated tRNA (Figure 6B). For the 3243A>G mutation, this is in contrast to the work of (16), who however used HeLa cybrids. Differences in phenotypic consequences of the same mutation in different nuclear backgrounds have been discussed extensively (22).
Figure 6.
Determination of the half-life of mitochondrial tRNAs in cybrid clones treated with EtBr for up to 48 h. (A) Decay of mitochondrial tRNAs was analysed by high-resolution PAGE followed by blot hybridization in 143B control cells, in the cybrid clone containing 98% of the 3302A>G mutation, as well as in the clone containing the 3243A>G mutation (3243 100%). RNA (0.5 μg) was loaded in triplicate per sample in order to minimize data acquisition errors of the signals. (B) Densitometric data for tRNALeu(UUR), tRNAVal and tRNALys were normalized to nuclear encoded 5S rRNA.
In conclusion, the 3302A>G mutation leads to decreased levels of the tRNA, but this is mostly due to decreased release from its precursor RNA19 and not due to decreased stability caused by the mutation.
Aminoacylation levels and structural changes
To investigate the possibility of additional, qualitative changes in mt-tRNALeu(UUR) function, we analysed the level of tRNA aminoacylation. RNA was extracted under acidic conditions and separated on a 6.5% acidic, denaturing polyacrylamide containing 8 M urea (Figure 7). Deacylated RNA samples of all cell lines plus a mixture of native and deacylated RNA from 143B cells were also loaded to facilitate identification of the two species. Surprisingly, we found that the tRNALeu(UUR) carrying the 3302A>G mutation migrated considerably slower than the wt-tRNA or the tRNA carrying the 3243A>G mutation. In the clone containing the mutation at 77%, the small amounts of deacylated wt-tRNA are not visible, thus only 3 bands can be seen. Another fast migrating RNA species of unknown identity (?) is always seen in the clone containing the 3243A>G mutation, in 143B cells after alkaline treatment, but never in the clones containing the 3302A>G mutation.
Figure 7.
Aminoacylation levels in cybrid clones. Degree of aminoacylation of mt-tRNALeu(UUR) in 143B cells, in the cybrid clones containing 77 or 98% of the 3302A>G mutation as well as in the clone containing the 3243A>G mutation (3243 100%). RNA was isolated from cells and separated by PAGE under acidic conditions. Deacylated RNA samples of all cell lines and a mixture of acylated and deacylated RNA from 143B cells (first lane) were also loaded in order to facilitate identification of the two species.
The ratio of aminoacylated to non-acylated tRNA is considerably lower in both the 77 and 98% clone for the 3302A>G mutation (∼50:50), compared with wt-143B cells (∼85:15, ratios calculated from three independent gels). It has been shown before that the level of aminoacylation of the tRNA carrying the 3243A>G mutation is also decreased (15), and this is confirmed by our data.
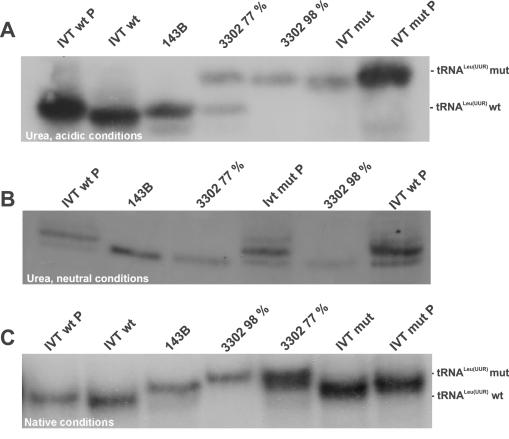
The retarded migration induced by the 3302A>G mutation could be due to (i) altered length resulting from misprocessing, (ii) altered charge or (iii) altered structure, resulting either directly from the change of the primary sequence or from changes in post-transcriptional modifications. To distinguish between these possibilities, RNA was isolated under neutral conditions to split off the amino acid and analysed under different gel conditions. In addition, run-off transcripts with the wt or the 3302A>G sequence were generated in vitro, were phosphorylated at the 5′ end and run together with RNA from cybrid cells (Figure 8). Acidic, urea containing gels showed that introducing the mutation into in vitro transcripts alone was sufficient to cause the retarded migration, hence the 3302A>G change of the primary sequence in tRNALeu(UUR) leads already to a structural change relevant for its electrophoretic mobility (Figure 8A). Cellular tRNAs comigrate with the 5′-phosphorylated in vitro transcripts. The 5′-phosphate group, which increases the molecular weight of the tRNA and adds negative charge, is sufficient to slow down migration, but not to the extent induced by the 3302A>G mutation. This, as well as the fact that the 3243A>G mutation causes a similar change in nucleotide composition, but causes no difference in migration seen in the aminoacylation experiment (Figure 7), argues against altered charge being the reason for different mobility.
Figure 8.
Analysis of tRNA structure, length and charge by different PAGE methods and northern blotting. RNA samples isolated under neutral conditions from cell lines in order to split off the amino acid were run together with run-off transcripts with the wt or 3302A>G sequence, phosphorylated (IVT P) at the 5′ end or non-phosphorylated (IVT) under different conditions. Gels were blotted and probed with a probe for tRNALeu(UUR). (A) Urea containing, acidic conditions. (B) Urea containing, neutral conditions. (C) Native, neutral conditions.
Under neutral, urea containing conditions, no migration difference between wt and mutated tRNA can be observed, and the clone containing 77% of mutated tRNA displays only one band, arguing against different lengths being the reason for different mobility (Figure 8B). Cellular tRNAs migrate slightly faster than in vitro transcripts, thus they seem to be more compact under these conditions.
Finally, non-denaturing gels which do not contain urea allowing at least partial folding of the tRNAs show again a retardation introduced by the mutation, and consequently the clone containing 77% of mutated tRNA displays two bands again (Figure 8C). Similar shifts in acidic, urea containing as well as neutral gels have been found recently in tRNALys containing the 8296A>G mutation (23).
In conclusion, the 3302A>G mutation causes structural changes which may be the reason for the impairment of charging with amino acid.
DISCUSSION
Our data shows that, at levels close to homoplasmy, the 3302A>G mutation in mt-tRNALeu(UUR) leads to a severe biochemical defect in cybrid cells, as it did in the patient's muscle. This defect appears due to a combination of factors including (i) a processing defect causing accumulation of the precursor RNA19, probably leading to (ii) a decreased steady-state level of mt-tRNALeu(UUR), (iii) a change in tRNA structure, and (iv) a partial decrease in aminoacylation, possibly due to this structural change.
The decreased steady-state level of mt-tRNALeu(UUR) is not due to destabilization of the mutated tRNA, as shown for mutations in the tRNASer(UCN) (24) or tRNAIle (25). Instead, it is due to a lowered rate of release from the precursor RNA19 that accumulates in cybrid cells, albeit to a lesser extent than was seen in the patient's muscle. Whether RNA19 induces ‘gain-of-function’ abnormalities such as becoming part of ‘pseudoribosomes’ due to incorporation of its 16S moiety (18), or being misread as an mRNA and thus blocking ribosomes (26) remains uncertain. The fact that RNA19 also accumulates in the clone containing the 3302A>G mutation at 77% (Figure 3), which however showed no defect in respiration, rather argues against a gain-of-function, at least at these steady-state levels of this precursor.
The 3302A>G mutation is close to the 3′ terminus of the tRNA, raising the possibility that it may impair recognition and/or cleavage by an RNase Z-like activity perhaps similar to that identified recently (27). Proximity to the 3′ cleavage site is not, however, a requirement for impairing the endonucleolytic step, since it was shown that the mutations 3243A>G, 3271T>C and 3302T>C all lead to the accumulation of RNA19 in vivo in biopsy samples of patients presenting with the mitochondrial myopathy, encephalopathy, lactacidosis, stroke (MELAS) syndrome (28). These authors showed that RNA19 molecules containing these mutations are more abundant in RNA samples than was predicted from the levels of DNA heteroplasmy suggesting that processing may be affected by mutations throughout the tRNA. Finally, artificial RNA molecules containing these and other mutations were tested in vitro as substrates for a 3′-tRNAse activity purified from HeLa cell mitoplasts. The 3302A>G mutation led to a decrease in Vmax, but not for the Km of the cleavage process (29). Such in vitro studies had also shown earlier that the 3243A>G mutation leads to severe impairment of processing of an artificial RNA19 at the 5′ end by mtRNAse P as well as nRNAse P, whereas RNA19 containing the 3302A>G construct was much less affected (30). Thus, both mutations cause processing defects, at both termini for 3243A>G and at the 3′ end for 3302 A>G.
Alteration of secondary and/or tertiary structure by the 3302A>G mutation was identified using a combination of electrophoretic techniques as a second possible mechanism involved in pathogenesis, besides lowered levels of mt-tRNALeu(UUR). Altered length due to misprocessing, altered charge due to the change in primary sequence or altered post-transcriptional modifications were excluded by comparing running properties of the native tRNAs containing the 3302A>G with those containing the 3243A>G mutation or in vitro transcripts, respectively (Figure 8). An altered structure caused by the 3302A>G mutation was rather unexpected, since although 3302A>G weakens the second (U2/A71) base-pair in the acceptor stem, leading to a G/U wobble pair, the stabilizing effect of the G1/C72 base pair should prevent important ‘breathing’ of this region (29). Unfortunately, detailed structural probing assays on in vitro transcribed substrates using various endonucleases was difficult for technical reasons at sites so close to RNA termini.
We postulate that the structural change in mt-tRNALeu(UUR) interferes with charging of leucine by the Leucyl-tRNA-synthetase (LeuRS). This leads to a lower ratio of acylated versus deacylated tRNA (Figure 7), which results in decreased availability of Leu-tRNALeu(UUR) molecules in the 98% clone at levels ∼30% of that seen in control cells, when taking into account the decreased levels of the tRNA as well. In addition, although these molecules are charged, they still may have a structure that cannot be recognized appropriately by the ribosome. The charging defect could either be due to a directly impaired interference of the LeuRS with the structurally altered tRNA (31) or primarily due to impaired post-transcriptional modification followed by impaired recognition by LeuRS. Our finding that introducing the mutation into in vitro transcripts is sufficient to alter the structure of the tRNA (Figure 8) supports the first possibility, as well as the fact that the small amounts of wt-Leu-tRNALeu(UUR) in the 77% clone (Figure 7) can obviously rescue the defect.
Poor aminoacylation has also been shown for the 3243A>G mutation in both, lung carcinoma cybrids (21) and 143B cells (15). Secondary and/or tertiary structural alterations with poor aminoacylation, alone or in combination with deficiencies of methylation of 3239G (32) and modification of uridine at the first position of the anticodon (16) were suggested to be the ultimate reason for impaired translation initiation, causing a general reduction in mitochondrial protein synthesis rate (15). Finally, it was also shown in muscle fibres in vivo, that deficient aminoacylation is important in pathogenesis of the MELAS syndrome (33).
Whether post-transcriptional modification defects are also present in mt-tRNALeu(UUR) containing the 3302A>G mutation remains to be determined, however, such a defect could provide another important disease mechanism, namely an incorporation of the wrong amino acid into the growing polypeptide chain. For example, studies in HeLa cybrids carrying the 3243A>G mutation showed that mt-tRNALeu(UUR) is charged properly with leucine, but that the altered modification of the wobble base could also lead to mistranslation of leucine for phenylalanine (16,34). Indeed, formation of truncated peptides and altered peptide fingerprint patterns of mitochondrial proteins have been demonstrated, for both the 3243A>G mutation in mt-tRNALeu(UUR) (35–37) and the 7445T>C in tRNASer(UCN) (38). Also, a number of pathogenic mutations in tRNALeu(UUR) was reported to impede post-transcriptional modification of the wobble-position uridine 34 (34,39).
In conclusion, a decrease in steady-state levels of mt-tRNALeu(UUR) carrying the 3302A>G mutation is caused by inefficient cleavage of the precursor RNA19. This, together with a defect in aminoacylation due to an altered structure results in a marked reduction of available Leu- tRNALeu(UUR) and a severe biochemical defect. Cellular dysfunction occurs, however, only when the mutation is present at almost homoplasmic levels, confirming that the threshold for this mutation is high. The small accumulation of RNA19 may contribute to the pathology in cybrid cells, however, this may be much more important in the patients muscle, where RNA19 accumulated massively, probably due to a different processing pathway in this tissue.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
The authors appreciate the skilful technical assistance of M. Bust, and we would like to thank Dr Karl Morton for generation of the initial 3302 cybrids and Dr Eric Shoubridge for his gift of 3243 clones. This work was supported by the Center for Molecular Medicine (CMMC) of the University of Köln (J.-C.v.K.R. and R.J.W.); the Köln Fortune Program, Faculty of Medicine, University of Köln (K.M.-W., K.E., M.M. and J.-C.v.K.R.); Maria Pesch Stiftung (S.E. and R.J.W.); Deutsche Gesellschaft für Muskelkranke—MitoNet (K.E.) and Deutsche Forschungsgemeinschaft (HE 3397/3 to M.H.). The authors also acknowledge the financial support of the European Commission (QLG1-CT-2001-00966). Funding to pay the Open Access publication charges for this article was provided by Center for Molecular Medicine Cologne (CMMC).
Conflict of interes statement. None declared.
REFERENCES
- 1.Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nature Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bindoff L.A., Howell N., Poulton J., McCullough D.A., Morten K.J., Lightowlers R.N., Turnbull D.M., Weber K. Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. A potential disease mechanism. J. Biol. Chem. 1993;268:19559–19564. [PubMed] [Google Scholar]
- 3.Van Den Bosch B.J., De Coo I.F., Hendrickx A.T., Busch H.F., De Jong G., Scholte H.R., Smeets H.J. Increased risk for cardiorespiratory failure associated with the A3302G mutation in the mitochondrial DNA encoded tRNA(Leu(UUR)) gene. Neuromuscul. Disord. 2004;14:683–688. doi: 10.1016/j.nmd.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Helm M., Brule H., Friede D., Giege R., Putz D., Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King M.P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 6.Blau H.M., Webster C. Isolation and characterization of human muscle cells. Proc. Natl Acad. Sci. USA. 1981;78:5623–5627. doi: 10.1073/pnas.78.9.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goto Y., Nonaka I., Horai S. A mutation in the tRNA(Leu)(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–653. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 8.Rustin P., Chretien D., Bourgeron T., Gerard B., Rotig A., Saudubray J.M., Munnich A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta. 1994;228:35–51. doi: 10.1016/0009-8981(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 9.Enriquez J.A., Chomyn A., Attardi G. MtDNA mutation in MERRF syndrome causes defective aminoacylation of tRNA(Lys) and premature translation termination. Nature Genet. 1995;10:47–55. doi: 10.1038/ng0595-47. [DOI] [PubMed] [Google Scholar]
- 10.Sohm B., Frugier M., Brule H., Olszak K., Przykorska A., Florentz C. Towards understanding human mitochondrial leucine aminoacylation identity. J. Mol. Biol. 2003;328:995–1010. doi: 10.1016/s0022-2836(03)00373-5. [DOI] [PubMed] [Google Scholar]
- 11.Fechter P., Rudinger J., Giege R., Theobald-Dietrich A. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett. 1998;436:99–103. doi: 10.1016/s0014-5793(98)01096-5. [DOI] [PubMed] [Google Scholar]
- 12.Helm M., Brule H., Degoul F., Cepanec C., Leroux J.P., Giege R., Florentz C. The presence of modified nucleotides is required for cloverleaf folding of a human mitochondrial tRNA. Nucleic Acids Res. 1998;26:1636–1643. doi: 10.1093/nar/26.7.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasukawa T., Suzuki T., Ishii N., Ueda T., Ohta S., Watanabe K. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNA(Lys) with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000;467:175–178. doi: 10.1016/s0014-5793(00)01145-5. [DOI] [PubMed] [Google Scholar]
- 14.Watmough N.J., Bindoff L.A., Birch-MacHin M.A., Jackson S., Bartlett K., Ragan C.I., Poulton J., Gardiner R.M., Sherratt H.S., Turnbull D.M. Impaired mitochondrial beta-oxidation in a patient with an abnormality of the respiratory chain. Studies in skeletal muscle mitochondria. J. Clin. Invest. 1990;85:177–184. doi: 10.1172/JCI114409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomyn A., Enriquez J.A., Micol V., Fernandez-Silva P., Attardi G. The mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episode syndrome-associated human mitochondrial tRNALeu(UUR) mutation causes aminoacylation deficiency and concomitant reduced association of mRNA with ribosomes. J. Biol Chem. 2000;275:19198–19209. doi: 10.1074/jbc.M908734199. [DOI] [PubMed] [Google Scholar]
- 16.Yasukawa T., Suzuki T., Ueda T., Ohta S., Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000;275:4251–4257. doi: 10.1074/jbc.275.6.4251. [DOI] [PubMed] [Google Scholar]
- 17.MacDonald R.J., Swift G.H., Przybyla A.E., Chirgwin J.M. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- 18.Schon E.A., Koga Y., Davidson M., Moraes C.T., King M.P. The mitochondrial tRNA(Leu)(UUR)) mutation in MELAS: a model for pathogenesis. Biochim. Biophys. Acta. 1992;1101:206–209. [PubMed] [Google Scholar]
- 19.Kaufmann P., Koga Y., Shanske S., Hirano M., DiMauro S., King M.P., Schon E.A. Mitochondrial DNA and RNA processing in MELAS. Ann. Neurol. 1996;40:172–180. doi: 10.1002/ana.410400208. [DOI] [PubMed] [Google Scholar]
- 20.Koga Y., Davidson M., Schon E.A., King M.P. Fine mapping of mitochondrial RNAs derived from the mtDNA region containing a point mutation associated with MELAS. Nucleic Acids Res. 1993;21:657–662. doi: 10.1093/nar/21.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Meziane A., Lehtinen S.K., Holt I.J., Jacobs H.T. Mitochondrial tRNALeu isoforms in lung carcinoma cybrid cells containing the np 3243 mtDNA mutation. Hum. Mol. Genet. 1998;7:2141–2147. doi: 10.1093/hmg/7.13.2141. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs H.T., Holt I.J. The np 3243 MELAS mutation: damned if you aminoacylate, damned if you don't. Hum. Mol. Genet. 2000;9:463–465. doi: 10.1093/hmg/9.4.463. [DOI] [PubMed] [Google Scholar]
- 23.Bornstein B., Mas J.A., Patrono C., Fernandez-Moreno M.A., Gonzalez-Vioque E., Campos Y., Carrozzo R., Martin M.A., Del Hoyo P., Santorelli F.M., et al. Comparative analysis of the pathogenic mechanisms associated with the G8363A and A8296G mutations in the mitochondrial tRNA Lys gene. Biochem. J. 2005;387:773–778. doi: 10.1042/BJ20040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mollers M., Maniura-Weber K., Kiseljakovic E., Bust M., Hayrapetyan A., Jaksch M., Helm M., Wiesner R.J., von Kleist-Retzow J.C. A new mechanism for mtDNA pathogenesis: impairment of post-transcriptional maturation leads to severe depletion of mitochondrial tRNASer(UCN) caused by T7512C and G7497A point mutations. Nucleic Acids Res. 2005;33:5647–5658. doi: 10.1093/nar/gki876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasukawa T., Hino N., Suzuki T., Watanabe K., Ueda T., Ohta S. A pathogenic point mutation reduces stability of mitochondrial mutant tRNA(Ile) Nucleic Acids Res. 2000;28:3779–3784. doi: 10.1093/nar/28.19.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.King M.P., Koga Y., Davidson M., Schon E.A. Defects in mitochondrial protein synthesis and respiratory chain activity segregate with the tRNA(Leu(UUR)) mutation associated with mitochondrial myopathy, encephalopathy, lactic acidosis, and strokelike episodes. Mol. Cell. Biol. 1992;12:480–490. doi: 10.1128/mcb.12.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan H., Zareen N., Levinger L. Naturally occurring mutations in human mitochondrial pre-tRNASer(UCN) can affect the tRNase Z cleavage site, processing kinetics and substrate secondary structure. J. Biol. Chem. 2006;281:3926–3935. doi: 10.1074/jbc.M509822200. [DOI] [PubMed] [Google Scholar]
- 28.Koga A., Koga Y., Akita Y., Fukiyama R., Ueki I., Yatsuga S., Matsuishi T. Increased mitochondrial processing intermediates associated with three tRNA(Leu(UUR)) gene mutations. Neuromuscul. Disord. 2003;13:259–262. doi: 10.1016/s0960-8966(02)00267-5. [DOI] [PubMed] [Google Scholar]
- 29.Levinger L., Oestreich I., Florentz C., Morl M. A Pathogenesis-associated mutation in human mitochondrial tRNA(Leu(UUR)) leads to reduced 3′-end processing and CCA addition. J. Mol. Biol. 2004;337:535–544. doi: 10.1016/j.jmb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 30.Rossmanith W., Karwan R.M. Impairment of tRNA processing by point mutations in mitochondrial tRNA(Leu)(UUR) associated with mitochondrial diseases. FEBS Lett. 1998;433:269–274. doi: 10.1016/s0014-5793(98)00928-4. [DOI] [PubMed] [Google Scholar]
- 31.Sohm B., Sissler M., Park H., King M.P., Florentz C. recognition of human mitochondrial tRNA(Leu(UUR)) by its cognate Leucyl-tRNA synthetase. J. Mol. Biol. 2004;339:17–29. doi: 10.1016/j.jmb.2004.03.066. [DOI] [PubMed] [Google Scholar]
- 32.Helm M., Florentz C., Chomyn A., Attardi G. Search for differences in post-transcriptional modification patterns of mitochondrial DNA-encoded wild-type and mutant human tRNA(Lys) and tRNA(Leu(UUR)) Nucleic Acids Res. 1999;27:756–763. doi: 10.1093/nar/27.3.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borner G.V., Zeviani M., Tiranti V., Carrara F., Hoffmann S., Gerbitz K.D., Lochmuller H., Pongratz D., Klopstock T., Melberg A., et al. Decreased aminoacylation of mutant tRNAs in MELAS but not in MERRF patients. Hum. Mol. Genet. 2000;9:467–475. doi: 10.1093/hmg/9.4.467. [DOI] [PubMed] [Google Scholar]
- 34.Kirino Y., Goto Y.I., Campos Y., Arenas J., Suzuki T. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl Acad. Sci. USA. 2005;102:7127–7132. doi: 10.1073/pnas.0500563102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunbar D.R., Moonie P.A., Zeviani M., Holt I.J. Complex I deficiency is associated with 3243G:C mitochondrial DNA in osteosarcoma cell cybrids. Hum. Mol. Genet. 1996;5:123–129. doi: 10.1093/hmg/5.1.123. [DOI] [PubMed] [Google Scholar]
- 36.Flierl A., Reichmann H., Seibel P. Pathophysiology of the MELAS 3243 transition mutation. J. Biol. Chem. 1997;272:27189–27196. doi: 10.1074/jbc.272.43.27189. [DOI] [PubMed] [Google Scholar]
- 37.Janssen G.M., Maassen J.A., van Den Ouweland J.M. The diabetes-associated 3243 mutation in the mitochondrial tRNA(Leu(UUR)) gene causes severe mitochondrial dysfunction without a strong decrease in protein synthesis rate. J. Biol. Chem. 1999;274:29744–29748. doi: 10.1074/jbc.274.42.29744. [DOI] [PubMed] [Google Scholar]
- 38.Reid F.M., Vernham G.A., Jacobs H.T. A novel mitochondrial point mutation in a maternal pedigree with sensorineural deafness. Hum. Mutat. 1994;3:243–247. doi: 10.1002/humu.1380030311. [DOI] [PubMed] [Google Scholar]
- 39.Yasukawa T., Kirino Y., Ishii N., Holt I.J., Jacobs H.T., Makifuchi T., Fukuhara N., Ohta S., Suzuki T., Watanabe K. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005;579:2948–2952. doi: 10.1016/j.febslet.2005.04.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.