Abstract
Bacterial IIC introns are a newly recognized subclass of group II introns whose ribozyme properties have not been characterized in detail. IIC introns are typically located downstream of transcriptional terminator motifs (inverted repeat followed by T's) or other inverted repeats in bacterial genomes. Here we have characterized the self-splicing activity of a IIC intron, B.h.I1, from Bacillus halodurans. B.h.I1 self-splices in vitro through hydrolysis to produce linear intron, but interestingly, additional unexpected products were formed that were highly dependent on ionic conditions. These products were determined to represent alternative splicing events at the 5′ junction and cleavages throughout the RNA transcript. The alternative splicing and cleavage events occurred at cryptic splice sites containing stem–loop and IBS1 motifs, suggesting that the 5′ exon is recognized by both elements. These results provide the first example of a group II intron that uses 5′ splice sites nonadjacent to the ribozyme structure. Furthermore, the data suggest that IIC introns differ from IIA and IIB introns with respect to 5′ exon definition, and that the terminator stem–loop substitutes in part for the missing IBS2–EBS2 (intron and exon binding sites 2) interaction.
INTRODUCTION
Group II introns are catalytic RNAs and retroelements found in bacterial genomes and in organellar genomes of plants, fungi and algae. The catalytic RNA portions of the introns fold into a conserved structure that is capable of self-excision from precursor transcript. In addition to this ribozyme portion, many group II introns encode reverse transcriptase (RT) ORFs within domain IV of the RNA structure, which allow the introns to act as retroelements (1–5).
Group II ribozymes are classified as IIA, IIB and IIC, based on conserved features of their RNA secondary structures (6–9). While all three classes have the same general secondary structure elements, there are significant differences both structurally and functionally. IIA and IIB introns, which are the most extensively characterized, share a common mechanism for 5′ exon definition, but differ in 3′ exon definition. In both types of introns, the 5′ exon is recognized through two Watson–Crick base pairing interactions, IBS1–EBS1 (intron and exon binding sites 1) and IBS2–EBS2 (intron and exon binding sites 2). Each interaction consists of ∼6 bp between the 5′ exon and a region in domain ID of the intron. Together these interactions anchor the 5′ exon to the ribozyme for the two splicing steps, during which the 5′ exon is cleaved from the intron in the first step, and attacks the 3′ intron–exon junction in the second step (10,11). For 3′ exon definition, IIA and IIB introns use a different single Watson–Crick base pairing. For IIA introns, δ' is the first nucleotide of the 3′ exon, and pairs with the δ position in domain ID3 (12). For IIB introns, the first nucleotide of the 3′ exon is called IBS3, and pairs with EBS3 in domain ID(III) instead of with δ′ (13). Another significant structural difference between IIA and IIB introns is the ɛ′ motif, which is a ∼14 nt bulge in IIA introns and a 4 nt bulge in IIB introns; in this case the difference does not correspond with a known functional variation. Other minor structural differences can be found in (8,9).
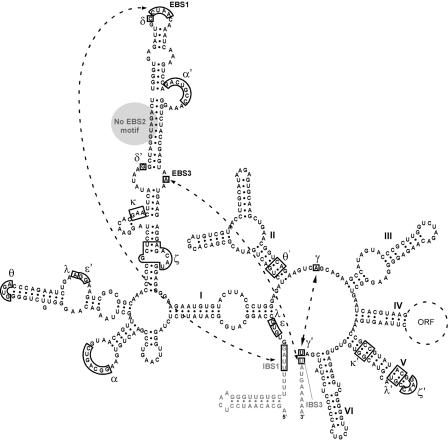
IIC introns differ fairly dramatically from IIA and IIB secondary structures (3,8,14). IIC introns are abbreviated in size, and while domain V is highly conserved, its distal stem is 2 bp shorter than for IIA and IIB introns, and its basal stem has a CGC in place of the highly conserved AGC triad (8,14) (Figure 1). The ɛ′ region of IIC introns has a unique 11 nt bulge motif with a highly conserved AGGAA sequence, while the EBS3 motif is the same as for IIB introns, presumably along with the mechanism of 3′ exon recognition. Like IIA and IIB introns, IIC introns have an IBS1–EBS1 interaction; however, the pairing is 3–4 bp rather than 6 bp as for IIA and IIB introns (Figure 1). Importantly for this study, IIC introns lack an EBS2 motif in the ribozyme structure, as well as a corresponding IBS2 motif in the 5′ exon. Instead, a stem–loop of an intrinsic terminator is located at the usual position of IBS2 (Figure 1). It is not known if the missing IBS2–EBS2 interaction is compensated functionally by another, as yet uncharacterized interaction. The presence of the terminator motifs in 5′ exons of IIC introns is best rationalized by the retroelement properties of IIC introns. All known IIC introns are components of retroelements, and introns of this class are observed to insert after transcriptional terminators or similar stem–loop motifs (15,16). This insertion specificity is a consequence of the biochemical mechanism of the mobility reaction (A. R. Robart, W. Seo and S. Zimmerly, manuscript submitted).
Figure 1.
Secondary structure of the B.h.I1(B) intron, a representative of group IIC introns. The six domains of the intron RNA structure are shown with black lettering, and exons with gray lettering. Domain IV encodes a 1.3 kb ORF, which is denoted by a dotted line. Tertiary interactions within the intron structure are indicated with Greek letters. Of these, the non-Watson–Crick interactions κ, θ, λ and ζ are predicted by analogy with other introns, but have not been confirmed for any IIC introns. The pairing interactions IBS1–EBS1, IBS3–EBS3 and γ–γ′ are involved in 5′ and 3′ exon recognition, and are highlighted by dotted lines. Like other IIC introns, B.h.I1(B) lacks an EBS2 motif in domain ID (gray circle), and the 5′ exon contains a terminator stem–loop in the position where the IBS2 is located in IIA and IIB introns.
Our understanding of group II ribozymes comes mainly from the study of IIA and IIB introns found in mitochondria. Characterized introns include the yeast mitochondrial introns aI5γ (cox1I5) (IIB RNA structure) (17), cob1I1 (IIB) (18), cox1I1 (IIA) (19), the Pylaiella littoralis mitochondrial intron LSUI2 (IIB) (20) and Podospora anserina mitochondrial intron cox1I1 (IIA) (21). In contrast, IIC introns have yet to be characterized extensively, and they are only found in bacteria. The only IIC intron tested is the GBSi1 intron of Streptococcus agalactiae (14). This intron was reported to have the unusual property of self-splicing through the hydrolysis pathway rather than the branching pathway, yielding linear intron rather than lariat. The possibility was considered that the terminator motif might affect splicing, but its deletion was not found to affect the reaction (14).
Interestingly, several bacterial introns have exhibited novel self-splicing properties compared to mitochondrial introns. A group II intron of Azotobacter vinelandii, which was alternately designated Avi.groEL (6) or AV (22), has been shown to self-splice at unexpectedly high temperatures. Mitochondrial introns generally self-splice at 42–45°C (11,23), but the Avi.groEL intron version tested by (6) had a splicing optimum of 55°C. The more streamlined AV construct tested by Adamidi et al. (22) had an optimum of 65°C. The high temperature optimum was speculated to be involved in the function of its host heat-shock gene groEL, although curiously the intron is inserted into the stop codon and does not affect the ORF sequence. In another example, an intron of Bacillus anthracis, B.a.I2, was shown to undergo alternative splicing, both in vitro and in vivo, which presumably controls expression of the downstream exon ORF (24). The RmInt1 intron of Sinorhizobium meliloti has been shown to produce noncognate cleavages in in vitro reactions (25,26), while the Bacillus cereus intron B.ce.I4 has an in vivo splice site 56 nt downstream of the expected 3′ boundary (27). The most studied of bacterial group II introns, Ll.ltrB of Lactococcus lactis, has been characterized for its maturase-assisted splicing reaction, but its self-splicing properties have not been reported to be unusual (28–30).
Here we have characterized the ribozyme properties of the IIC intron B.h.I1 of Bacillus halodurans. The B.h.I1 intron is present in the B.halodurans genome in five copies having >99.8% identity. Four copies are in analogous locations at the end of ribosomal RNA (rrn) operons (31), and are denoted here as B.h.I1(B), B.h.I1(D), B.h.I1(F) and B.h.I1(G) because they are located after rrn operons B, D, F and G, respectively. The fifth copy, B.h.I1(X), is located downstream of a terminator-like motif, but its exons have no sequence identity with the other four copies. Self-splicing assays show B.h.I1(B) to be an active ribozyme in vitro, but unexpected products are produced that are due to noncognate recognition of cryptic 5′ exons, thus resulting in alternative ligation of exons and cleavages at cryptic 5′ exon-intron boundaries. The data suggest that the 5′ exon stem–loop motif strongly influences exon recognition, and that exon recognition of IIC introns has features distinct from that of IIA and IIB introns.
MATERIALS AND METHODS
Growth of cells and preparation of genomic DNA
Escherichia coli (DH5α) was grown and DNA was prepared by standard methods (32). B.halodurans C-125 (obtained from Hideto Takami, Japan Marine Science and Technology Center) was grown in alkaline medium (ATCC medium #18) overnight at 37°C with shaking (31). Genomic DNA was prepared from 1.5 ml stationary phase culture. Pelleted cells were washed and resuspended in 0.45 ml of disruption buffer [30 mM Tris–HCl (pH 7.4), 100 mM NaCl and 5 mM EDTA], followed by the addition of 0.1 mg of lysozyme and 50 μg of RNase A and incubation at 37°C for 10 min. Lysis was by the addition of 0.1 mg proteinase K and SDS to a final concentration of 1% with incubation at 37°C for 45 min. The sample was repeatedly extracted with an equal volume of phenol-CIA (25:24:1 of phenol:chloroform:isoamyl alcohol) until there was a clear interface upon centrifugation, followed by ethanol precipitation in the presence of 0.3 M NaOAc (pH 5.2). The pellet was washed with cold 75% ethanol, air-dried and dissolved in TE [10 mM Tris–HCl (pH 7.4) and 1 mM EDTA].
Plasmid constructs
The B.h.I1 intron was PCR amplified from genomic DNA in two pieces to delete 1208 bp of the ORF, using the primers BhAS1 (5′-CCTCTCTTCGTGTCCCCAGCGAGAT) and Bh5′NGS1 (5′CGGGATCCAGCGACGACAGACCGGAGCGTACAC), and BhS1 (5′-CGAAGAGAGGGTCGCTCACAGCCCGATCATGGATT) and Bh3′NGAS1 (5′-CCATCGATTCGGGCATTGCCTCAGCTTCCTCGA). The two pieces were ligated by recombinant PCR, and the DNA was cloned into the BamHI and ClaI sites of pBluescript KS+ (Stratagene, La Jolla, CA) to yield the plasmid pBh6. The 5′ exon in this construct is identical to B.h.I1(B, D, F, G) genomic copies, while the 3′ exon corresponds to the B.h.I1(B). The B.h.(X) intron copy was similarly amplified from genomic DNA in two amplifications with the primers BhAS1 and 5′-CGGGATCCGAGTAAGAGGCGCGGTTCTTGGCGG, and BhS1 and 5′-CCATCGATCAGATCACCTTCCTGCCGCTGAACC, followed by recombinant PCR and cloning into the SmaI site of pBluescript KS+. For all remaining constructs, the inserts were cloned into the EcoRV site of the polylinker. The pΔterminator insert was amplified from pBh6 with the primers 5′-AAGCGATAAATTTTTTAGGTGTGCCAGGCATGCGC and Bh3′NGAS1; the pΔterminatorΔIBS1 insert was amplified from pBh6 with the primers 5′-AAAAAAAAGTGTGCCAGGCATGCGCATATTCTC and Bh3′NGAS1; and the pΔIBS1 insert was amplified from pΔterminatorΔIBS1 with the primers 5′-GCACAATCCAATCGGGTTGTGCAAAAAAAAGTGTGCCAGGCATGC and Bh3′NGAS1. All plasmid constructs were confirmed by sequencing.
In vitro transcription and self-splicing reactions
The templates for in vitro transcription were linearized with ClaI, except for the 3′ exon extension experiments, where plasmid was linearized with PvuII. Transcription was in a volume of 40 μl of 40 mM Tris–HCl (pH 8.0), 8 mM MgCl2, 2 mM spermidine, and 50 mM NaCl, 1 mM NTPs, 5 mM DTT, and 0.05% Triton X-100, and with 1 μg template and 2 μl T7 RNA polymerase (noncommercial source, undetermined activity). The reaction was incubated at 37°C for 1 h followed by phenol-CIA (25:24:1) extraction and ethanol precipitation in the presence of 2.5 M NH4OAc. For radiolabeled transcript, reactions contained 2 μl of [α-32P]UTP (10 μCi/μl; 3000 Ci/mmol; Amersham, Piscataway, NJ). Transcript was resuspended in TE.
For self-splicing reactions, 32P-labelled (27 000 c.p.m.) or cold transcript (200 ng) was first prefolded in a PCR machine (Perkin Elmer GeneAmp 2400, Norwalk, CT) with incubations at 90°C for 1 min, 75°C for 5 min and slow cooling to 45°C over 15 min. Buffer was added to a final volume of 50 μl of 100 mM MgCl2, 2 M NH4Cl and 50 mM Tris–HCl (pH 7.5), and the reaction was incubated at 55°C for 15 min. Alternative reaction conditions were with 0.5, 1.0, or 2.0 M NaCl substituted for 2 M NH4Cl, or with 10 mM MgCl2 or 10 mM MnCl2 substituted for 100 mM MgCl2. The splicing reaction was ethanol precipitated with 0.3 M NaOAc (pH 5.2) and resuspended in formamide dye [95% formamide, 5 mM EDTA (pH 8.0), 0.1% of xylene cyanol and 0.1% of bromophenol blue], heated to 85°C for 2 min and resolved on a 4% polyacrylamide (19:1 acrylamide:bisacrylamide ratio)/8 M urea gel. For the unspliced control, reactions were done in parallel without MgCl2 or monovalent salts. For the oligonucleotide-directed disruption of the terminator stem, 2.5–400 ng of the oligonucleotide 5′ GCACAACCCGATTGGATTGTGCTTTATCGCTTGGC was added to 54 000 c.p.m transcript in a volume of 4 μl during the prefolding step to allow annealing of the oligo to the exon. The substrate was diluted into a splicing reaction of 50 μl with 10 mM MgCl2, and splicing was otherwise under standard conditions.
Primer extension, PCR and RT–PCR
Primer extension and RT–PCR were with Superscript II (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. Primer extension was with the 5′ end-labeled primers BhASPE1 (5′ end mapping; 5′ TTAGGCAATCTCACCCAAGTCTAC), and BhI13′endAS (3′ ICP cleavage mapping; 5′ ATCGAGTAGGAGGGGGTGACTAA). RT–PCR was with the first strand primer Bh3′NGAS1, followed by PCR with primers Bh5′NGS1 and Bh3′NGAS1. Products were resolved on 1% agarose gels with 1× TBE, and subcloned into pBluescript for sequencing, all by standard methods (32).
RESULTS
The B.h.I1 intron produces unexpected self-splicing products
To examine self-splicing properties of B.h.I1, the B.h.I1(B) sequence and its flanks were cloned into pBluescript in two segments to delete the ORF in domain IV (Methods). The intron was transcribed in vitro and subjected to self-splicing under conditions suitable for other introns [50 mM Tris (pH 7.5), 2 M NH4Cl and 100 mM MgCl2]. Like previous results for the related group II intron of S.agalactiae (14), self-splicing was observed without lariat formation (Figure 2A lane 2). Instead, products corresponded in size to linear intron, ligated exons, and unligated exons, and there were also bands of unknown composition that are further investigated below.
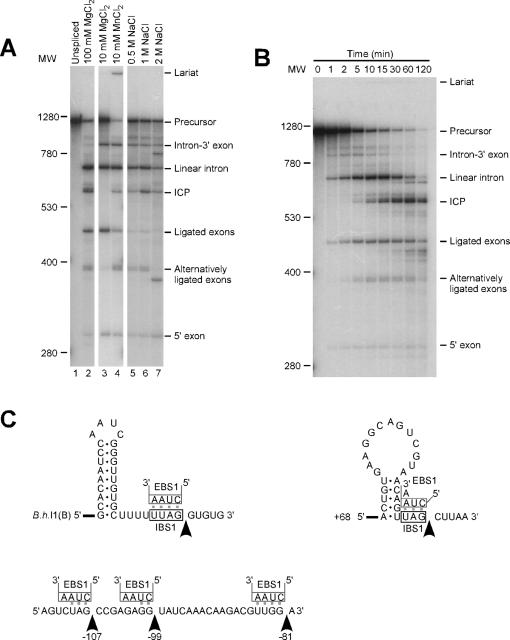
Figure 2.
Self-splicing products under different conditions and identification of bands. (A) B.h.I1(B) transcript was self-spliced in standard conditions of 100 mM MgCl2, 50 mM Tris (pH 7.5) and 2 M NH4Cl at 55°C for 15 min, or in alternative conditions with substitutions of 10 mM MgCl2 or 10 mM MnCl2 for 100 mM MgCl2, or 0.5, 1.0 or 2 M NaCl for 2 M NH4Cl. RNA size markers are indicated to the left in nucleotides, and product identifications to the right are explained in the text. (B) Time course of the self-splicing reaction under standard conditions, illustrating the initial formation of linear intron and ligated exons, and subsequent accumulation of other products. (C) Sequences and structures of the RNA cleavages produced in the self-splicing reaction. Arrows indicate cleavage sites at: the exon–intron junction of B.h. I1(B); the ICP cleavages at −107, −99 and −81 relative to the 3′ end of the intron; and the ICP cleavage at +68 relative to the 5′ end of the intron. EBS1 pairings of the intron are shown above the exon sequences.
A variety of reaction conditions were tested, which showed that self-splicing occurs over a range of conditions, but that products vary. Monovalent salt concentrations of 0.5–2 M were suitable, with NH4Cl, (NH4)2SO4 and KCl producing equivalent reactions (not shown). These salt requirements are consistent with reactions of several other group II introns (19–22), but different from aI5γ, which is inhibited by high concentrations of monovalent cations (33). Surprisingly, reactions with NaCl produced multiple novel bands that fluctuated with NaCl concentration (lanes 5–7). Suitable MgCl2 concentrations were 10–100 mM, with maximal activity at 100 mM (lanes 2–3). Interestingly, the products of 10 mM MgCl2 reactions differed from those of 100 mM reactions; 10 mM reactions produced substantially greater intron-3′ exon levels, and none of the bands shown below to be internal cleavage product (ICP) and alternatively ligated exons (lane 3). Thus, 10 mM MgCl2 supports greater fidelity in substrate recognition, while 100 mM MgCl2 supports greater reactivity. The divalent ions Ca2+ and Co2+ did not support activity (data not shown), but importantly 10 mM Mn2+ resulted in significant levels of a band migrating as lariat (6% putative lariat, 94% linear intron; lane 4), thus suggesting that B.h.I1 is capable of splicing via the branching pathway under some conditions. To our knowledge this is the first group II intron capable of using Mn2+ as a divalent cation. Long exposures of the 100 mM MgCl2 reaction also revealed a small amount of putative lariat (data not shown). Unlike some other group II introns, the addition of spermidine had no effect on the splicing reaction, and did not enhance splicing at magnesium concentrations under 10 mM (data not shown). The temperature optimum was relatively high, at 55°C, while activity was supported from 43–65°C (data not shown). Based on these data, we chose a standard splicing condition of 50 mM Tris (pH 7.5) 2 M NH4Cl, and 100 mM MgCl2 at 55°C for 15 min. In alternative reaction conditions, 100 mM MgCl2 was substituted with 10 mM MgCl2, or NH4Cl was substituted with 0.5, 1.0 or 2.0 M NaCl.
A time course of the self-splicing reaction under standard reaction conditions showed that the products formed within the first two minutes were linear intron and ligated exons, while other bands accumulated subsequently (Figure 2B). This suggests that linear intron and spliced exons are the initial reaction products, while some of the other bands may be secondary products formed from the initial products. One of these products has a size of ∼ 600 nt (termed ICP; see below) and accumulated over 2 h to become the major RNA in the reaction.
To identify the product bands, three methods were used: (i) altering the length of the 3′ exons in the precursor RNA to shift product sizes in the gel; (ii) blotting of the splicing products onto membranes and hybridizing with probes specific to either intron, 5′ exon or 3′ exon sequences; and (iii) primer extension using intron- or exon-specific primers, and with either total reaction products or gel-purified RNAs. Bands readily identified were linear intron, ligated exons, lariat and intron-3′exon. The lariat's branch point was not mapped and is assumed to be the bulged A in domain VI (Figure 1); however, other circular forms could comigrate at the same position in the gel and cannot be excluded as products (26,34). The alternatively ligated exon bands were confirmed by RT–PCR and sequencing of gel-purified RNAs (below). The ICP was more difficult to identify, and was ultimately determined to represent a mixture of truncated RNAs comigrating on the gel, which were formed by either 5′ or 3′ truncations of linear intron. This conclusion was initially supported by hybridization and 3′ exon lengthening experiments, which showed the ICP to contain only intron and not 3′ exon sequence (data not shown). Primer extension of gel-purified ICP showed two 5′ termini. Ninety-one percent of 5′ ends were the native 5′ GUGUG start of the intron, while 11% represented an internal truncation at +68 relative to the start of the intron (data not shown). The +68 truncation is consistent with the ICP gel migration, while the native 5′ end is not, and would require a 3′ truncation to account for its gel migration. Truncations from the 3′ ends were determined by primer extension of total reaction products, using a primer complementary to intron domain 6. Cleavages were detected at positions −107, −99 and −81 relative to the intron 3′ end (Figure 2C). The occurrence of these cleavages corresponds to the doublet formed under NH4Cl conditions (81, 99 nt truncations yielding 594 and 577 nt products; see Figure 2A lane 2) and with the single band under NaCl conditions (107 nt truncation, 566 nt RNA; Figure 2A lanes 5–7). In comparison, the +68 cleavage produces a 607 nt band, which may be too faint to detect in Figure 2A; however, the +68 cleavage is very important because it corresponds to an alternative ligation product (below). Importantly, the positions of the ICP cleavages correspond to cryptic 5′ exon sites (Figure 2C). The +68 cleavage occurs at a potential IBS1 sequence directly downstream of a stem–loop (domain IB in the secondary structure, Figure 1), while the −107, −99, and −81 cleavages are positioned after potential IBS1 sequences, but without obvious upstream stem–loops (Figure 2C).
The mechanism of cleavage may be an SER-related (spliced exon reopening) reaction, in which incomplete reverse splicing of excised intron into an exon junction results in cleavage of the two exons without covalent linkage of intron to the exons (33). Alternatively, the +68 cleavage could be due to the aborted usage of the internal site as a 5′ exon; although this possibility seems unlikely, it is supported by an alternative ligation product at the +68 site in a subsequent experiment (below). Additional bands in Figure 2A and B were not mapped, but may correspond to analogous cleavages after other cryptic IBS1 sequences. Similar noncognate product bands have been reported for ribozyme reactions of the yeast cox1I1 intron in buffers containing KCl, but not NH4Cl. These RNAs were secondary products of the spliced intron, and at least some were shown to be internal cleavages within spliced intron (19). We did not examine whether noncognate products were affected by the deletion of sequence in domain IV.
To address whether these unexpected reaction products are general to bacterial class C introns, we tested splicing of three other bacterial class C introns. The S.meliloti intron S.me.I3 self-spliced accurately in vitro to produce linear products but not lariat (data not shown). In contrast, the A.vinelandii intron A.v.I2 produced mainly lariat, with a small amount of linear intron. The Streptococcus pneumonia intron S.p.I1, which is 90% identical to the splicing-competent GBSi1 ribozyme (14), failed to splice in vitro under any conditions tested (data not shown). Together, the four self-splicing examples (B.h.I1, GBSi1, S.me.I3, A.v.I2) suggest that self-splicing through hydrolysis will be a common property for bacterial class C introns, but will not be the exclusive pathway. In addition, the introns S.me.I3 and A.v.I2 showed multiple bands upon self-splicing that might correspond to noncognate splicing products, analogous to B.h.I1, but these products were not characterized further.
Alternative ligation events involving the 5′ exon
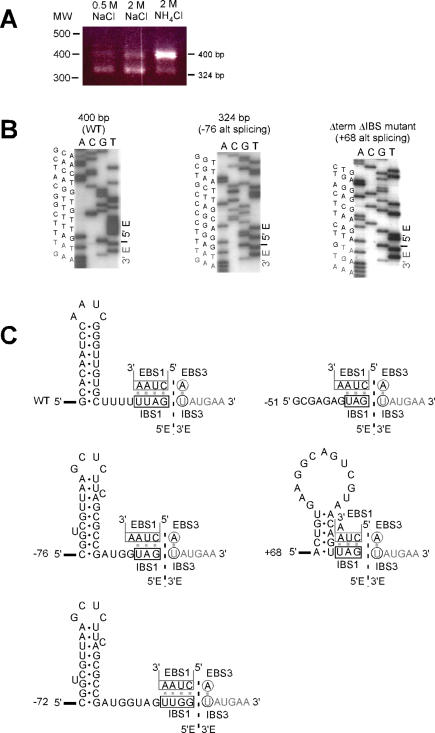
Accurate ligation of exons was tested by RT–PCR amplification of total reaction products and sequencing. Surprisingly, DNAs of two sizes were amplified, whose ratios were dependent on salt conditions in the splicing reaction (Figure 3A). Splicing reactions with 2M NH4Cl produced a predominant RT–PCR product of 400 bp, which is the expected size for accurate exon ligation, and which was confirmed by sequencing (Figure 3B). However, reactions with 0.5 or 2M NaCl produced a 324 bp band at substantial levels, which upon sequencing was found to represent an alternative splicing event in which the normal 3′ exon is spliced to a cryptic 5′ splice site located 76 bp upstream of the intron (Figure 3B and C). At this point, the band labeled ‘alternatively ligated exons’ in Figure 2 was excised from a gel, RT–PCR amplified, and sequenced to confirm its identity. Interestingly, examination of the −76 site showed it to contain an IBS1 sequence preceded by a stem–loop (Figure 3C). The stem–loop it is predicted to form by MFOLD (35) despite its two bulges (data not shown).
Figure 3.
Cognate and alternative exon ligation events during in vitro splicing reactions. (A) RT–PCR of exon junctions produced in self-splicing reactions. Self-splicing was under standard conditions with 2 M NH4Cl, or with 0.5 or 2 M NaCl. RT–PCR detected two splicing products of 400 bp and 324 bp, whose proportions varied with ionic conditions. (B) Sequencing of ligated exon products. Sequences are shown for the exon-exon junctions of the 400 bp and 324 bp bands in Panel A, and also for the major ligation product formed for the ΔterminatorΔIBS1 mutant described in Figure 4. (C) Sequences and structures of ligated exons. Structures are shown for: the exon–exon products of a cognate splicing reaction (WT); the upstream alternative splicing events at −76, −72, and −51 (relative to the exon–intron junction); and the internal splicing event +68 (relative to the exon–intron junction). EBS1 and EBS3 interactions are drawn above the exon sequences. The +68 stem–loop is present in the native secondary structure, although recognition of the stem may require unpairing of α–α′.
To expand sampling of alternative splicing events, approximately 100 individually cloned RT–PCR products were examined for insert size by PCR, using products from all three reaction conditions in Figure 3A. Nearly all PCR products comigrated with the 324 and 400 bp bands; however, 17 bands did not, and were sequenced. Four of these (representing four of the ∼100 initial clones) in fact had a new 5′ splice site, either at position −72 (3 clones), or position −51 (1 clone) (Figure 3C). The other 13 junctions were wild-type. Both of the minor ligation sites followed IBS1 sequences (UAG and UUGG), with the −72 position being downstream of a stem–loop. A fourth alternative splice site was later found for an intron having a mutated IBS1 motif (ΔtermΔIBS1, described below). Together, these data show that B.h.I1 can utilize 5′ splice sites that are not adjacent to the 5′ GUGYG start of the intron, and ligate the cryptic 5′ sites to a 3′ exon. This is a highly unexpected property not reported for other group II introns.
Mutations and sequence variations in the 5′ exon
Because of the implied role of the stem–loop motif in 5′ exon recognition, we examined its contribution more thoroughly through mutagenesis and sequence variations. First, another intron copy was tested, B.h.I1(X), because it has different flanking exons compared to the other four B.h.I1 copies. In the 5′ exon of B.h.I1(X) is a mispaired stem–loop lacking an extended run of U's, and the IBS1 provides only 3 bp with EBS1 (Figure 4A). The 3′ exon, although different from B.h.I1(B), is not expected to affect the reaction because it has the IBS3 pairing that is critical for 3′ exon recognition. Self-splicing of B.h.I1(X) transcript produced linear intron and other products, but notably no ligated exons (Figure 4B). Importantly, even RT–PCR failed to detect ligated exon products (data not shown). These observations suggest that the 5′ exon sequence has a profound effect on the splicing reaction, and that there are determinants in addition to the IBS1–EBS1 pairing that specify the 5′ exon, since the IBS1–EBS1 pairing is only mildly weakened in this variant. Failure to produce ligated exons is most likely due to dissociation of the 5′ exon from the intron after the first step of splicing.
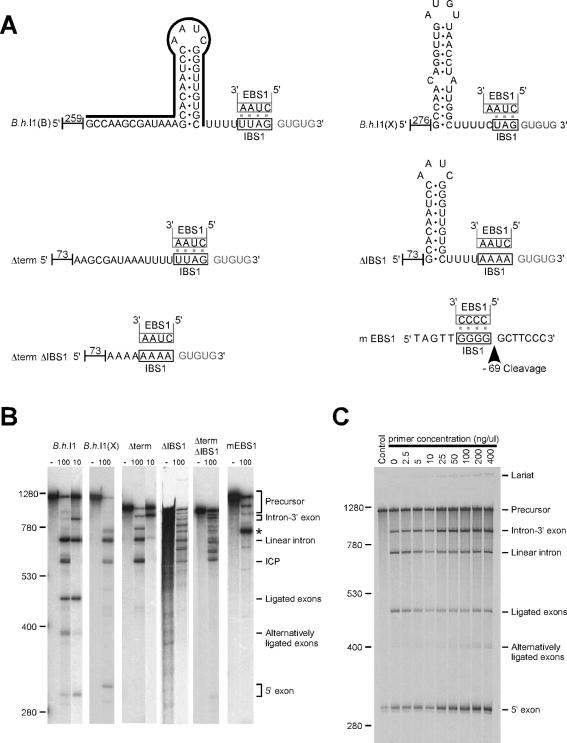
Figure 4.
Mutations and sequence variations in motifs involved in 5′ exon recognition. (A) Diagram of variations tested: B.h.I1(B), wild-type exons; B.h.I1(X), exons from the B.h.I1(X) site with a mispaired stem rather than a canonical rho-independent terminator motif; Δterm, deleted terminator stem–loop; ΔIBS1, mutated IBS1 motif; ΔtermΔIBS1, deleted terminator stem–loop and mutated IBS1 motif; mEBS1, mutated EBS1 motif in the intron. Numbers indicate the length of the 5′ leader (vector sequence and native exon combined). The line above the B.h.I1(B) sequence shows the positions corresponding to the complementary oligonucleotide added to the reaction in Panel C. The arrow for mEBS1 indicates the cleavage produced during the splicing reaction. (B) Self-splicing reactions of the variants tested. Each set of lanes shows unspliced transcript incubated without magnesium (left) and intron transcript spliced under standard conditions with either 10 mM or 100 mM MgCl2. The asterisk denotes the cleavage at −69 in the 5′ exon, which was determined by primer extension. Unligated 5′ exons cannot be seen in panel B for the constructs Δterm, ΔIBS1, ΔtermΔIBS1 and mEBS1 because the exons are too short, and unligated 3′ exons are too short to be seen for all constructs. (C) Self-splicing in the presence of an oligonucleotide that pairs with the terminator stem–loop. An oligonucleotide complementary to the terminator stem–loop of B.h.I1 was added to intron transcript during the denaturation/renaturation step, to disrupt the stem–loop structure (Panel A). The disruption resulted in an increase in unligated 5′ exon, and intron-3′ exon intermediates.
Next, a mutant of B.h.I1(B) was tested that lacks the terminator stem–loop but has an intact IBS1 sequence (Δterm; Figure 4A). Splicing products of this mutant are similar to B.h.I1(X) in that no ligated exons are produced (Figure 4B). Importantly, self-splicing of Δterm RNA under conditions of 10 mM MgCl2 produced a dramatic accumulation of intron-3′ exon intermediate compared to wild-type intron under the same conditions. This finding strengthens the argument that the stem–loop affects the anchoring of the 5′ exon to the intron after the first step of splicing, and that the 5′ exon may dissociate if this interaction is disrupted.
Mutations of IBS1 sequence had a different, more drastic effect. Very similar products were seen for two constructs mutated either for IBS1 alone (ΔIBS1), or for IBS1 and the terminator together (ΔtermΔIBS1) (Figure 4B). Many unexpected products were formed, presumably due to the hydrolysis at cryptic IBS1 sites in the absence of a proper IBS1. The exact nature of the bands has not been determined, but primer extension experiments showed that at least some products have native 5′ intron termini (data not shown). Most important is the fact that no correctly ligated exons were detected in gels (Figure 4B), and RT–PCR amplification revealed only an alternative ligation corresponding to the joining of the correct 3′ exon to the +68 site internal to the intron, as shown in Figure 3B and C. In this mutant, the absence of a usable IBS1 sequence resulted in recognition of a cryptic 5′ site within the intron, at +68, which is the same position as the +68 cleavage in Figure 2B and C. Therefore, the intron can use an internal site as a 5′ exon, as well as the distant upstream site at −76.
A converse mutation was made, in which the EBS1 sequence was mutated to CCCC while leaving the IBS1 unchanged. Again the normal splicing reaction was blocked, and the major product was a cleavage in the 5′ exon (starred band in Figure 4B), which was identified by primer extension to occur at −69 in the 5′ exon, and to correspond to the cryptic IBS1 sequence GGGG (Figure 4A). This cleavage reaction is analogous to ICP formation, in that the intron recognizes and cleaves at an IBS1 sequence, but the cryptic exon is not ligated to a 3′ exon. RT–PCR did not detect any exon ligation for this mutant, indicating that a strong IBS1 interaction alone is not sufficient for ligation to the 3′ exon. Notably, there is no ICP formation for the EBS1 mutant, because the altered EBS1 sequence no longer pairs with the cryptic IBS1 targets in the intron, thus corroborating the deduced mechanism of ICP formation. Together, these IBS1 mutations underscore the primary importance of the IBS1–EBS1 interaction, and suggest that when a proper interaction cannot form, the EBS1 interacts with cryptic IBS1 sequences to produce cleavages.
As a final investigation of the effect of the terminator structure on splicing, an oligonucleotide complementary to the stem–loop was added to the splicing reaction to inhibit formation of the stem–loop (Figure 4A). RNase H digestion of the primer-annealed RNA confirmed formation of the RNA–DNA heteroduplex (data not shown). Using this assay, it was found that increasing levels of oligonucleotide resulted in substantial increases in both unligated 5′ exon and intron-3′ exon intermediate (Figure 4C), suggesting that disruption of the stem–loop causes a barrier after the first step of splicing. Unexpectedly, ligated exon levels did not change greatly, but this is explained by the carrier effect of the oligonucleotides during ethanol precipitation, which produced greater recovery in lanes with more oligonucleotide, but which should not produce differential recovery of RNA species. Consistent with these explanations, phosphorimaging quantitation of bands in the gel showed that the two products of the first step of splicing (5′ exon + intron-3′ exon signals combined) increased from 16.7% of the five bands in lane 2, to 34.9% of the five bands in lane 10. These data support the idea that disruption of an interaction between the ribozyme and stem–loop antagonizes the ability of the ribozyme to anchor the 5′ exon after the first step of the reaction, and further implicates the stem–loop in 5′ exon recognition.
DISCUSSION
We have characterized a IIC ribozyme for its self-splicing reaction and uncovered novel properties, including the first examples of alternative 5′ splicing events for a group II intron, and evidence that IIC introns differ from IIA and IIB introns in recognition of their 5′ exons. To our knowledge, this is the first example of a wild-type group II intron that can utilize 5′ splice sites nonadjacent to the canonical GUGYG start of the intron. In fact, four nonadjacent splice sites were detected in our reactions, and one of these occurred at high efficiencies under certain ionic conditions. The flexibility of the ribozyme to act on distant exons has important implications for 5′ exon recognition for IIC introns, and also suggests new approaches in engineering IIA and IIB introns to act in trans on other RNA molecules.
Significance of the noncognate splicing products
Although the unexpected ribozyme reactions are remarkable and informative with regard to 5′ exon recognition, they probably do not occur in vivo. In vivo, the reaction is facilitated by the maturase (RT) protein, which is expected to affect product fidelity. Indeed, recent experiments reconstituting a maturase-assisted reaction for the B.h.I1 intron detected none of the unusual products found in this study, but only the expected canonical splicing products (A. R. Robart, W. Seo and S. Zimmerly, manuscript submitted). In addition, the in vivo spliced exons detected by RT–PCR amplification and sequencing of RNA isolated from B.halodurans cells showed only correct splicing events for B.h.I1 (W. Seo and S. Zimmerly, unpublished data). B.h.I1 has been determined to be expressed in vivo by ∼1% termination readthrough; W. Seo and S. Zimmerly, unpublished data. Therefore, splicing of B.h.I1 in vivo is expected to exhibit the accuracy seen for other group II introns, rather than the unusual products seen in the RNA-alone reaction.
Nevertheless, the ability to utilize nonadjacent splice sites should be kept in mind when considering the diversity of group II introns in nature. Already there are two reports of unexpected 3′ splice sites, one of which resembles recognition in trans. The B.ce.I4 intron utilizes a 3′ splice site 56 nt downstream of the expected splice site, as detected by RT–PCR and sequencing, which permits the downstream ORF to be translated in frame with the upstream exon (27). In a less dramatic example, in vivo splicing was found to occur to a low extent at a minor site 4 nt downstream of the expected 3′ boundary, which again fuses upstream and downstream exon ORFs (24). Unexpected 5′ splice sites are also suggested for introns that lack strong IBS1 pairings. For example, a nonadjacent 5′ splice site is suggested by LSUI5 of Agrocybe aegerita mitochondria, which lacks IBS1 and IBS2 pairings directly upstream of the 5′GUGYG, but has a good IBS1 match 23 nt upstream (36).
Mechanism of 5′ exon recognition
It is perplexing that IIC introns recognize their 5′ exons through only 3–4 bp (IBS1–EBS1), as predicted based on their secondary structures (Figure 1). Thus the 5′ exon-intron interactions are reduced from 12 bp for IIA and IIB introns to 3–4 bp for IIC introns. This drastic difference raises the question of how the 5′ exon is recognized, and whether there is a compensating interaction that substitutes for the IBS2–EBS2 pairing. A second possibility is that IIC introns are simply less stringent in their 5′ exon recognition compared to IIA or IIB introns. The latter possibility should be considered seriously because IIC introns do not reside within ORFs, and consequently host gene expression should not be affected by the accuracy of exon ligation, or even whether splicing occurs. Along this line of reasoning, the only purpose of splicing for IIC introns is to generate an RNP complex capable of carrying out mobility, which is a property demonstrated biochemically for this intron (A. R. Robart, W. Seo and S. Zimmerly, manuscript submitted). As long as some amount of this RNP complex is produced, the element should be able to survive as a selfish DNA element. Nevertheless, only accurate exon ligation events are detected in vivo (W. Seo and S. Zimmerly, unpublished data) or in a maturase-assisted reaction (A. R. Robart, W. Seo and S. Zimmerly, manuscript submitted), indicating that exon definition occurs accurately in vivo for IIC introns in spite of the lower information content in the 5′ exon pairings.
We suggest here that a second 5′ exon–intron interaction occurs for IIC introns and contributes to 5′ exon recognition, namely a non-Watson–Crick interaction between intron and the terminator stem–loop in the 5′ exon. This interaction is clearly less critical than IBS1, because some splicing events occur without the stem–loop motif (Figure 2), whereas this is not true for the IBS1 motif. Furthermore, high magnesium concentrations can suppress the absence of the stem–loop to some extent, while IBS1 mutations abolish correct splicing (e.g. Δterm, ΔIBS1 in Figure 4B). The paramount importance of IBS1 is consistent with properties of IIA and IIB introns. Some IIA and IIB introns have weak or missing IBS2–EBS2 pairings, and mutagenesis of IBS1 sequences has much more drastic effects than mutations of IBS2 sequences (9,37,38).
The involvement of the stem–loop in 5′ exon recognition is supported by several accumulated lines of evidence. Most striking is the finding that in reactions with NaCl, the preferred ligation event occurs at a cryptic site at position −76 having a stem–loop and IBS1 motif, rather than the wild-type site. This perplexing observation indicates that under certain conditions, the intron prefers a distant, noncognate 5′ exon over the normal sequence and position. Two other alternative splicing products (−72 and +68; Figure 3) show the same phenomenon of cryptic exon ligations occurring at stem–loop/IBS1 motifs, together suggesting that both elements are involved in 5′ exon recognition.
In a second line of evidence, cleavages were detected at noncognate stem–loop/IBS1 motifs, again identifying these motifs as imperfect 5′ exons. Such cleavages were seen at the +68 internal cleavage site (Figure 2C), and in self-splicing reactions of B.h.(X) (Figure 4A). Both of these reactions can be seen as the failure to ligate cleaved exons after the first step of a splicing reaction. In both reactions, a cryptic 5′ exon motif is recognized by the ribozyme, but the ribozyme cannot react fully to ligate the 5′ exon, and so a break is produced in the RNA. The +68 cleavage could alternately be seen as an SER-type cleavage reaction, although the fact that ligation can occur at that position indicates that it is recognized as a 5′ exon at least in some instances. The B.h.I1(X) reaction is particularly important in this argument, because there is no detectable exon ligation, even using RT–PCR. The IBS1–EBS1 pairing is weakened by only 1 bp in this variation, and it seems unlikely that this by itself would obliterate exon ligation. Hence the 5′ exon sequences upstream of IBS1 are implicated in the failure of exon ligation in this context.
The third line of evidence implicating the stem–loop comes from the experimental disruption of the motif. Deletion of the terminator stem (Δterm mutation) results in the inability of the intron to ligate the exons (Figure 4), an unexpected result if IBS1 alone is required for exon ligation. Further, at 10 mM magnesium concentrations for this mutant, there is an accumulation of unligated intron-3′ exon intermediate, indicating a block in the second step that is rationalized as the dissociation of the 5′ exon because of disrupted contacts. In contrast, wild-type intron is still able to produce ligated exons at 10 mM (Figure 4B). The oligonucleotide inhibition experiment also supports an interaction between the stem–loop and ribozyme, because the 5′ exon sequence is unchanged.
Together, these lines of evidence clearly show that the splicing reaction is influenced by 5′ exon sequence in addition to IBS1, and strongly suggest that the stem–loop is recognized by the ribozyme in an interaction that substitutes to some extent for the missing IBS2–EBS2 interaction in IIC introns. The data also suggest that the spacing between the stem–loop and IBS1 is not critical, because among the different reactions, splice sites are observed in the range of 3–11 nt downstream of the stem–loop. It is notable that not all stem–loops are equivalent in the ribozyme reactions. For example, it is not readily obvious why the bulged −76 stem–loop competes with wild-type exon for exon ligation, while the B.h.(X) stem–loop interferes with exon ligation. Further insight into the nature and significance of these interactions awaits more thorough mutagenesis and biochemical experiments.
Evolutionary implications
There now appear to be three different modes of exon recognition for group II introns. IIA introns rely on IBS1, IBS2 and δ′ pairings; IIB introns rely on IBS1, IBS2 and IBS3 pairings; and IIC introns rely on IBS1 and IBS3 pairings, and putatively on a new nonpairing interaction with a stem–loop. The question arises as to what form of exon recognition was ancestral, and how the three varieties of ribozymes arose.
While it is not possible to answer this issue at present, the best line of evidence is based on the observation that the RNAs and ORFs of group II introns largely coevolved (8), thus allowing one at least in principle to follow evolution of the ribozyme by following evolution of the RT ORF. By this criterion, the earliest branching class of introns is most likely to be bacterial class C (IIC RNA structure), which is inferred from RT trees that were outgroup rooted with RTs of retrons or non-LTR elements [(39); D. Simon and S. Zimmerly, unpublished data]. Even if this conclusion is borne out with the accumulation of more evidence, one still cannot infer decisively the properties of the ancestral intron, because all lineages have diverged since their split. For example, the ancestral intron might have had IBS1 and IBS2 interactions, with IIC introns losing IBS2 and gaining a stem–loop interaction. Or, the ancestral intron might have had only IBS1 and IBS3 interactions, with IIA and IIB introns acquiring the IBS2 interaction and IIC introns acquiring a stem–loop interaction. Hence, what seems clear now is that IIC introns are the most divergent and unusual of known self-splicing group II ribozymes.
Acknowledgments
This work was supported by funds from Natural Sciences and Engineering Research Council (NSERC). In addition, A.R.R. was supported by a NSERC PGS-B studentship, and S.Z.'s salary was from Alberta Heritage Foundation for Medical Research (AHFMR). Funding to pay the Open Access publication charges for this article was provided by NSERC grant 203717-98.
Conflict of interest statement. None declared.
REFERENCES
- 1.Robart A.R., Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 2005;110:589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 3.Toro N. Bacteria and archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 4.Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M. Mobile introns: pathways and proteins. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 761–783. [Google Scholar]
- 5.Bonen L., Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- 6.Ferat J.L., Le Gouar M., Michel F. A group II intron has invaded the genus Azotobacter and is inserted within the termination codon of the essential groEL gene. Mol. Microbiol. 2003;49:1407–1423. doi: 10.1046/j.1365-2958.2003.03649.x. [DOI] [PubMed] [Google Scholar]
- 7.Toro N., Molina-Sánchez M.D., Fernández-López M. Identification and characterization of bacterial class E group II introns. Gene. 2002;299:245–250. doi: 10.1016/s0378-1119(02)01079-x. [DOI] [PubMed] [Google Scholar]
- 8.Toor N., Hausner G., Zimmerly S. Coevolution of group II intron RNA structures with their intron-encoded reverse transcriptases. RNA. 2001;7:1142–1152. doi: 10.1017/s1355838201010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel F., Umesono K., Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 10.Qin P.Z., Pyle A.M. The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol. 1998;8:301–308. doi: 10.1016/s0959-440x(98)80062-6. [DOI] [PubMed] [Google Scholar]
- 11.Michel F., Ferat J.L. Structure and activities of group II introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 12.Jacquier A., Jacquesson-Breuleux N. Splice site selection and role of the lariat in a group II intron. J. Mol. Biol. 1991;219:415–428. doi: 10.1016/0022-2836(91)90183-7. [DOI] [PubMed] [Google Scholar]
- 13.Costa M., Michel F., Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–5018. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granlund M., Michel F., Norgren M. Mutually exclusive distribution of IS1548 and GBSi1, an active group II intron identified in human isolates of group B streptococci. J. Bacteriol. 2001;183:2560–2569. doi: 10.1128/JB.183.8.2560-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai L., Toor N., Olson R., Keeping A., Zimmerly S. Database for mobile group II introns. Nucleic Acids Res. 2003;31:424–426. doi: 10.1093/nar/gkg049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai L., Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 2002;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyle A.M., Lambowitz A.M. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland R.F., Cech T.R., Atkins J.F., editors. The RNA World, 3rd edn. NY: Cold Spring Harbor Press, Cold Spring Harbor; 2006. pp. 469–506. [Google Scholar]
- 18.Mörl M., Niemer I., Schmelzer C. New reactions catalyzed by a group II intron ribozyme with RNA and DNA substrates. Cell. 1992;70:803–810. doi: 10.1016/0092-8674(92)90313-2. [DOI] [PubMed] [Google Scholar]
- 19.Hebbar S.K., Belcher S.M., Perlman P.S. A maturase-encoding group IIA intron of yeast mitochondria self-splices in vitro. Nucleic Acids Res. 1992;20:1747–1754. doi: 10.1093/nar/20.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa M., Fontaine J.M., Loiseaux-de Goër S., Michel F. A group II self-splicing intron from the brown alga Pylaiella littoralis is active at unusually low magnesium concentrations and forms populations of molecules with a uniform conformation. J. Mol. Biol. 1997;274:353–364. doi: 10.1006/jmbi.1997.1416. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt U., Riederer B., Mörl M., Schmelzer C., Stahl U. Self-splicing of the mobile group II intron of the filamentous fungus Podospora anserina (COI I1) in vitro. EMBO J. 1990;9:2289–2298. doi: 10.1002/j.1460-2075.1990.tb07400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamidi C., Fedorova O., Pyle A.M. A group II intron inserted into a bacterial heat-shock operon shows autocatalytic activity and unusual thermostability. Biochemistry. 2003;42:3409–3418. doi: 10.1021/bi027330b. [DOI] [PubMed] [Google Scholar]
- 23.Perlman P.S., Podar M. Reactions catalyzed by group II introns in vitro. Methods Enzymol. 1996;264:66–86. doi: 10.1016/s0076-6879(96)64010-5. [DOI] [PubMed] [Google Scholar]
- 24.Robart A.R., Montgomery N.K., Smith K.L., Zimmerly S. Principles of 3′ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA. 2004;10:854–862. doi: 10.1261/rna.5246804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa M., Michel F., Toro N. Potential for alternative intron-exon pairings in group II intron RmInt1 from Sinorhizobium meliloti and its relatives. RNA. 2006;12:338–341. doi: 10.1261/rna.2240906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa M., Michel F., Molina-Sánchez M.D., Martinez-Abarca F., Toro N. An alternative intron-exon pairing scheme implied by unexpected in vitro activities of group II intron RmInt1 from Sinorhizobium meliloti. Biochimie. 2006;88:711–717. doi: 10.1016/j.biochi.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Tourasse N.J., Stabell F.B., Reiter L., Kolstø A.B. Unusual group II introns in bacteria of the Bacillus cereus group. J. Bacteriol. 2005;187:5437–5451. doi: 10.1128/JB.187.15.5437-5451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saldanha R., Chen B., Wank H., Matsuura M., Edwards J., Lambowitz A.M. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 29.Noah J.W., Lambowitz A.M. Effects of maturase binding and Mg2+ concentration on group II intron RNA folding investigated by UV cross-linking. Biochemistry. 2003;42:12466–12480. doi: 10.1021/bi035339n. [DOI] [PubMed] [Google Scholar]
- 30.Cui X., Matsuura M., Wang Q., Ma H., Lambowitz A.M. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J. Mol. Biol. 2004;340:211–231. doi: 10.1016/j.jmb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Takami H., Nakasone K., Takaki Y., Maeno G., Sasaki R., Masui N., Fuji F., Hirama C., Nakamura Y., Ogasawara N., et al. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J., Russell D.W. Molecular cloning: A Laboratory Manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 33.Jarrell K.A., Peebles C.L., Dietrich R.C., Romiti S.L., Perlman P.S. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- 34.Murray H.L., Mikheeva S., Coljee V.W., Turczyk B.M., Donahue W.F., Bar-Shalom A., Jarrell K.A. Excision of group II introns as circles. Mol. Cell. 2001;8:201–211. doi: 10.1016/s1097-2765(01)00300-8. [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toor N., Zimmerly S. Identification of a family of group II introns encoding LAGLIDADG ORFs typical of group I introns. RNA. 2002;8:1373–1377. doi: 10.1017/s1355838202023087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacquier A., Michel F. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group II self-splicing introns. J. Mol. Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- 38.Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987;50:17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- 39.Rest J.S., Mindell D.P. Retroids in archaea: phylogeny and lateral origins. Mol. Biol. Evol. 2003;20:1134–1142. doi: 10.1093/molbev/msg135. [DOI] [PubMed] [Google Scholar]