Abstract
Multiple sequence alignments are often used to reveal functionally important residues within a protein family. They can be particularly useful for the identification of key residues that determine functional differences between protein subfamilies. We present a new entropy-based method, Sequence Harmony (SH) that accurately detects subfamily-specific positions from a multiple sequence alignment. The SH algorithm implements a novel formula, able to score compositional differences between subfamilies, without imposing conservation, in a simple manner on an intuitive scale. We compare our method with the most important published methods, i.e. AMAS, TreeDet and SDP-pred, using three well-studied protein families: the receptor-binding domain (MH2) of the Smad family of transcription factors, the Ras-superfamily of small GTPases and the MIP-family of integral membrane transporters. We demonstrate that SH accurately selects known functional sites with higher coverage than the other methods for these test-cases. This shows that compositional differences between protein subfamilies provide sufficient basis for identification of functional sites. In addition, SH selects a number of sites of unknown function that could be interesting candidates for further experimental investigation.
INTRODUCTION
In the quest for knowledge about protein function, understanding differences between protein families is essential. It is therefore not surprising that a large number of methods have already been introduced for the positional comparison of amino acid compositions between different protein families or subtypes [(1–9); for a review see Whisstock and Lesk (10)]. Though these methods have contributed greatly to the understanding of the relation between sequence and function (11), coverage of known sites of functional differences has been limited.
An early method called AMAS by Livingstone and Barton (1) analyses conservation patterns using a number of physicochemical properties of amino acids. The method assigns sites to either of three classes: globally conserved, conserved in subfamilies, or not conserved, based on arbitrarily set conservation threshold values. This method was intended primarily to ‘allow the residue-specific similarities and differences in physicochemical properties between groups of sequences to be identified quickly’ (1).
Several methods have been based on evolutionary trace analysis (2) and rely on residue conservation within subfamilies to construct subfamily-specific consensus sequences. These sequences are then aligned to reveal the variation between the different subfamilies. Kuipers et al. (3) relaxed the requirement of intra-group conservation and allowed the selection of ‘residues that are conserved in one class of proteins with a certain function but are different in other classes’.
Del Sol Mesa et al. (6) introduced a method called TreeDet that uses mutational behaviour analysis of so-called ‘tree-determinant’ residues. The method uses an internal algorithm for unsupervised grouping of the input sequences. It then selects residues that follow mutation patterns similar to that of the overall phylogeny. For each alignment position, this is measured using the correlation coefficient between the substitution matrix derived for the position considered and that for the whole protein. Residues selected tend to be conserved within each subfamily but different between them. The method aims to find ‘the most appropriate way of dividing a protein family into subfamilies in order to associate the tree-determinants with sites, which are likely to be responsible for functional differences between these subfamilies’.
The method SDP-pred by Kalinina et al. (7) selects residues that ‘are well conserved within specificity groups but differ between these groups’. The method is based on mutual information analysis and complex statistical treatment. It extends an earlier method (5) by using residue frequencies smoothed and weighted by the blosum average substitution scores. Significance is estimated by Z-scores of expected mutual information obtained from column shuffling. Subsequently, an appropriate Z-score threshold for selection of high-ranking sites is determined using the Bernoulli estimator. Recently, Donald and Shakhnovich (8) added an automated procedure for functional grouping.
The above methods focus on sites that are conserved in one or both groups and subsequently select those sites that are different between these groups (10). Unfortunately, this scenario excludes sites that are not highly conserved within each of the groups. This may not seem a serious problem at first hand, but let us consider an example. Take a group A comprising a protein subfamily binding a certain molecule (e.g. a ligand or receptor; this group represents the ‘binders’) and group B comprising proteins that do not (the ‘non-binders’). Certainly, one can expect sites that are crucial for binding to be conserved in group A (the ‘binders’). However, for group B to avoid binding, the corresponding site would need to avoid the conserved residue of group A. It seems therefore imprudent to expect conservation throughout group B (the ‘non-binders’) as well (3). Moreover, if group A contains binders to different (but related) molecules (ligands or receptors), even the requirement of conservation in group A may not apply.
To address these restrictions here, we introduce an alternative similarity measure for comparing groups of sequences within a multiple sequence alignment, which we name Sequence Harmony (SH). The SH measure is derived from Shannon's general information entropy (12) as applied to biomolecular sequences by Shenkin et al. (13). We will show that SH has well-defined properties and is easily calculated. The values obtained fall within a convenient fixed interval and correspond intuitively to differences in the amino acid compositions as observed in the alignment. Moreover, no additional residue substitution matrices, calculations of mutual information or computations of statistical significance are needed (5,7).
We evaluate the SH method using four benchmark sets comprising experimental data on specificity-switching residues and compare its performance against three other state-of-the-art methods. From this, we obtain new insights about the principles that govern the accurate detection of functional sites. We will show that SH is able to identify sites associated with subfamily specificity systematically and with relatively few errors.
THEORY
Comparing sequences by relative entropy
Relative entropy (rE) is commonly used in sequence comparison to quantify the degree of conservation (14,15). It is derived from Shannon's general information entropy (12) as applied to biological sequences by Shenkin et al. (13):
| 1 |
where and are the observed probabilities of amino acid type x at a position i in the alignment of groups A and B, respectively. Relative entropy measures the difference in information content between both distributions of amino acid types. Interestingly, for sites to be maximally different between the two groups, amino acid types in one group should be absent in the other or vice versa. This leads to a degenerate result (singularity) whenever the entropy function of Equation 1 is used. Inclusion of so-called ‘pseudo-counts’ (16) solves the degeneracy but not the unbounded, asymptotic behaviour of Equation 1. Also taking, for instance, the relative entropy with respect to both groups, as , does not solve the unbounded behaviour. Moreover, the upper limit depends on the ratio of the number of sequences in both groups.
Calculating differences by SH
We address the degeneracy of Equation 1 by defining SH as
| 2 |
This can be viewed as the relative entropy of group A relative to the sum of the probabilities of both groups ( pA + pB). As a consequence of the separate weighting of both groups, we eliminate the dependence on relative group sizes. Since, in general, SHA/B ≠ SHB/A, we remedy this non-commutativeness by taking the average. Using Equation 2 this leads to:
| 3 |
Using Shannon's information entropy:
| 4 |
and writing
| 5 |
as Shannon's entropy weighting groups A and B equally, we can rewrite Equation 3 as
| 6 |
In other words, SH can also be conveniently expressed as a simple linear combination of entropies.
The SH function juxtaposes relative entropy since it becomes zero for maximally different sites and one for sites with identical distributions. We therefore coined the phrase Sequence Harmony (SH) as it indicates to what extent amino acid compositions between two groups of sequences are in harmony.
To illustrate how SH works in practice, we present a hypothetical alignment in Table 1. Positions 1, 2 and 3 all have a SH value of zero. In fact, the formula becomes zero any time the amino acid composition in one group is non-overlapping with that in the other group, regardless of conservation. At position 4, the amino acid of group A (Ala) also occurs once in group B, yielding the lowest possible non-zero score for this number of sequences. For positions 5 and 6, there is an increasing overlap and hence growing SH values. Position 8 is conserved overall in the whole family and is therefore maximally harmonious with an SH value of one. Note that unconserved sites have SH = 1 whenever the two groups have identical compositions. This is illustrated in position 7, where equal proportions of R and K are shown.
Table 1 .
Hypothetical alignment of two subfamilies A (4 sequences) and B (6 sequences)
| Alignment position | ||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Group A | ||||||||
| seq1 | R | E | L | A | A | A | K | K |
| seq2 | R | E | L | A | A | F | K | K |
| seq3 | R | E | A | A | A | Y | R | K |
| seq4 | R | E | A | A | A | F | R | K |
| Group B | ||||||||
| seq1 | H | N | V | A | A | Y | R | K |
| seq2 | H | N | V | F | F | Y | R | K |
| seq3 | H | N | F | F | A | F | K | K |
| seq4 | H | S | F | F | F | Y | R | K |
| seq5 | H | S | M | F | F | F | K | K |
| seq6 | H | T | M | F | F | Y | K | K |
| SH | 0.00 | 0.00 | 0.00 | 0.35 | 0.54 | 0.79 | 1.00 | 1.00 |
For each position in the alignment the SH score between the subfamilies is calculated.
The SH measure is implemented in a simple online server for calculating SH from an alignment. It can be accessed at www.ibi.vu.nl/programs/seqharmwww.
Ranking identically scoring SH sites
The example of Table 1 shows that sites with different compositions can have identical SH values, which therefore cannot be ranked. This is particularly important for sites with SH = 0, as these are potentially discriminative for function. To address this issue, we derived a simple but effective ranking for identically scoring sites based on the distribution of low SH values (SH ≤ 0.2) over the alignment positions. Groups of sequentially adjacent low-harmony sites can have one intermediate high-harmony site in between, i.e. the sequence distance is two or less. The ranking is first on increasing SH (low harmony first), and then on decreasing group size (larger sequential groups first). Finally, sites that have equal SH scores and group sizes are ranked on the total entropy of the sites in both subfamilies. This scenario is implemented in the SH method which we will label ‘SH’.
We further explored the performance of an even simpler ranking on just SH values and entropy of the site, i.e. without ranking on sequential groups. This approach we will refer to as ‘SH/E’.
Benchmarking
We compared predictions by SH with those obtained from three other methods over several protein families. For this purpose, we have used the online server of AMAS (http://barton.ebi.ac.uk/servers/amas_server.html) (1), the ‘mutational behaviour’ method available from the TreeDet server, which we will refer to as TreeDet/MB (http://somosierra.cnb.uam.es/Servers/treedetv2/) (6) and the SDP-pred server (http://math.genebee.msu.ru/~psn/) (5,7). The recent method by Donald and Shakhnovich (8) (see Introduction) could not be evaluated in this study due to lack of an online server.
The methods were evaluated over a number of test-sets (see below) using receiver–operator characteristics (ROC) plots of coverage (% recovered known functional sites) versus error (% wrong predictions). These were constructed from ranked lists of sites for SH and for SDP-pred. For AMAS, the conservation threshold was varied from 0 to 10 (see Introduction). For TreeDet/MB, the cut-off value was varied from 0 to 1.0. In both cases, small steps were taken so that the addition of single sites to the selection could be observed.
DATASETS FOR VALIDATION
We have selected three relevant protein families, for which experimental evidence is available on subfamily-specific sites. These include families that have been used for construction, testing and/or validation of the other prediction methods and one family for which we have assembled an extensive dataset.
In many cases, the available experimental evidence concerns the exchange of a sequence segment, e.g. a loop or a helix. Positions within such segments that are conserved over both subfamilies were not regarded as subfamily specific. Although such positions are likely to be important for the function of the family, they are unable to explain functional differences between subfamilies at the residue level.
TGF-β-associated transcription factors (Smad family)
The Smad family of transcription factors plays a crucial role in the transforming growth factor-β (TGF-β) signalling pathway. Smads are also critical for determining the specificity between alternative TGF-β pathways [for recent extensive reviews, see Feng and Derynck (17) and Massagué et al. (18)]. This complex signalling network is involved in the regulation of many cellular processes such as division and differentiation, motility, adhesion and programmed cell death. The TGF-β family of growth factors induces Type-I transmembrane receptors to phosphorylate and activate the receptor-regulated Smads (R-Smads) (18). The R-Smads can be subdivided into two major groups: the AR-Smads, which are mainly induced by TGF-β-type receptors (TβR-I), and the BR-Smads, which are mainly induced by the BMP-type receptors (BMPR-I and ALK1/2). Subsequent associations among Smads are responsible for control of TGF-β target genes in the nucleus. It has been shown that most of the above interactions involve the so-called Mad Homology 2 (MH2) domain of the Smad proteins (17). From an extensive literature search, we have identified 29 specific sites in the MH2 domain that are experimentally validated to be important for Smad specificity, as listed in Table 2.
Table 2 .
Summary of all known functional sites and sites of low SH (SH ≤ 0.2) in the MH2 domain of Smad
| Position | Sec. struc. | Consensus | SH | SH group Size | Other methods | Function | References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Align | Smad2 | AR | BR | AMAS | Tree Det | SDP pred | |||||
| 2 | (L263) | B1′ | La | Vfm | 0 | 1 | + | — | — | SARA | (29) |
| 3 | (Q264) | B1′ | Qa | Qrh | 0.81 | — | — | — | — | SARA | (29) |
| 6 | T267 | B1′ | Tm | Acen | 0 | 2 | — | — | — | SARA | (29) |
| 8 | S269 | loop | CSh | Eq | 0 | 2 | — | — | — | ?(SARA) | — |
| 11 | A272 | loop | A | Kqls | 0 | 2 | — | — | — | ? (SARA) | — |
| 12 | F273 | loop | F | Hy | 0 | 2 | — | — | — | ? (SARA) | — |
| 23 | Q284 | B2 | Qt | N | 0 | 1 | — | — | — | TβR-I | (30) |
| 33 | Q294 | loop | Q | Sq | 0.16 | 4 | — | — | — | c-Ski/SnoN | (31) |
| 34 | P295 | B3 | P | Trl | 0 | 4 | — | 0.85 | — | c-Ski/SnoN | (31) |
| 36 | L297 | B3 | LMi | Vi | 0.11 | 4 | — | — | — | c-Ski/SnoN | (31) |
| 37 | T298 | B3 | T | Li | 0 | 4 | — | 0.88 | — | c-Ski/SnoN | (31) |
| 47 | S308 | L1 | Sa | N | 0 | 3 | — | — | — | c-Ski/SnoN | (31) |
| 48 | — | L1 | — | Nsd | 0 | 3 | — | — | — | c-Ski/SnoN | (31) |
| 49 | E309 | L1 | E | Krs | 0 | 3 | — | — | — | c-Ski/SnoN | (31) |
| 63 | A323 | H1 | Ae | S | 0 | 3 | — | 0.84 | — | BMPR-I/ALK1/2 | (32) |
| 65 | V325 | H1 | V | I | 0 | 3 | — | 0.87 | 2.17 | BMPR-I/ALK1/2 | (32) |
| 67 | M327 | H1 | LMq | N | 0 | 3 | + | 0.83 | — | BMPR-I/ALK1/2 | (32) |
| 74 | R334 | loop | Rk | K | 0.18 | 1 | — | — | — | ? (c-Ski/SnoN) | — |
| 77 | R337 | B5 | R | H | 0 | 1 | — | 0.87 | 2.25 | not SARA (c-Ski/SnoN) | (29) |
| 81 | I341 | B5 | I | V | 0 | 1 | — | 0.87 | 2.24 | SARA/Mixer | (29,33) |
| 86 | F346 | B6 | F | Y | 0 | 1 | — | 0.87 | 2.14 | SARA/Mixer | (29,33) |
| 94 | A354 | B7 | As | S | 0.18 | 1 | — | — | — | ? | — |
| 100 | P360 | H2 | P | R | 0 | 1 | + | 0.87 | 2.21 | FAST1 | (34) |
| 104 | Q364 | H2 | Q | Yf | 0 | 4 | — | — | — | Mixer/FAST1 | (34,35) |
| 105 | R365 | H2 | R | Hq | 0 | 4 | — | — | — | Mixer/FAST1 | (34,35) |
| 106 | Y366 | H2 | Y | H | 0 | 4 | — | 0.86 | 2.02 | SARA/Mixer/FAST1 | (29,33–35) |
| 108 | W368 | loop | W | F | 0 | 4 | — | 0.87 | 2.22 | SARA/Mixer/FAST1 | (29,33,34) |
| 118 | P378 | loop | P | Sp | 0.16 | 1 | — | — | — | ? | — |
| 121 | N381 | B9 | N | S | 0 | 1 | — | 0.87 | — | SARA/Mixer | (29,33) |
| 136 | A392 | H3 | A | Qeh | 0 | 1 | — | 0.85 | — | ? (FAST1/Mixer) | — |
| 144 | Q400 | loop | Q | H | 0 | 1 | + | 0.87 | 2.03 | ? (FAST1/Mixer) | — |
| 151 | Q407 | H4 | Qr | E | 0 | 1 | — | 0.83 | — | not receptor binding (FAST1/Mixer) | (28) |
| 154 | R410 | H4 | R | K | 0 | 1 | — | 0.87 | 2.28 | ? (FAST1/Mixer) | — |
| 171 | R427 | L3 | R | H | 0 | 1 | — | 0.87 | 2.01 | TβR-I/BMPR-I/ALK1/2 | (28) |
| 174 | T430 | L3 | T | D | 0 | 1 | — | 0.87 | 2.08 | TβR-I/BMPR-I/ALK1/2 | (28) |
| 184 | L440 | B11 | L | Iv | 0 | 1 | — | 0.84 | — | ? | — |
| 187 | N443 | H5 | N | Hn | 0.16 | 1 | — | — | — | ? (SARA) | — |
| 204 | S460 | C-tail | Snr | Hlr | 0.06 | 4 | — | — | — | TβR-I/BMPR-I | (28) |
| 205 | V461 | C-tail | Ivl | N | 0 | 4 | + | 0.83 | — | TβR-I/BMPR-I | (28) |
| 206 | R462 | C-tail | Rp | P | 0.17 | 4 | — | — | — | TβR-I/BMPR-I | (28,36) |
| 207 | C463 | C-tail | C | I | 0 | 4 | + | 0.86 | 2.30 | TβR-I/BMPR-I | (28,36) |
| 210 | M466 | C-tail | MV | V | 0.69 | — | — | — | — | TβR-I/BMPR-I | (28,36) |
Sequence positions are indicated relative to the alignment (Align) as well as to Smad2, according to PDB 1KHX (29). Secondary structure elements are indicated according to Chen et al. (34). SH scores and corresponding size of sequential groups of low SH sites are listed. In addition, predictions for the other methods using default settings are shown (see text for details). The consensus patterns for the AR-Smads and BR-Smads are shown. All amino acid types are listed in order of decreasing frequency. Those of half or less than the frequency of the dominant type are in lower case. The known functions, with corresponding reference(s), are indicated. Putative functions corresponding to structural clustering shown in Figure 5A are indicated in brackets (see text for more detail).
Small GTPases (RAS superfamily)
Members of the Ras superfamily of GTPases are implicated in the regulation of growth, survival, differentiation and other processes in haematopoietic cells (19). They comprise six families. Experimental evidence for functional sites is available from the literature for the Ras versus Ral families (6,20). This set was used by Del Sol Mesa et al. (6) for the development of the TreeDet method. In addition, we include a test set of Rab5/6-specific sites obtained from (21,22).
Integral membrane transporters (MIP family)
Members of the MIP family are mainly involved in facilitating the transport of both water and small neutral solutes through the cellular membrane in all domains of life. There are about six MIP subfamilies, the two major being the aquaporins (AQPs) and the glycerol-uptake facilitators (GLPs) (23). The AQP and GLP subfamilies were used for the initial validation of the SDP-pred method (7). An arbitrary measure for functional significance of a site used by Kalinina et al. (7) was the proximity to the glycerol molecules that are bound inside the GLP pore channel in the crystal structure 1FX8 (24). Sites that were conserved in the training set of sequences were excluded. This scenario yielded a set of 23 putative functional sites closer than 5 Å to any of the three glycerol molecules (7).
Sequence retrieval and alignment
R-Smad protein sequences were collected using the NCBI query for sequence retrieval (www.ncbi.nih.gov). This resulted in 15 non-redundant sequences for AR-Smads and 17 for BR-Smads. All sequences were aligned using the PSI-Praline multiple sequence alignment online server (www.ibi.vu.nl/programs/pralinewww) (25,26). From the alignment obtained, the MH2 domain was selected for further analysis.
For the Ras superfamily, as used by Del Sol Mesa et al. (6), sequences and alignments were directly obtained from Pfam-B (27). Selection of sequences for Ras, Ral, Rab5 and Rab6 families was simply performed by matching sequence names on Ras, Ral, Rab5 and Rab6, respectively yielding 69, 20, 4 and 6 sequences. The hypervariable termini of Rab5 and Rab6 were not present in the Pfam alignment.
For the MIP family, we took all bacterial AQP and GLP protein sequences, as defined in Figure 1 of Kalinina et al. (7). For the other sequences mentioned in this figure, the classification was less obvious. We therefore decided not to take these into account. This scenario yielded 12 sequences for the AQP subfamily and 48 for both GLP subfamilies (one GLP identifier ‘YA17_HAEIN’ could not be resolved). The sequences were aligned using PSI-Praline as mentioned above for the Smads.
RESULTS
TGFβ-associated transcription factors (Smad family)
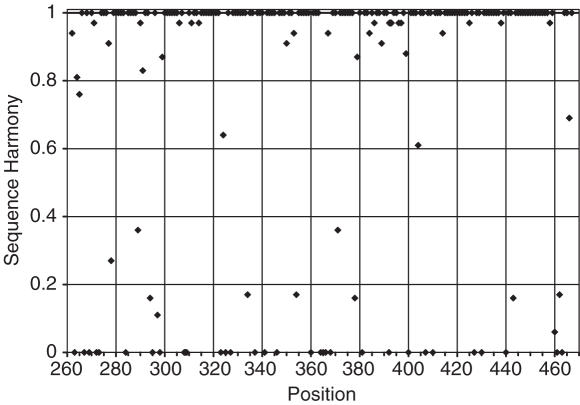
SH between AR- and BR-Smads was calculated using Equation 3 for all 211 positions along the Smad MH2 alignment, as shown in Figure 1. It is clear that the vast majority is conserved overall, i.e. 135 sites have SH = 1. On the other hand, relatively few sites are completely non-harmonious, i.e. 32 have SH = 0. Out of these, only 13 are conserved in both groups, as can been seen in Table 2. The other 19 sites show a more variable composition but the subgroups still have non-overlapping compositions. Further, a relatively small fraction of sites show intermediate SH values, i.e. 44 are in between 0 and 1. In the range of 0–0.2, eight sites are found, and between 0.2 and 0.8 only seven. This means that the choice of a cut-off value for determining sites of ‘low’ SH is not very critical in this case.
Figure 1 .
SH for AR-Smads versus BR-Smads along the sequence of the MH2 domain of Smads. 40 sites have SH zero. Most sites have SH one, meaning these have the same composition in each subgroup. Relatively few sites have intermediate SH values.
Out of the 211 residues in the Smad MH2 sequence alignment, only 40 have a low SH (SH ≤ 0.2). In Table 2, these sites are listed together with their known interactions. It is clear that the vast majority of low-harmony sites also have a known function. Table 3 provides a further summary of the number of low-harmony sites associated with a particular function. Out of the 40 low-harmony sites, 27 have a known function (68%), while of the 32 non-harmonious sites (SH = 0) 23 have a known function (72%). In total, there are 29 known sites of functional specificity in the Smad dataset. These include several with rather high compositional variation in one or both groups (Table 2). Of the 171 remaining sites (SH > 0.2), only two sites are known to be important for the specificity of the Smad–receptor interactions (1%).
Table 3 .
Summary of functional sites and sites of unknown function with no (SH zero) or low (SH ≤ 0.2) SH and specificity of the functional prediction
| Function | SH = 0 | SH ≤ 0.2 |
|---|---|---|
| TβRI/BMPRI/ALK1/2 binding | 8 | 10 |
| c-Ski/SnoN binding | 5 | 7 |
| SARA/Mixer/FAST1 binding | 10 | 10 |
| Total ‘functional’ | 23 | 27 |
| Putative function | 8 | 10 |
| Unknown function | 1 | 3 |
| Total ‘unknown’ | 9 | 13 |
| Total | 32 | 40 |
| Functional versus total (%) | 72 | 68 |
| Functional + putative versus total (%) | 97 | 93 |
The AMAS method selects six sites that are different with respect to one or more physicochemical properties (Table 2). Consequently, these sites have non-overlapping amino acid compositions between the groups. Three are conserved in both groups, two are conserved in one group and one is not conserved in either group. Five out of the six selected sites are of known function, but clearly the majority of the 29 known functional sites are missed.
TreeDet/MB selects 21 sites that are completely conserved in at least one group but show no intergroup overlap (Table 2). Seven additional sites show the same characteristics but are not selected. The 21 selected sites contain 16 known functional sites. The other 7 sites contain 5 known functions.
SDP-pred selects 12 sites that are conserved in both groups and show no overlap (Table 2). Of these, 9 have known functions. One site of known function is conserved in both groups, but is not selected by the method. SDP-pred reaches a maximum coverage of ∼80%. All sites selected by SDP-pred are also selected by TreeDet/MB (see above).
All sites selected by AMAS, TreeDet/MB or SDP-pred are also selected by SH. SH selects 18 additional sites, 11 of which have a known function. None of the 18 sites are conserved in both groups, which explains why the other methods have difficulties finding them. Two further known functional sites remain undetected by any of the included methods.
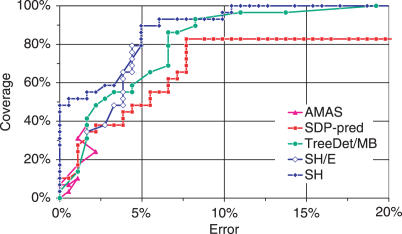
The ROC plot shows that TreeDet/MB, SDP-pred and SH/E all reach ∼40% coverage with similar error (Figure 2). At higher coverage, TreeDet and SH/E yield lower error than SDP-pred. AMAS does not reach higher coverage and therefore it has not been applied to the other test-sets. It is clear that SH outperforms its counterparts at all coverage/error combinations. Notably, the first 14 sites selected by SH are all validated functional sites.
Figure 2 .
ROC plots for AR-Smads versus BR-Smads using the different prediction methods. For SH and SDP-pred the ranked results were used. One point for each unique rank value is drawn. Coverage is calculated as TP/(TP + FN), and Error as FP/(TN + FP). Note that the error rate is shown up to 20%. Note also that no method reaches a higher coverage for higher error rates.
Small GTPases (Ras superfamily)
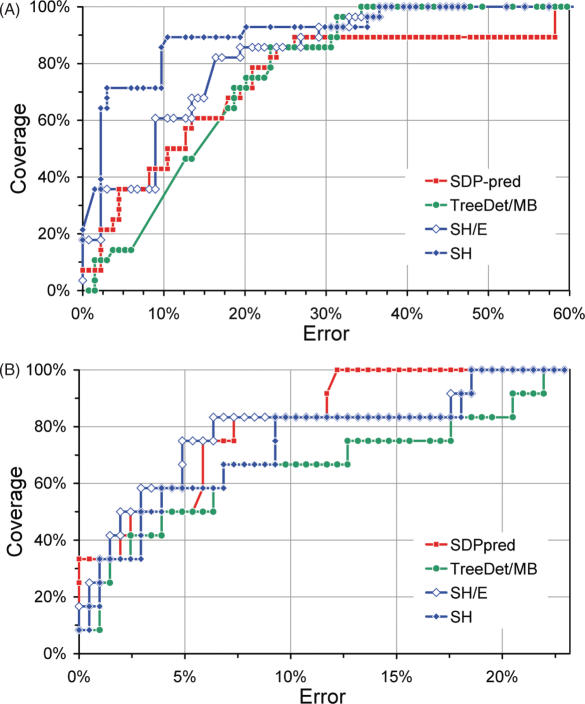
In Figure 3, ROC-curves are shown for two sets of families from the Ras superfamily of small GTPases; Rab 5 versus Rab 6 (21,22), and Ras versus Ral (6,20).
Figure 3 .
ROC plots for (A) Rab 5/6, and (B) Ras/Ral-specific sites using SH, SDP-pred and TreeDet/MB (see text and Figure 2 caption for details). Validation of Rab5/6-specific sites was taken from Stenmark and co-workers (21,22), and for Ras/Ral specificity from Bauer et al. (6,20) and Del Sol Mesa et al. (6). Note that error rate is shown up to 60% and 23% for (A and B), respectively.
For Rab5/6 specificity (Figure 3A), the SH predictions show a very high coverage, even at low error rates. Overall, TreeDet/MB, SDP-pred and SH/E achieve somewhat similar coverage and error. Nevertheless, SH/E reaches 35% coverage at only half the error of SDP-pred. At that coverage, TreeDet/MB shows a 2-fold error relative to SDP-pred. SH overall outperforms all other methods by a significant margin. The first 20 sites selected contain only two unknowns, while 70% coverage is attained within 10% error (Figure 3A).
For the Ras/Ral test-set, however, the prediction methods perform more similarly (Figure 3B). The first four sites (33%) selected by SDP-pred are validated. At higher coverage, SH/E performs slightly better than SDP-pred. Nevertheless, SDP-pred reaches 100% coverage at ∼12% error, while at this error rate SH/E and SH are just above 80% coverage and TreeDet/MB attains 65% coverage.
Integral membrane transporters (MIP family)
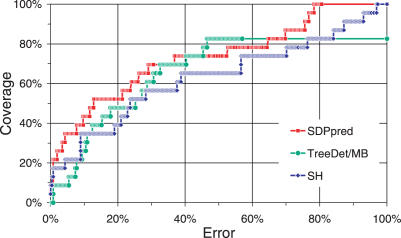
Figure 4 shows the relative performance over the MIP family. Predictions with the SDP-pred server were obtained using our own alignment and are comparable with those reported by Kalinina et al. (7).
Figure 4 .
ROC plots for MIP specificity using SH, TreeDet/MB and SDP-pred (see text and Figure 2 caption for details). MIP subfamily-specific sites were selected based on a minimum distance of 5 Å from the glycerol molecules bound in the pore channel in the glycerol uptake protein crystal structure 1FX8 (24).
At 20% error, all methods only achieve medium coverage (∼50%, Figure 4). In contrast, for the other test sets at 20% error all other methods achieve higher coverage (between 75% and 100%). SH achieves somewhat higher coverage at the lowest error rates (the first 3 selected sites are <5 Å from their nearest ligand). At higher error rates, SDP-pred generally outperforms the other methods. TreeDet/MB performs similar to SH at lower coverage and more similar to SDP-pred at higher coverage. For this dataset, ranking for SH was dominated by the harmony score and only minute differences were seen between SH and SH/E (data for SH/E not shown).
Spatial and functional clustering
In Figure 5, the SH data are projected onto representative crystal structures of the four protein families included in the benchmark.
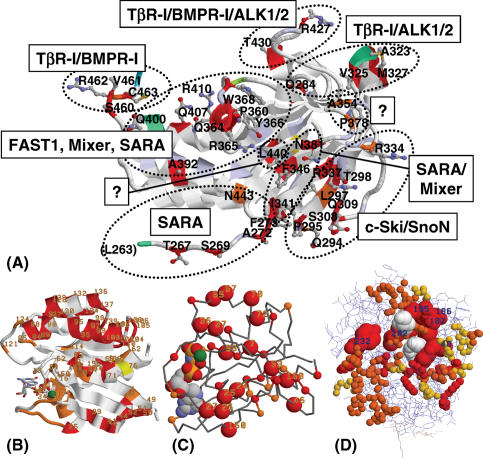
Figure 5 .
SH in a representative crystal structure for each of the test-sets. Non-harmonious sites (SH zero) are red and low harmony (SH ≤ 0.2) orange. Residue numbers for the low-harmony sites (SH ≤ 0.2) are indicated. (A) AR-Smads versus BR-Smads colour-coded onto the crystal structure of the MH2 domain of Smad2 (1KHX) (29) The spatial clustering of low-harmony sites is indicated with dotted ellipses, and clusters are labelled with corresponding known functions. Intermediate values go from white to light blue for maximum harmony (SH one). (B) SH for Rab5/6 using the crystal structure 5P21 and a representation and orientation similar to Figure 3a in Stenmark and co-workers (21,22). (C) id. for Ras/Ral using 5P21 and similar to Figure 4 in Del Sol Mesa et al. (6). (D) id. for MIP using 1KHX and similar to Figure 5 in Kalinina et al. (7).
For the Smad2 MH2 domain, we identified a limited number of spatial clusters of low-harmony sites as indicated in Figure 5A. Taking membership of these clusters as a guideline, we assign putative functions to 10 out of the 13 unknowns, as indicated in Table 2. It is clear that most of these could not have been assigned from the sequence alone. Unknowns 392, 400, 407 and 410 can be assigned a putative function in FAST1 and/or Mixer binding. The two SARA-binding residues (366 and 368) in this FAST1/Mixer/SARA-binding cluster are furthest away from the unknowns. Importantly, 407 is known not to be involved in receptor interactions (28). Unknowns 334 and 337 can be assigned a putative function for Co-repressor (c-Ski/SnoN) binding. Position 337 is known not to be involved in SARA binding (29). Unknowns 269, 272, 273 and 443 can be assigned a putative function in SARA binding. In all, only three sites remain that cannot be assigned a putative function, out of a total of 40 low-harmony sites.
For the other test-sets, we have chosen layouts in Figure 5 analogous to those used in the corresponding papers (see Figure 5 caption). For Rab5/6 (Figure 5B) and Ras/Ral (Figure 5C), it can be seen that selected sites form localized groups in the structure. For the MIPs (Figure 5D), this trend is much less salient.
DISCUSSION
In this paper, we have introduced the SH as a means to pinpoint putative functionally different sites and compared the results with other methods that rely mostly on conservation.
Performance varies for methods and datasets
For the Smad test-set, all five methods reach >80% coverage at <10% error (Figure 2). Nonetheless, differences among the methods are substantial and SH maintains highest coverage virtually throughout. For the Rab5/6 test-set, all four methods perform at least moderately well (Figure 3A). The differences among the methods are somewhat larger than for the Smad test-set, with coverage ranging from 40% to ∼80% at 10% error. Also here, SH performs best.
As to the Ras/Ral test-set, all four methods perform well and reach >80% coverage at 10% error (Figure 3B). The differences among the predictions are small and there is no method that outperforms the others over a pronounced range in coverage or error.
For the MIP test-set, overall prediction quality attained by the four methods is dramatically lower than for the other test-sets, with coverages ranging from 20% to 40% at 10% error (Figure 4). Although the differences are relatively small, SH performs slightly worse at a coverage >35% than its counterparts for this dataset.
In summary, for two of the four test-sets all predictions are very accurate while for another set only SH achieves high accuracy. In contrast, for the fourth set none of the methods give accurate predictions. Furthermore, while predictions for two of the four test-sets show only small performance differences, the differences are larger for the remaining two test-sets and here SH achieves highest coverage throughout.
Different methods select different sites
AMAS focuses on conservation of physicochemical properties. This principle leads to very specific predictions but the existing method seems to be overly conservative.
SDP-pred, TreeDet/MB and SH/E focus on conservation and yield good predictions in general. Differences among these methods are relatively small. This suggests that the signal arising from conservation dominates over possible differences arising from methodology or ranking schemes. However, there appears to be a margin for improvement that could indicate that other factors are more important than conservation for determining functional specificity.
SDP-pred uses the Bernoulli estimator to automatically determine an optimal cut-off and yields highly specific but conservative predictions. TreeDet uses an internal algorithm for unsupervised grouping of sequences that may not always lead to finding differences of interest.
SH focuses on non-overlapping composition between subfamilies and yields good predictions with very high coverage in several cases. This suggests that subfamily differences and sequence context are crucial determinants for functional specificity.
The examples studied here indicate that emphasis on conservation is not sufficient to specifically detect known functional sites. Shifting the focus completely to differences as we have implemented in the SH method, seems to give better predictions overall, at least on the datasets tested here. However, it remains to be seen whether other factors may be involved and what relative weight should be attached to conservation and compositional differences.
The difference between our SH and SH/E methods is the use of sequence context by SH for the ranking of selected sites. Generally, SH performs better than SH/E, which indicates that sequence context may be an important indicator to select regions of interest. It is interesting to note that several of the sequential ranges identified in the Smads (Table 2) are located inside helices. This may seem counter-intuitive since first and second neighbours are on opposite sides in a helix. Nevertheless, the average distance between the neighbouring Cβs in a helix is only ∼5 Å and the flexibility of the sidechain would allow them to approach close enough to participate in the same function. The alternating pattern of β-strands is accommodated by the inclusion of second neighbours, as already described. An interesting refinement of the method could integrate knowledge of the secondary structure, e.g. selecting only first neighbours for loops, only second neighbours for β-strands, and first, third or fourth neighbours for helices.
Performance varies for different datasets
For the protein families presented here, the functional specificities of many sites have been investigated. Validation based on point mutations (Smad, Ras/Ral) is the most specific, but ignores the possible cooperative role of additional sites. Validation based on the exchange of sequence segments (Rab5/6) does include the possible cooperative role of sites, but makes it difficult to assess the discriminatory effect of individual sites. Validation based on distance-to-ligand (MIP) provides no information about the individual role of sites. In all test-cases included, many sites remain that have not been investigated experimentally and it is likely that many additional functional sites exist. This would lead to an underestimation of the number of true positives and complicates the evaluation of different prediction results.
For the two test-sets based on point-mutants (Smad, Ras/Ral), we see a generally high performance of the methods. The abundance of direct and high quality experimental validation for the Smad test-set allows an accurate assessment of the quality of the predictions. For the other two test-sets (Rab5/6, MIP), performance is generally lower. The methods perform well in identifying specific sites, but experience more difficulty in delineating regions corresponding to swapped segments or residues that are close to bound ligands.
The degree of conservation differs significantly between sites, but the available experimental evidence does not necessarily cover this distribution uniformly. Notably, conserved sites are often likely to be functionally relevant and this has led to a likely bias in studying mutations of relatively conserved sites. Such a bias would lead to an overestimation of the importance of conservation.
For further development of this field of research, a well-crafted collection of high-quality experimental data for a variety of protein families would be of great value. Preferably, experimental validation would be based on a representative mix of sites with different degrees of conservation and include many specific point mutations as well as swapped segments to assess supporting roles of sites and strengthen confidence in true negatives.
Functional sites can be grouped by spatial clustering
Functionally related residues tend to form spatial clusters in a protein structure. SH selects sites of unknown function for the protein families considered, and most of these cluster with sites of known function. This provides a way of grouping selected sites and can be used to transfer functional annotation for residues assigned to the same cluster. Preliminary data seem to indicate that coarse-grained structures of medium or even low quality, e.g. by homology modelling or ab initio prediction, may also prove sufficient for this type of clustering.
For the Smad protein family many important questions about the specific interactions with other factors in the TGF-β and BMP-associated pathways are still open. The sites selected by SH are likely candidates for supporting these specific interactions. The putative functions assigned to these sites based on the spatial clustering may provide important guidance for future experiments.
CONCLUSION
We have shown that SH achieves predictions of high quality. While some other methods use sophisticated statistical analysis methods, this does not appear to lead to an increased quality of the predictions. The simplicity of SH makes the results easy to understand. SH achieves high coverage in general and selects additional sites of unknown function. The location of these sites in the crystal structures associated with the benchmark sets used here indicates most of them as promising candidates for further investigation.
The current study provides, to the best of our knowledge, a first attempt of a systematic comparison of prediction methods for functional differences between protein subfamilies. From this analysis, we conclude that exploiting conservation alone is not sufficient, and that more emphasis on sequence differences and context could be the crucial factor for identification of sites of functional specificity.
The SH web-server is available at www.ibi.vu.nl/programs/seqharmwww.
Acknowledgments
K.A.F. and W.P. acknowledge financial support from the Netherlands Bioinformatics Centre, BioRange Bioinformatics research programme (SP 2.3.1 and SP 3.2.2, respectively). We thank the two anonymous referees for their thoughtful and thorough comments that have lead to significant improvements in the paper. We thank Willie Taylor for a critical reading of the manuscript. We also thank Mikhail Gelfand for sharing alignment data on the MIP family. Funding to pay the Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Livingstone C.D., Barton G.J. Identification of functional residues and secondary structure from protein multiple sequence alignment. Methods Enzymol. 1996;266:497–512. doi: 10.1016/s0076-6879(96)66031-5. [DOI] [PubMed] [Google Scholar]
- 2.Lichtarge O., Bourne H.R., Cohen F.E. An evolutionary trace method defines binding surfaces common to protein families. J. Mol. Biol. 1996;257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 3.Kuipers W., Oliveira L., Vriend G., Ijzerman A.P. Identification of class-determining residues in G protein-coupled receptors by sequence analysis. Recept. Channels. 1997;5:159–174. [PubMed] [Google Scholar]
- 4.Hannenhalli S.S., Russell R.B. Analysis and prediction of functional sub-types from protein sequence alignments. J. Mol. Biol. 2000;303:61–76. doi: 10.1006/jmbi.2000.4036. [DOI] [PubMed] [Google Scholar]
- 5.Mirny L.A., Gelfand M.S. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J. Mol. Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 6.Mesa A.D., Pazos F., Valencia A. Automatic methods for predicting functionally important residues. J. Mol. Biol. 2003;326:1289–1302. doi: 10.1016/s0022-2836(02)01451-1. [DOI] [PubMed] [Google Scholar]
- 7.Kalinina O.V., Mironov A.A., Gelfand M.S., Rakhmaninova A.B. Automated selection of positions determining functional specificity of proteins by comparative analysis of orthologous groups in protein families. Protein Sci. 2004;13:443–456. doi: 10.1110/ps.03191704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donald J.E., Shakhnovich E.I. Predicting specificity-determining residues in two large eukaryotic transcription factor families. Nucleic Acids Res. 2005;33:4455–4465. doi: 10.1093/nar/gki755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye K., Lameijer E.W., Beukers M.W., Ijzerman A.P. A two-entropies analysis to identify functional positions in the transmembrane region of class A G protein-coupled receptors. Proteins. 2006;63:1018–1030. doi: 10.1002/prot.20899. [DOI] [PubMed] [Google Scholar]
- 10.Whisstock J.C., Lesk A.M. Prediction of protein function from protein sequence and structure. Quart. Rev. Biophys. 2003;36:307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 11.Obenauer J.C., Denson J., Mehta P.K., Su X., Mukatira S., Finkelstein D.B., Xu X., Wang J., Ma J., Fan Y., et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 12.Shannon C.E. A Mathematical Theory of Communication. Bell System Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- 13.Shenkin P.S., Erman B., Mastrandrea L.D. Information-theoretical entropy as a measure of sequence variability. Proteins. 1991;11:297–313. doi: 10.1002/prot.340110408. [DOI] [PubMed] [Google Scholar]
- 14.Heger A., Lappe M., Holm L. Accurate detection of very sparse sequence motifs. J. Comput. Biol. 2004;11:843–857. doi: 10.1089/cmb.2004.11.843. [DOI] [PubMed] [Google Scholar]
- 15.Mihalek I., Res I., Lichtarge O. A family of evolution-entropy hybrid methods for ranking protein residues by importance. J. Mol. Biol. 2004;336:1265–1282. doi: 10.1016/j.jmb.2003.12.078. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff J.G., Henikoff S. Using substitution probabilities to improve position-specific scoring matrices. Comput. Appl. Biosci. 1996;12:135–143. doi: 10.1093/bioinformatics/12.2.135. [DOI] [PubMed] [Google Scholar]
- 17.Feng X.H., Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 18.Massagué J., Seoane J., Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 19.Reuther G.W., Der C.J. The Ras branch of small GTPases: Ras family members don't fall far from the tree. Curr. Opin. Cell. Biol. 2000;12:157–165. doi: 10.1016/s0955-0674(99)00071-x. [DOI] [PubMed] [Google Scholar]
- 20.Bauer B., Mirey G., Vetter I.R., Garcia-Ranea J.A., Valencia A., Wittinghofer A., Camonis J.H., Cool R.H. Effector recognition by the small GTP-binding proteins Ras and Ral. J. Biol. Chem. 1999;274:17763–17770. doi: 10.1074/jbc.274.25.17763. [DOI] [PubMed] [Google Scholar]
- 21.Stenmark H., Olkkonen V.M. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenmark H., Valencia A., Martinez O., Ullrich O., Goud B., Zerial M. Distinct structural elements of rab5 define its functional specificity. EMBO J. 1994;13:575–583. doi: 10.1002/j.1460-2075.1994.tb06295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zardoya R., Villalba S. A phylogenetic framework for the aquaporin family in eukaryotes. J. Mol. Evol. 2001;52:391–404. doi: 10.1007/s002390010169. [DOI] [PubMed] [Google Scholar]
- 24.Fu D., Libson A., Miercke L.J., Weitzman C., Nollert P., Krucinski J., Stroud R.M. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 25.Simossis V.A., Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. doi: 10.1093/nar/gki390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simossis V.A., Heringa J. The PRALINE online server: optimising progressive multiple alignment on the web. Comput. Biol. Chem. 2003;27:511–519. doi: 10.1016/j.compbiolchem.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo R.S., Chen Y.G., Shi Y., Pavletich N.P., Massague J. The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-beta receptors. EMBO J. 1998;17:996–1005. doi: 10.1093/emboj/17.4.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G., Chen Y.G., Ozdamar B., Gyuricza C.A., Chong P.A., Wrana J.L., Massague J., Shi Y. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science. 2000;287:92–97. doi: 10.1126/science.287.5450.92. [DOI] [PubMed] [Google Scholar]
- 30.Huse M., Muir T.W., Xu L., Chen Y.G., Kuriyan J., Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol. Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 31.Mizuide M., Hara T., Furuya T., Takeda M., Kusanagi K., Inada Y., Mori M., Imamura T., Miyazawa K., Miyazono K. Two short segments of Smad3 are important for specific interaction of Smad3 with c-Ski and SnoN. J. Biol. Chem. 2003;278:531–536. doi: 10.1074/jbc.C200596200. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y.G., Massague J. Smad1 recognition and activation by the ALK1 group of transforming growth factor-beta family receptors. J. Biol. Chem. 1999;274:3672–3677. doi: 10.1074/jbc.274.6.3672. [DOI] [PubMed] [Google Scholar]
- 33.Randall R.A., Germain S., Inman G.J., Bates P.A., Hill C.S. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J. 2002;21:145–156. doi: 10.1093/emboj/21.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y.G., Hata A., Lo R.S., Wotton D., Shi Y., Pavletich N., Massague J. Determinants of specificity in TGF-beta signal transduction. Genes Dev. 1998;12:2144–2152. doi: 10.1101/gad.12.14.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Germain S., Howell M., Esslemont G.M., Hill C.S. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- 36.Yakymovych I., Heldin C.H., Souchelnytskyi S. Smad2 phosphorylation by type I receptor: contribution of arginine 462 and cysteine 463 In the C terminus of Smad2 for specificity. J. Biol. Chem. 2004;279:35781–35787. doi: 10.1074/jbc.M404377200. [DOI] [PubMed] [Google Scholar]