Abstract
Specificity of mammalian gene regulatory regions is achieved to a large extent through the combinatorial binding of sets of transcription factors to distinct binding sites, discrete combinations of which are often referred to as regulatory modules. Identification and subsequent characterization of gene regulatory modules will be a key step in assembling transcriptional regulatory networks from gene expression profiling data, with the ultimate goal of unravelling the regulatory codes that govern gene expression in various cell types. Here we describe the new bioinformatics tool, Composite Motif Discovery (CoMoDis), which streamlines computational identification of novel regulatory modules starting from a single seed motif. Seed motifs represent binding sites conserved across mammalian species. CoMoDis facilitates novel motif discovery by automating the extraction of DNA sequences flanking seed motifs and streamlining downstream motif discovery using a variety of tools, including several that utilize phylogenetic conservation criteria. CoMoDis is available at http://hscl.cimr.cam.ac.uk/CoMoDis_portal.html.
INTRODUCTION
The identification of gene regulatory elements is integral to the reconstruction of the regulatory networks that determine the spatiotemporal control of gene expression. Individual genes within transcriptional regulatory networks are connected through regulatory modules, typically multi-protein complexes bound to cis-regulatory regions containing multiple transcription factor binding sites (TFBSs). However, identification of mammalian regulatory modules represents a formidable task because (i) the individual DNA sequence motifs recognized by transcription factors are often short and degenerate, so that they will occur by chance and (ii) unlike in lower model organisms regulatory elements can be located many kilobases away from the proximal promoter in distal 5′ or 3′ enhancers or in introns (1). Methods such as phylogenetic footprinting for the identification of evolutionary conserved sites and the use of whole genome statistics, such as the regulatory potential score (2,3) can improve the discovery of motifs bound by transcription factors in vivo. In addition, strategies aimed at identifying clusters of motifs also improve the chances of finding ‘real’ sites (4–8). Only a small number of combinatorial regulatory codes have been identified, such as those controlling liver and muscle tissue specific gene expression (9,10). In light of the rapidly increasing generation of expression profiling datasets, the identification of additional combinatorial regulatory codes will be essential to gain a mechanistic understanding of the molecular controls that generate differential expression patterns. This will be important to understand developmental/differentiation time courses and differences between normal/pathological states. It may also enable the development of future therapies that would, e.g. reverse malignant gene expression patterns or permit the reprogramming of differentiated cells to immature progenitors to regenerate aged and/or damaged tissue.
We describe here a new bioinformatics tool Composite Motif Discovery (CoMoDis) to aid in the discovery of composite regulatory modules ‘seeded’ by a single known motif that is thought to be important in the regulation of a set of genes. Given a list of genes from either human or mouse genomes, CoMoDis extracts the sequence surrounding all conserved seed motifs in the vicinity of these genes and streamlines downstream motif discovery. CoMoDis has some similarities to a number of other tools, notably the Composite Module Analyst (11,12), POXO (13) and CRSD (14). All three of these web accessible tools accept lists of coregulated genes and attempt to generate hypotheses about the factors controlling their common expression pattern, including TFBSs and, in the case of CRSD, microRNA. However, an important difference is that CoMoDis can utilize the sequence of entire gene loci, whereas the other methods focus on the promoter region of input genes (CRSD also searches the 3′-untranslated region) potentially missing important regulatory elements. Moreover, CoMoDis provides integration with eight different motif discovery programs, facilitating the prioritization of those motifs that are computationally predicted by multiple methods.
METHODOLOGY OF CoMoDis
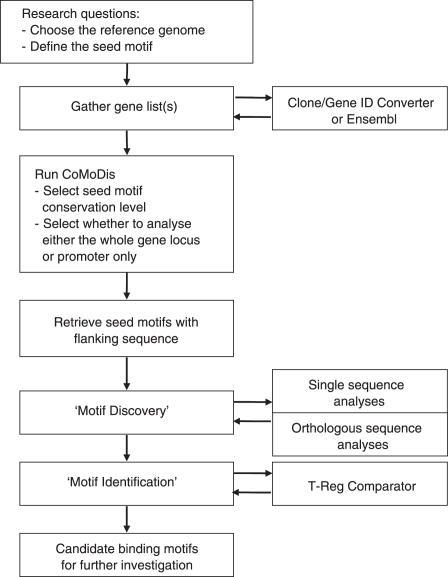
The flow of data from the conception of a motif discovery experiment, through the processing of a gene list by CoMoDis, and the final analysis of seed motif associated sequences generated by CoMoDis is summarized in Figure 1. A typical motif discovery experiment using CoMoDis begins with a list of genes that are thought to be controlled by the same transcription factor that has a known DNA sequence binding motif. CoMoDis locates all conserved motifs for this factor (‘seed motifs’) within the loci of the presumed target genes and outputs seed motif flanking sequences for subsequent motif discovery. The figure highlights questions that should be considered when using CoMoDis and also external tools that will aid the user in completing the analysis.
Figure 1.
The flow of data in a typical motif discovery experiment using CoMoDis. The user begins with a list of genes thought to be controlled by the same transcription factor with a known DNA sequence binding motif. CoMoDis locates all conserved motifs for this factor (‘seed motifs’) within the loci of the presumed target genes and outputs seed motif flanking sequences for subsequent motif discovery. Questions are highlighted that should be considered when using CoMoDis. External tools are also shown that will aid the user in completing the analysis.
Seed motif datasets
The seed motifs are stored in a set of datafiles each of which contains the genomic positions of a given motif, conserved across whole-genome alignments. In addition to sequence conservation, regulatory potential score and in vivo promoter mapping datasets can be used to help distinguish likely functional binding sites (true positives) from the background noise of non-functional sites (false positives).
Genome-wide positions for three sets of IUPAC code defined TFBS consensus sequences and one set of positional weight matrices can be used as seed motifs. The first IUPAC code set consists of 41 consensus sequences (see Table 1) curated from the literature and the databases TRANSFAC (15) and JASPAR (16,17). Background information and references for each IUPAC consensus sequence can be found at http://hscl.cimr.cam.ac.uk/TFBScluster_genome_35_filters_background.html. Five datafiles containing genome-wide collections of matching conserved sites with increasing levels of sequence conservation have been generated for all 41 IUPAC consensus sequences. The first datafile contains ‘non-exact’ matches to the core sequence; both sequences match the IUPAC consensus, but degenerate IUPAC codes are allowed to differ between species. The second datafile contains ‘exact’ matches, where degenerate IUPAC codes must be identical between species, thus requiring degenerate consensus sequences to be aligned in regions with higher levels of sequence identity. The last three datafiles also require an exact match and extend the overall length of sequence identity. To achieve this, the IUPAC code ‘N’ (any nucleotide) is added to both ends of the consensus, resulting in three files with two, four and six conserved nucleotides flanking the core sequence. Functional binding sites are often located in highly conserved sequence regions. Therefore, increasing the degree of conservation in TFBS datafiles should enrich functional binding sites whilst decreasing the number of false positive sites. The identification of completely or near completely conserved binding sites is a method that has been successfully used in a variety of other approaches (18).
Table 1.
Transcription factor seed motifs currently available in CoMoDis
| In-house curated motifs | Xie et al. motifs (19) | Ettwiller et al. (CpG) motifs (20) | |||
|---|---|---|---|---|---|
| Name | Consensus | Identifier | Consensus | Identifier | Consensus |
| AML1 | TGYGGT | #1 (NRF-1) | RCGCANGCGY | #1 (CAAT) | CCAATC |
| AP1 | NNNSTCA | #2 (MYC) | CACGTG | #2 (SP1) | GGGCGG |
| CRE | TGACGTCA | #3 (ELK-1) | SCGGAAGY | #3 (CRE) | TGACGTCA |
| CRE | TGACG (half) | #4 (Novel) | ACTAYRNNNCCCR | #4 (ETS) | CGGAAG |
| CEBP | SYAAY | #5 (NK-Y) | GATTGGY | #5 (Ebox) | CACGTG |
| EBF | CCCNNGRG | #6 (SP1) | GGGCGGR | #6 | ACTACA |
| Ebox | CANNTG | #7 (AP-1) | TGANTCA | #7 (CRE-like) | GTGACG |
| Ebox-GATA | CANNTG-GATA | #8 (Novel) | TMTCGCGANR | #8 | CTTTGT |
| c-Myc | CAYGYG | #9 (ATF3) | TGAYRTCA | #9 (SP1-like) | CCCTCCCCC |
| MyoD | CANCWG | #10 (YY1) | GCCATNTTG | #10 | GCGCAGGCGC |
| ETS | GGAW | #11 (GABP) | MGGAAGTG | #11 | GCGCGC |
| GATA | GATA | #12 (E12) | CAGGTG | #12 | AACTTT |
| GLI1 | GACCACCCA | #13 (LEF1) | CTTTGT | #13 | CCTTTAA |
| HMG | WWCAAWG | #14 (ATF3) | TGACGTCA | #14 | TGCGCA |
| HNF1 | GTTAAT | #15 (AP-4) | CAGCTG | #15 | CTCGCGAGA |
| HNF3 | TRTTTRY | #16 (C-ETS-2) | RYTTCCTG | #16 | TTGGCT |
| HNF4 | CAAAGK | #17 (IRF1) | AACTTT | #17 (TATA) | TATAAA |
| Ikaros | HRGGAW | #18 (SREBP-1) | TCANNTGAY | #18 | AAGATGGCGG |
| Iroquois | ACANNTGT | #19 (Novel) | GKCGCN(7)TGAYG | #19 | TTTGTT |
| MEF2 | CTAWWWWTAR | #20 (E4F1) | GTGACGY | #20 | ATGCAAAT |
| MEIS1 | TGACAS | #21 (Novel) | GGAANCGGAANY | #21 | TAATTA |
| MYB | YAACNG | #22 (Novel) | TGCGCANK | #22 | TTTAAG |
| NBOX | CACNAG | #23 (CHX10) | TAATTA | #23 | CGCATGCG |
| NANOG | SATTANS | #24 (MAZ) | GGGAGGRR | #24 | ATAAAT |
| NFAT | GGAAA | #25 (ESRRA) | TGACCTY | #25 | TTTAAA |
| NFAT-AP1 | WGGAAA-TGASTCA | #26 (E4BP4) | TTAYRTAA | #26 | GCCATTTT |
| NFAT-AP1 | WGGAAA-STCA (half) | #27 (Novel) | TGGN(6)KCCAR | #27 | ATAAAA |
| NFKB | GGGRNNYYY | #28 (RSRFC4) | CTAWWWATA | #28 | TAAATA |
| NKX2.5 | CAMTTNR | #29 (Novel) | CTTTAAR | #29 (HTH) | CAGGTG |
| OCT3/4 | ATGMWWVW | #30 (Novel) | YGCGYRCGC | #30 | CTAGCAAC |
| OTX | TAATCY | #31 (Novel) | GGGYGTGNY | #31 (CRE) | TGACGC |
| p53 | RCNWGYNN*0-1*NNRCAWGY | #32 (NF-E2) | TGASTMAGC | #32 | CATTGT |
| PAX5 | RNKMANBSNWGNRKRMM | #33 (MEF-2) | YTATTTTNR | #33 | GCCATCTT |
| RE1 | NTYAGMRCCNNRGMSAG | #34 (Novel) | CYTAGCAAY | #34 | ATTTAT |
| SOX2 | CWTTGTD | #35 (MYOD) | GCANCTGNY | #35 | ATGAAT |
| SP1 (1) | GGGHGGG | #36 (FREAC-2) | RTAAACA | Ettwiller et al. (non-CpG) motifs (20) | |
| SP1 (2) | GGGSWGGG | #37 (Novel) | GTTRYCATRR | ||
| SP1 (3) | GGKGYGGG | #38 (ERR-alpha) | TGACCTTG | #1 | TAATTA |
| SRF | CCWWWWWWGG | (Novel) | TCCCRNNRTGC | #2 | CAGCTG |
| STAT5 | TTCYNRGAA | #40 (STAT5A) | TTCYNRGAA | #3 (TRE) | TGAGTCA |
| TEF | CATTCC | #41 (MEIS1) | TGACAGNY | #4 (ETS) | CAGGAAGT |
| #42 (Novel) | TGACATY | #5 | CCCTCCC | ||
| #43 (Novel) | GTTGNYNNRGNAAC | #6 | AATAAA | ||
| #44 (OCT-X) | YATGNWAAT | #7 (Homeo-like) | AATTAA | ||
| #45 (Novel) | CCANNAGRKGGC | #8 | AGAAAA | ||
| #46 (Novel) | WTTGKCTG | #9 | ATAAAA | ||
| #47 (NF-1) | TGCCAAR | #10 | TTTCCA | ||
| #48 (C-REL) | GCGNNANTTCC | #11 (TATA-box) | TATAAATAG | ||
| #49 (SOX-9) | CATTGTYY | #12 | AGGAAA | ||
| #50 (PU.1) | RGAGGAARY | #13 | TTTCCT | ||
| #14 | TTCAAA | ||||
| #15 | TGACCT | ||||
| #16 | ATTTGCAT | ||||
| #17 | TTGTTT | ||||
| #18 | TTTAAA | ||||
| #19 | TTTCAG | ||||
The second set of IUPAC codes was taken from a recently published study (19), which identified common regulatory motifs conserved in human, dog, mouse and rat genomes. We have now taken the top 50 IUPAC consensus sequences from this study and have determined their positions in whole genome alignments (human–mouse, human–dog, human–opossum, mouse–human, mouse–dog and mouse–opossum). The majority of these sites have been assigned to be the binding sites for specific transcription factors or transcription factor families (see Table 1). The third set of IUPAC codes was taken from a similar study (20) that identified a ‘dictionary’ of conserved consensus sequences in the promoter regions of orthologous human and mouse genes. Again, we have determined the genome-wide positions of the non-degenerate IUPAC consensus sequences detailed in this second study, which differentiated between those located in CpG-rich regions (35 consensus sequences) and those in non-CpG-rich regions (19 consensus sequences). Finally for the human version of CoMoDis, we have incorporated the genome-wide positions of TFBSs matching 410 positional weight matrices conserved in human, mouse and rat whole genome alignments. We obtained these from the University of California at Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu/). The positional weight matrices originate from the TRANSFAC database v8.3.
Whole genome alignments
Human and mouse versions of CoMoDis have been implemented to serve these two large research communities. Both versions incorporate the genome-wide positions of the motifs (described above) conserved in a series of pair-wise genome alignments, where either the human or mouse genome is the reference sequence. The pair-wise genome comparisons were downloaded from Genome Bioinformatics at the UCSC (http://hgdownload.cse.ucsc.edu/downloads.html). For the human centric version (NCBI35/hg17) TFBSs conserved in the mouse genome (NCBI33/mm5) represent the default level of conservation. Positions of TFBSs have also been catalogued in human–dog (canFam1) and human–opossum (monDom1) alignments to produce additional sets of human–mouse conserved sites that retain only those sites that are also conserved in dog or opossum. For the mouse version (NCBI34/mm6) the default level of conservation uses sites conserved in the human (NCBI35/hg17) genome and filtered sets have been produced using mouse–dog (canFam1) and mouse-opossum (monDom1) alignments. Positional information for human and mouse genes was extracted from version 37 of the Ensembl database using the Ensembl API (21).
INPUT
The user first chooses the reference genome (human or mouse) to determine the relevant genome annotation and external data used for the subsequent analysis of candidate regulatory regions. The first screen specifies which dataset of TFBSs should be used, either in-house consensus sequences, conserved regulatory motifs from two other studies (19,20) or TRANSFAC v8.3 conserved matrices. The final option screen requires specific information about the seed motif. The motif itself is selected from a drop-down menu, together with the degree of sequence conservation surrounding the core motif. By default CoMoDis will use the binding sites that are conserved between human–mouse or mouse–human genomes. The degree of conservation and therefore the stringency of the search can be increased by restricting analysis to only those sites that are also conserved in dog or opossum.
In the human version of CoMoDis we have implemented additional filters to increase the likelihood that TFBSs represent functional sites. Regulatory potential scores have been shown by others (2,3) to provide significant enrichment of regulatory sequences. A filter has therefore been implemented that can restrict CoMoDis to use only those sites located in areas of regulatory potential with scores greater than zero (based on the threshold suggested by UCSC http://hgdownload.cse.ucsc.edu/goldenPath/hg17/regPotential/). It is also possible to consider only those motifs that are present within experimentally determined active promoters, using the fibroblast cell line IMR90. This dataset is based on a recent study that identified active promoters using a microarray-based chromatin immunopreciptation method to detect all RNA polymerase II preinitiation complexes assembled on DNA throughout the human genome (22).
The next step is to select a list of genes that are thought to be controlled by the transcription factor binding the specified seed motif. Human or mouse Ensembl identifiers (prefixed by ENSG or ENSMUSG, respectively) can be input directly or via a plain-text file. This allows the user to easily interrogate a single gene of interest or a whole list of genes. The user must then specify whether the entire span of each gene locus will be scanned for occurrences of the seed motif, or whether seed motif searches will be limited to the 5′ region of each gene locus. For the first method the user specifies the nucleotide distance from either side of the genes (limited to 100 kb either side). This will always include motifs present in the introns of a gene. For the second method, the user specifies the nucleotide distance up and downstream of the transcription start site; we define this as the start of the most 5′ exon of all transcripts for a particular gene annotated in Ensembl (limited to 50 kb upstream and 10 kb downstream).
Having defined the parameters for locating seed motifs, the next step is to specify the sequence space for subsequent motif discovery. First, the user chooses the number of nucleotides to be extracted either side of the core motif, including the core sequence itself; this is limited to 500 nt either side. Second, the user specifies whether sequences will be extracted relative to the reference sequence only, or whether orthologous sequence pairs will be extracted. The CoMoDis output for orthologous sequence pairs includes several formats required by different downstream analysis tools, including PhyloCon, PhyloGibbs and PhyME (see Table 2).
Table 2.
Summary of motif discovery and motif scanning tools. The addresses link to the author's website
| Tool | Web site | Reference |
|---|---|---|
| Motif Discovery—single sequence output | ||
| BioProspector | http://ai.stanford.edu/~xsliu/BioProspector/ | (33) |
| DME | http://rulai.cshl.edu/dme/index.shtml | (34) |
| GAME | http://mail.med.upenn.edu/~zhiwei/GAME/ | (35) |
| nMICA | http://www.sanger.ac.uk/Software/analysis/nmica/ | (36) |
| Weeder | http://159.149.109.16:8080/weederWeb/ | (37) |
| YMF | http://wingless.cs.washington.edu/YMF/YMFWeb/YMFInput.pl | (38) |
| Motif Discovery—orthologous sequence output | ||
| PhyloCon | http://ural.wustl.edu/~twang/PhyloCon/ | (39) |
| PhyloGibbs | http://www.imsc.res.in/~rsidd/phylogibbs/ http://www.phylogibbs.unibas.ch/cgi-bin/phylogibbs.pl | (40) |
| PhyME | http://edsc.rockefeller.edu/cgi-bin/phyme/download.pl | (41) |
| MotifScanning | ||
| Clover | http://biowulf.bu.edu/MotifViz/ | (42) |
| MotifScanner | http://homes.esat.kuleuven.be/~thijs/Work/MotifScanner.html | Unpublished |
| PROMO | http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promo.cgi?dirDB=TF_8.3&calledBy=alggen | (43,44) |
CoMoDis OUTPUT
After the job has finished, the results will appear as a series of web links to files that can be viewed or downloaded. The ‘job summary’ file contains information about the parameters that were selected, the genomic coordinates that were used, and the number of motifs localized to each gene of the initial input list. This is important to gauge whether more or less stringent parameters should be used. The user should bear in mind that many downstream analysis tool have sequence input limits; 100 sequences is a good estimate. Motif discovery tools can be broadly grouped into those that utilize a set of sequences extracted from a single genome and those that utilize the sequence similarities found between two sets of phylogenetically orthologous sequences. To this end CoMoDis can format the output sequences, so they can be directly used with the chosen method of downstream analysis. Single reference genome sequences are output in the FASTA format, whereas orthologous sequence pairs are output in four different formats. The first is a general multiple FASTA format and the remaining files are compatible with specific tools. If two motif sequence regions overlap, they will be merged to prevent the same sequence being represented more than once. The seed core motif sequence is masked using the IUPAC character ‘N’ to prevent the contamination of any downstream analysis by the seed motif sequence. Repeat sequences are also masked, and the format this takes depends on the output file type. Finally, a UCSC custom track file ‘motif regions in BED format’ can be used to view the positions of the seed motif regions (including flanking sequence) in relation to all the other chromosomal features that can be displayed on the UCSC genome browser via the custom track utility (http://genome.ucsc.edu/goldenPath/help/customTrack.html#format). When using the UCSC genome browser the correct assembly of the human or mouse genomes must be selected to match those used by Ensembl version 37; these are human build 35 (hg17/May 2004) and mouse build 34 (mm6/March 2005).
CoMoDis has been designed with motif discovery in mind. Regulatory elements are often composed of binding site clusters and CoMoDis facilitates the identification of any other motifs that are over-represented in the vicinity of the seed motif. The results page provides access to another page containing web links to tools that are useful in motif discovery. The details of these tools are summarized in Table 2. Motif discovery tools that use the single reference genome sequence output include: BioProspector, DME, GAME, nMICA, Weeder and YMF. nMICA is a JAVA program that must be installed and run by the user. Motif discovery programs that use the orthologous sequence output include: PhyloCon, PhyloGibbs and PhyME. These three programs together with DME and GAME have been installed on our server where we have now made them publicly available as web tools; the interfaces have been specifically designed to work with the output of CoMoDis. Apart from PhyloGibbs, there have not previously been any publicly accessible web versions of these tools. The selection of tools available via CoMoDis has been chosen as a useful cross-section of the available tools to date in light of a study that assessed the effectiveness of 13 different motif discovery tools (23). We have also provided links to motif scanning tools that will search for known motifs in the single sequence output; these include Clover, MotifScanner and PROMO ‘MultiSearchSites’, summarized in Table 2.
EXAMPLE OF USE
To demonstrate the use of CoMoDis in composite motif discovery we used the results of a published microarray experiment (24) that provided us with well-characterized lists of genes to analyse, the results of which is detailed below. The aim of the mouse study was to investigate how the zincfinger transcription factor GATA-1 regulates cell proliferation. Therefore, GATA-1 expression was induced in a GATA-1 null cell line. Genes that were up and downregulated as a consequence of GATA-1 expression were identified using microarray expression profiling. We used two groups of clustered genes from the whole experiment; upregulated GATA-1 target genes (‘target’) and Myc-related genes repressed by GATA-1 (‘repressed’). Our aim was to discover other motifs apart from the GATA motif itself that are recruited to promote either GATA-1 induced upregulation or repression.
We searched the Ensembl mouse database to retrieve the Ensembl gene identifiers corresponding to the gene symbols available in the paper. An automated tool called the ‘Clone/Gene ID Converter’ (http://idconverter.bioinfo.cnio.es/) can be used when gene identifiers from specified databases are available. We started with two lists of 9 and 8 Ensembl gene identifiers for the ‘target’ and ‘repressed’ groups, respectively. Given the subject of the published study our aim was to identify any over-represented motifs within 50 nt (up and downstream) of conserved GATA sites (consensus sequence ‘GATA’) located within 20 kb either side of the genes in the two lists. The mouse version of CoMoDis was run using the default settings, selecting GATA from the seed motif list and supplying the mouse gene lists as a plain-text file. The first time CoMoDis was run with each list it was clear that certain genes possessed many more seed motifs than the others in the same list. Therefore, in order to prevent an over-representation of sequences from any one gene, they were not used in the final analysis. For the ‘target’ group Ank1 and Gypa were excluded (with 17 and 19 seed motifs, respectively). For the ‘repressed’ group Myb and Kit were excluded (with 49 and 38 seed motifs, respectively). Moreover, certain genes did not possess any instances of the seed motif within the specified area and therefore did not contribute to the final analysis. For this reason Alas2 was excluded from the ‘target’ group. Hbb-b1 was not found in the Ensembl (version 37) database. For the ‘repressed’ group Tmk was excluded. Therefore, five genes were left in both groups. The CoMoDis summary output files can be viewed on our web site (http://hscl.cimr.cam.ac.uk/supplementary_comodis06.html). The tool was run twice to retrieve both single and orthologous sequence output files. CoMoDis generated 20 sets of motif sequences in the vicinity of five genes for the ‘target’ group and 24 sets of motif sequences in the vicinity of five genes for the ‘repressed’ group.
We focussed on motif discovery using the tools summarized in Table 2. The tools were run using the default settings, searching for motifs 6 nt (more numerous) and 8 nt (more information, encompassing longer motifs) in length, searching both strands of the input sequences and using the input sequences themselves to derive the background model, where requested. YMF required the correct genome to be chosen as the background (in this example Mus musculus). Weeder was set to allow the presence of more than one occurrence of a motif per sequence. Both BioProspector and PhyloGibbs were run three times as both tools utilize a Gibbs sampling strategy to find over-represented motifs. More information regarding the use of each tool, the raw output files generated for each tool, together with a summary of the candidate motifs can be downloaded from our web site.
The web tool T-Reg Comparator [(25); http://treg.molgen.mpg.de/cgi-bin/pfm_meme_form.pl] was used to help identify possible transcription factors that could bind to the motifs represented in the output files of the motif discovery tools. This tool is able to compare IUPAC consensus, positional weight matrix and aligned sequence representations of motifs with published libraries of TFBSs. All publicly available vertebrate datasets were searched. The dissimilarity cutoff was set to 0.5 to exclude weak matches. In the ‘target’ group we identified Ebox (Hen1, V$MYOD_Q6, Myf), helix–loop–helix (V$SREBP1_02), Rel-family (NFKB), zincfinger (V$SF1_Q6) and homeodomain binding motifs. SP1 motifs were also observed by us, but were not recognized by T-Reg Comparator. In the ‘repressed’ group we identified Ets-family (MA0081, V$ETS_Q4, V$ETS_Q6), Rel-family (p50) and MEF-2 binding motifs.
The usefulness of facilitating access to multiple motif discovery tools from a single web site is perfectly illustrated by the above example. Each tool on its own predicts several motifs and it is not necessarily obvious which predicted motifs should be prioritized for subsequent functional validation. Having easy access to multiple tools facilitates the comparison of output files so that motifs recurrently identified by several different tools can be prioritized for functional assays. DME, GAME, PhyloGibbs and PhyME all reported Ebox motifs in the GATA-1 ‘target’ set. This result was striking as a composite Ebox—GATA-1 binding motif had previously been shown to control activation of the several key erythroid genes, such as α-globin (26), glycophorin A (27) and band 4.2 (28).
We also looked at gene lists from two other gene expression profiling studies; analysis of Gata3−/− mice (29) and an analysis of upregulated candidate MyoD target genes in primary mouse muscle tissue (30). CoMoDis analysis of genes downregulated in Gata3 mutant embryos identified the clustering of conserved GATA sites with NFAT binding sites. Motifs identified by more than one discovery tool were V$NFAT_Q6, V$IRF2_01 and homeodomain family. Gata3 and NFAT transcription factors have previously been linked as important regulators of T-cell development and function (31). In the study of genes representing candidate MyoD targets, Ebox motifs (representing MyoD binding sites) were shown to be clustered together. Hen-1 (Ebox) was identified by more than one discovery tool. Functional Ebox clusters have been identified in other muscle genes, such as the promoter of the MyoD target gene myostatin (32). Taken together therefore, CoMoDis analysis of both of these datasets revealed clusters of motifs consistent with previous functional studies. More detailed results of these additional analyses can be seen on our website (http://hscl.cimr.cam.ac.uk/supplementary_comodis06.html).
SOFTWARE AND ACCESS
CoMoDis is comprised of scripts written in PERL and is accessible through a Perl CGI interface on a web server, hosted by the University of Cambridge. The Perl scripts are available on our web site (http://hscl.cimr.cam.ac.uk/supplementary_comodis06.html). The run time of a submitted job is typically <10 min. Throughout, all user input screens are designed to be simple and used in a step-wise manner; where appropriate, default values have been pre-entered as good starting points to run the analysis.
CONCLUSIONS
Genomic information is becoming available at an ever increasing rate. It is therefore imperative that user friendly computational tools are developed, e.g, to start reconstructing the transcriptional networks that govern gene expression in a variety of cell types and conditions. CoMoDis is designed to aid in generating candidates for experimental validation of new network connections, utilizing a wide selection of motif discovery and motif scanning tools. CoMoDis readily integrates eight different motif discovery programs and we provide easily accessible web interfaces for five motif discovery tools that previously required installation onto the user's computer. CoMoDis can also be used in a more simplistic way to quickly identify the locations of a particular conserved motif in relation to any gene of interest that is annotated in the Ensembl database, highlighting areas for further scrutiny. Although our tool is confined to using pre-processed seed motif positions (albeit a diverse set), additional motifs can be processed on request.
Acknowledgments
Work in the authors' laboratory is funded by the Cambridge MIT Institute, an SUR grant from IBM and the Leukemia Research Fund. Funding to pay the Open Access publication charges for this article was provided by Leukemia Research Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nobrega M.A., Ovcharenko I., Afzal V., Rubin E.M. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 2.King D.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolbe D., Taylor J., Elnitski L., Eswara P., Li J., Miller W., Hardison R., Chiaromonte F. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchette M., Bataille A.R., Chen X., Poitras C., Laganiere J., Lefebvre C., Deblois G., Giguere V., Ferretti V., Bergeron D., et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome. Res. 2006;16:656–668. doi: 10.1101/gr.4866006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donaldson I.J., Chapman M., Gottgens B. TFBScluster: a resource for the characterization of transcriptional regulatory networks. Bioinformatics. 2005;21:3058–3059. doi: 10.1093/bioinformatics/bti461. [DOI] [PubMed] [Google Scholar]
- 6.Donaldson I.J., Gottgens B. TFBScluster web server for the identification of mammalian composite regulatory elements. Nucleic Acids Res. 2006;34:W524–W528. doi: 10.1093/nar/gkl041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hallikas O., Palin K., Sinjushina N., Rautiainen R., Partanen J., Ukkonen E., Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 8.Ovcharenko I., Nobrega M.A. Identifying synonymous regulatory elements in vertebrate genomes. Nucleic Acids Res. 2005;33:W403–W407. doi: 10.1093/nar/gki466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krivan W., Wasserman W.W. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wasserman W.W., Fickett J.W. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- 11.Kel A., Konovalova T., Waleev T., Cheremushkin E., Kel-Margoulis O., Wingender E. Composite Module Analyst: a fitness-based tool for identification of transcription factor binding site combinations. Bioinformatics. 2006;22:1190–1197. doi: 10.1093/bioinformatics/btl041. [DOI] [PubMed] [Google Scholar]
- 12.Waleev T., Shtokalo D., Konovalova T., Voss N., Cheremushkin E., Stegmaier P., Kel-Margoulis O., Wingender E., Kel A. Composite Module Analyst: identification of transcription factor binding site combinations using genetic algorithm. Nucleic Acids Res. 2006;34:W541–W545. doi: 10.1093/nar/gkl342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kankainen M., Pehkonen P., Rosenstom P., Toronen P., Wong G., Holm L. POXO: a web-enabled tool series to discover transcription factor binding sites. Nucleic Acids Res. 2006;34:W534–W540. doi: 10.1093/nar/gkl296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C.C., Lin C.C., Chen W.S., Chen H.Y., Chang P.C., Chen J.J., Yang P.C. CRSD: a comprehensive web server for composite regulatory signature discovery. Nucleic Acids Res. 2006;34:W571–W577. doi: 10.1093/nar/gkl279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinemeyer T., Chen X., Karas H., Kel A.E., Kel O.V., Liebich I., Meinhardt T., Reuter I., Schacherer F., Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vlieghe D., Sandelin A., De Bleser P.J., Vleminckx K., Wasserman W.W., van Roy F., Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakash A., Tompa M. Discovery of regulatory elements in vertebrates through comparative genomics. Nat. Biotechnol. 2005;23:1249–1256. doi: 10.1038/nbt1140. [DOI] [PubMed] [Google Scholar]
- 19.Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ettwiller L., Paten B., Souren M., Loosli F., Wittbrodt J., Birney E. The discovery, positioning and verification of a set of transcription-associated motifs in vertebrates. Genome Biol. 2005;6:R104. doi: 10.1186/gb-2005-6-12-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stabenau A., McVicker G., Melsopp C., Proctor G., Clamp M., Birney E. The Ensembl core software libraries. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim T.H., Barrera L.O., Zheng M., Qu C., Singer M.A., Richmond T.A., Wu Y., Green R.D., Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 24.Rylski M., Welch J.J., Chen Y.Y., Letting D.L., Diehl J.A., Chodosh L.A., Blobel G.A., Weiss M.J. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roepcke S., Grossmann S., Rahmann S., Vingron M. T-Reg Comparator: an analysis tool for the comparison of position weight matrices. Nucleic Acids Res. 2005;33:W438–W441. doi: 10.1093/nar/gki590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anguita E., Hughes J., Heyworth C., Blobel G.A., Wood W.G., Higgs D.R. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lahlil R., Lecuyer E., Herblot S., Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z., Huang S., Chang L.S., Agulnick A.D., Brandt S.J. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Airik R., Karner M., Karis A., Karner J. Gene expression analysis of Gata3−/− mice by using cDNA microarray technology. Life Sci. 2005;76:2559–2568. doi: 10.1016/j.lfs.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 30.Zhao P., Seo J., Wang Z., Wang Y., Shneiderman B., Hoffman E.P. In vivo filtering of in vitro expression data reveals MyoD targets. C. R. Biol. 2003;326:1049–1065. doi: 10.1016/j.crvi.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 31.Kuo C.T., Leiden J.M. Transcriptional regulation of T lymphocyte development and function. Annu. Rev. Immunol. 1999;17:149–187. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- 32.Spiller M.P., Kambadur R., Jeanplong F., Thomas M., Martyn J.K., Bass J.J., Sharma M. The myostatin gene is a downstream target gene of basic helix-loop-helix transcription factor MyoD. Mol. Cell. Biol. 2002;22:7066–7082. doi: 10.1128/MCB.22.20.7066-7082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., Brutlag D.L., Liu J.S. BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001;6:127–138. [PubMed] [Google Scholar]
- 34.Smith A.D., Sumazin P., Zhang M.Q. Identifying tissue-selective transcription factor binding sites in vertebrate promoters. Proc. Natl. Acad. Sci. USA. 2005;102:1560–1565. doi: 10.1073/pnas.0406123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Z., Jensen S.T. GAME: detecting cis-regulatory elements using a genetic algorithm. Bioinformatics. 2006;22:1577–1584. doi: 10.1093/bioinformatics/btl147. [DOI] [PubMed] [Google Scholar]
- 36.Down T.A., Hubbard T.J. NestedMICA: sensitive inference of over-represented motifs in nucleic acid sequence. Nucleic Acids Res. 2005;33:1445–1453. doi: 10.1093/nar/gki282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavesi G., Mereghetti P., Mauri G., Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha S., Tompa M. YMF: A program for discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res. 2003;31:3586–3588. doi: 10.1093/nar/gkg618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T., Stormo G.D. Combining phylogenetic data with co- regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- 40.Siddharthan R., Siggia E.D., van Nimwegen E. PhyloGibbs: a Gibbs sampling motif finder that incorporates phylogeny. PLoS Comput. Biol. 2005;1:e67. doi: 10.1371/journal.pcbi.0010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha S., Blanchette M., Tompa M. PhyME: a probabilistic algorithm for finding motifs in sets of orthologous sequences. BMC Bioinformatics. 2004;5:170. doi: 10.1186/1471-2105-5-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu Y., Frith M.C., Haverty P.M., Weng Z. MotifViz: an analysis and visualization tool for motif discovery. Nucleic Acids Res. 2004;32:W420–W423. doi: 10.1093/nar/gkh426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farre D., Roset R., Huerta M., Adsuara J.E., Rosello L., Alba M.M., Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messeguer X., Escudero R., Farre D., Nunez O., Martinez J., Alba M.M. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]