Abstract
The deposition of proteins onto newly spliced mRNAs has far reaching consequences for their subsequent metabolism. We affinity-purified spliced human mRNPs under physiological conditions from HeLa nuclear extract and present the first comprehensive inventory of their protein composition as determined by mass spectrometry. Several proteins previously not known to be mRNP-associated were detected, including the DEAD-box helicases DDX3, DDX5, and DDX9, and the ELG, hNHN1, BCLAF1, and TRAP150 proteins. The association of some of the newly identified mRNP proteins was shown to be splicing-dependent, but not to require EJC formation. Initial recruitment of EJC proteins to the spliceosome did not require an EJC binding platform at the −20/24 region of the 5′ exon. Finally, while recruitment of EJC proteins and stable EJC formation were not dependent on the cap binding complex, several of the newly identified mRNP proteins required the latter for their association with mRNPs. These results provide novel insights into the composition of spliced mRNPs and the requirements for the association of mRNP proteins with the newly spliced mRNA.
Keywords: CBC, EJC, mass spectrometry, mRNP, TREX
INTRODUCTION
Splicing is a fundamental step in eukaryotic gene expression during which introns are excised from pre-mRNA and exons are ligated together to form a continuous reading frame (for review, see Will and Lührmann 2006). The mRNA product generated by splicing is complexed with a currently unknown number of proteins, some of which are deposited as a consequence of splicing. These proteins influence every aspect of the subsequent metabolism of the mRNA, including its export from the nucleus, translation efficiency, and stability in the cytoplasm (for review, see Moore 2005). In this way, splicing not only plays a crucial role in the maturation of an mRNA in the nucleus, but also has far reaching downstream effects on the ultimate fate of mRNAs in the cell.
Splicing is catalyzed by the spliceosome, which forms by the ordered association of the U1, U2, U4/U6, and U5 spliceosomal snRNPs and numerous non-snRNP proteins with the pre-mRNA (for review, see Will and Lührmann 2006). Distinct spliceosome assembly/functional intermediates can be isolated including the E, A, B, B*, and C complexes (forming in that order). Upon catalytic activation, the B complex (which contains all of the above-mentioned snRNPs), is converted to the activated B* complex, which lacks the U1 and U4 snRNPs. Complex C is formed after the first catalytic step of splicing and, after the second step, the spliced mRNA, in the form of an mRNP, and the excised intron (initially complexed with the U2, U5, and U6 snRNPs) are released.
Mass spectrometry (MS) analyses provided a comprehensive inventory of the protein composition of the A, B, B*, and C spliceosomal complexes (Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Deckert et al. 2006). They also revealed that a large number of proteins associate with the pre-mRNA, many in a highly dynamic manner. Our understanding of the protein composition of spliced mRNPs, in contrast, is far from complete. Initially, immunoprecipitation studies provided valuable information about proteins associated with mRNAs in cell extracts (for review, see Dreyfuss et al. 2002). More recently, affinity-selected mRNPs generated by in vitro splicing were subjected to MS and their protein composition identified. However, the latter mRNPs were isolated in the presence of heparin, which strips off all but the most tightly associated proteins (Reichert et al. 2002; Tange et al. 2005).
The best characterized group of proteins known to bind mRNAs are those of the exon junction complex (EJC), which in mammals binds in a sequence-independent manner to spliced mRNAs 20–24 nucleotides (nt) upstream of the site of exon–exon ligation (Le Hir et al. 2000; Tange et al. 2005; for review, see Tange et al. 2004). The EJC is a highly dynamic entity, consisting of several core proteins that stably associate with the mRNA, as well as other factors that bind in a weaker or transient manner. Core components of the human EJC include the proteins Y14 and Magoh, which form a tight heterodimer (Lau et al. 2003), as well as eIF4A3/DDX48 and MLN51/BTZ, which appear to be the only EJC components directly contacting the mRNA (Ballut et al. 2005; Tange et al. 2005; Stroupe et al. 2006). A stable EJC core RNP complex can be reconstituted in vitro from the latter four proteins in the presence of single-stranded RNA and ATP. Like the EJC generated by splicing, formation of this complex leads to the protection of an 8–9 nucleotide stretch of the mRNA from nucleolytic digestion (Ballut et al. 2005). Less stably associated or transiently associated EJC components include Aly/REF (also termed BEF and THOC4), UAP56/DDX39, TAP/NXF1, RNPS1, Pinin, Acinus, SAP18, SRm160, hUpf1, hUpf2, and hUpf3 (Tange et al. 2004 and references therein). Although there are some indications of a stepwise association of EJC components with the pre-mRNA (Reichert et al. 2002; Kataoka and Dreyfuss 2003), the manner in which these proteins are recruited during splicing remains unclear.
After its stable formation on the mRNA, the EJC undergoes extensive remodeling within the cell. Some EJC proteins, such as UAP56, TAP/NXF1, Acinus, and Pinin, are found solely in the nucleus (Li et al. 2003), dissociating prior to/during export of the mRNP from the nucleus, whereas others appear to associate exclusively in the cytoplasm (e.g., Upf1) (Applequist et al. 1997). During the first round of translation, EJCs are removed from the mRNA by the ribosome and most of the remaining components shuttle back to the cell nucleus (Dostie and Dreyfuss 2002). Interestingly, the association of other known components of mRNPs is also dynamic in nature. Although some proteins remain stably bound to the mRNA after its export into the cytoplasm, other factors are removed during or shortly after the passage of the mRNA through the nuclear pore. For example, CBP80 and CBP20 (cap binding protein) that form the m7G cap-binding complex (CBC), and the poly-A-binding protein 2 (PABP2) are replaced shortly after the emergence of the mRNP in the cytoplasm, by eIF4E and PABP1, respectively.
EJC-associated proteins play important roles in several steps of mRNA metabolism. For example, SRm160, RNPS1, and Pinin have been shown to act as splicing coactivators or alternative splicing factors (Mayeda et al. 1999; Wang et al. 2002). UAP56, Aly/REF, and NXF1/TAP are documented mRNA export factors, where UAP56 is thought to recruit Aly/REF to the mRNA which in turn recruits NXF1/TAP, a factor directly interacting with proteins of the nuclear pore complex (Luo et al. 2001). “Quality control” of spliced mRNAs is achieved in the cytoplasm by a process termed nonsense-mediated decay (NMD), which removes mRNAs with premature stop codons (PTCs) from the mRNA pool. Distinct functions in this process have been ascribed to the NMD factors hUpf1, hUpf2, and hUpf3, as well as to the EJC proteins Y14, eIF4A3, and MLN51 in hUpf2-dependent NMD, and to RNPS1 in hUpf2-independent NMD (Gehring et al. 2005). In Drosophila, homologs of Y14 and Magoh play a crucial role in the proper cytoplasmic localization of oskar mRNA in developing oocytes (Hachet and Ephrussi 2001). Finally, EJC formation increases mRNA translational yield, apparently by enhancing incorporation of spliced mRNPs into polysomes (Wiegand et al. 2003; Nott et al. 2004).
Recent, co-immunoprecipitation experiments with HeLa nuclear extracts revealed that components of the so-called THO/TREX (transcription- and export-related) complex not only associate with spliceosomes (Zhou et al. 2002), but also preferentially with spliced mRNAs as opposed to unspliced mRNAs (Masuda et al. 2005). In humans, the TREX complex is comprised of the EJC-associated proteins Aly/REF and UAP56 together with the THO proteins THOC1 (hHpr1/p84), THOC2 (hRlr1), THOC3 (hTex1), THOC5 (fSAP79), THOC6 (fSAP35), and THOC7 (fSAP24) (Masuda et al. 2005). Although much evidence indicates that the function of the TREX complex as an adaptor between the mRNA and components of the export machinery is conserved among eukaryotes, in Drosophila the majority of mRNAs can be exported from the nucleus independently of the THO complex (Rehwinkel et al. 2004).
Here we have isolated, spliced AdML and β-globin mRNPs under physiological conditions from HeLa nuclear extract. Analyses of their protein composition by MS techniques and also by Western blotting led to the identification of ∼45 proteins, including several shown for the first time to be mRNP-associated. Requirements for the association of these newly detected proteins with mRNA, including their dependency on splicing, EJC formation, and the presence of the cap binding complex (CBC) were subsequently analyzed. Likewise, 5′ exon and CBC requirements for the initial association of EJC components with the spliceosome, as well as stable EJC formation on spliced mRNAs, were also investigated. These studies reveal for the first time the composition of newly spliced mRNPs isolated under native conditions and enhance our understanding of those factors contributing to the recruitment of mRNP-associated proteins in the nucleus.
RESULTS
MS2 affinity selection of mRNPs spliced in vitro
To determine the protein composition of mRNPs isolated under native, low-stringency conditions (i.e., in the absence of heparin), we adopted the MBP-MS2 purification system previously used to isolate spliceosomal complexes (Jurica et al. 2002; Zhou et al. 2002; Deckert et al. 2006). For this purpose, three repeats of an RNA hairpin bound by the MS2 coat protein were introduced at the 3′ end of an adenovirus-derived pre-mRNA (AdML-M3) or the human β-globin pre-mRNA (β-Glo-M3). AdML-M3 lacking an intron (AdMLΔI-M3), which generates nonspliced mRNP, or AdML-M3 containing a truncated, 15-nt long exon 1 (AdML15-M3), which does not support EJC formation, were also constructed.
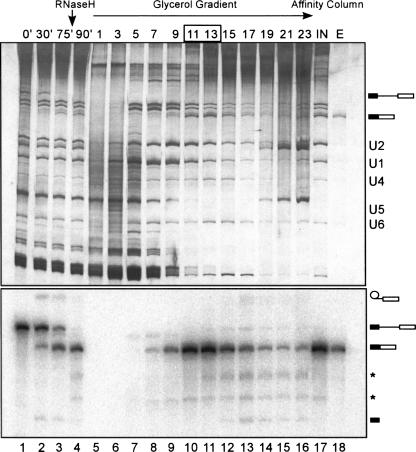
Prior to incubating under splicing conditions, m7G-capped, 32P-labeled pre-mRNA or mRNA from the intronless construct was incubated with an MBP-MS2 fusion protein. Splicing was carried out for 75 min, and the amount of unspliced or partially spliced pre-mRNA was reduced by targeted RNase H digestion of intron sequences. Glycerol gradient centrifugation was then performed to separate mRNPs from other RNP complexes. As shown in Figure 1 for β-globin-M3 pre-mRNA, the distribution of radioactivity across the gradient revealed the presence of only one peak in the 30S region, which corresponded to the spliced β-globin-M3 mRNP. Native full-length AdML-M3 mRNPs also sedimented in the 30S region of the gradient (data not shown), whereas both spliced AdML15-M3 and unspliced AdMLΔI-M3 mRNPs sedimented in the 20S–25S region (data not shown). Gradient fractions containing mRNPs were pooled and affinity selection with amylose beads was performed. Bound RNP complexes were eluted under native conditions with an excess of maltose.
FIGURE 1.
MS2 affinity selection of in vitro spliced mRNPs. MS2-tagged β-globin pre-mRNA was incubated under splicing conditions in HeLa nuclear extract for 0–75 min (lanes 1–3). After 75 min, intron containing RNA species were removed by RNAse H digestion (lane 4) and the splicing reaction was separated on a linear 10%–30% glycerol gradient (lanes 5–16). Fractions 11–13 were pooled and subjected to MS2 affinity selection (lane 17, where “IN” represents 10% of the input material). Bound complexes were eluted with maltose (lane 18, where 10% of eluted material was analyzed). RNA was recovered, separated by 12% denaturing PAGE, and visualized by silver staining (upper panel) or, to detect 32P-labeled species, by PhosphorImager analysis (lower panel). The positions of snRNA and/or the pre-mRNA and splicing intermediates and products is shown on the right. Asterisks mark cleavage products generated by RNase H digestion.
RNA was recovered from the eluate, analyzed by denaturing PAGE and visualized by silver staining or with a PhosphorImager. Apart from spliced mRNA, only faint amounts (<4%) of unspliced pre-mRNA, but no splicing intermediates could be detected in the β-globin-M3 and AdML-M3 eluates (Figs. 1, lane 18 and 2A, lane 1), indicating that these mRNP preparations are essentially free from spliceosomes or excised intron-containing complexes (Fig. 1, lower panel). Consistent with these results, silver staining confirmed the presence of a single major band corresponding either to the β-globin-M3 (Fig. 1, lane 18, upper panel) or AdML-M3 mRNA (Fig. 2A, lane 2), with only low amounts of the spliceosomal snRNAs detectable in each case.
FIGURE 2.
Characterization of affinity-selected AdML mRNPs. RNA (A) and protein (B) composition of MS2 affinity-purified AdML-M3 mRNPs. (A) RNA from 10% of the MS2 eluate was analyzed as in Figure 1. (B) Proteins from the entire eluate were separated by SDS-PAGE and stained with silver. A subset of proteins identified by MS are indicated at the right (alternative names in parentheses). Lane 1: molecular weight markers. Lane 2: control pulldown performed with untagged AdML pre-mRNA in the presence of MBP-MS2.
Mass spectrometry of spliced, wild-type AdML-M3 and β-Globin mRNPs
Proteins were recovered from the AdML-M3 eluate, fractionated by SDS-PAGE and analyzed by LC-MS/MS mass spectrometry (Fig. 2B, lane 3; Table 1). A total of ∼45 proteins were identified. As expected, most EJC-associated proteins (Y14, Magoh, eIF4A3, RNPS1, Acinus, Pinin, UAP56, and Aly/REF), with the highly notable exception of MLN51, were detected. UAP56 and Aly/REF are also components of the human THO/TREX complex. Consistent with recent reports (Masuda et al. 2005), the TREX-associated proteins THOC1, THOC2, THOC3, THOC5, and THOC7 were also found. Numerous SR proteins, as well as several members of the hnRNP protein family, were identified; likewise, the general RNA-binding proteins NFAR-1 (nuclear factor associated with ds-RNA, also termed ILF3), its binding partner NF45/ILF2 and YB-1, which were found in all mRNP preparations isolated in the absence of heparin (Table 1), the cap binding proteins CBP80 and CBP20, and ARS2 (Arsenite resistance protein 2), a previously identified mRNP protein, were found. MS analyses of spliced AdML mRNPs also revealed the presence of proteins with no known connection to spliced mRNPs: namely BCLAF1 (Bcl-2-associated transcription factor), ELG, hNHN1 (LOC124245), and TRAP150 (or Thrap3 for thyroid hormone receptor associated protein 3), as well as the DEAD-box proteins DDX3, DDX5/p68, and DDX9 (RNA helicase A), which henceforth will be designated “newly identified mRNP proteins.”
TABLE 1.
Protein composition of human mRNPs isolated under physiological conditions
A number of snRNP proteins were also identified in the AdML-M3 mRNP eluate, but several lines of evidence indicate that they do not represent bona fide mRNP proteins. First, several snRNP proteins, including U5–220K, U5–200K, U5–100K, SF3b130, and SF3b49 were detected in a mock affinity selection performed with AdML-M3 pre-mRNA lacking the MS2 tag (Table 1). Finally, immunoprecipitation experiments with antibodies against the U4/U6–61K protein (hPrp31) provided evidence that it does not associate with purified AdML-M3 mRNPs (Supplemental Fig. S-1; http://edoc.mpg.de/288481/fulltext/30456/0).
MS analyses of β-globin-derived spliced mRNPs revealed a very similar protein composition (Table 1). Significantly, DDX3 and DDX5, as well as the newly identified proteins BCLAF1, ELG, hNHN1, and TRAP150, were also detected, indicating that they are common components of all spliced mRNPs. In contrast, several AdML mRNP proteins (e.g., DDX9, THOC1, THOC3, THOC5, and THOC7) were not detected in the β-globin mRNP, suggesting they may associate with only a subset of mRNP species, or are loosely associated components that are readily lost and thus not consistently observed.
Surprisingly, MLN51, a core component of EJC complexes (Ballut et al. 2005; Tange et al. 2005), was not detected in either of our purified mRNPs. To determine whether it may have eluded detection by MS, we performed Western blotting. MLN51 was detected in HeLa nuclear extract, but not in purified AdML-M3 mRNPs (Fig. 3A). In contrast, Magoh, Aly/REF, and CBP80 were readily detected in both nuclear extract and purified mRNPs (Fig. 3A, cf. lanes 1,2). To test whether MLN51 is associated with partially purified mRNPs, we fractionated in vitro splicing reactions containing AdML-M3 mRNPs on glycerol gradients and analyzed the distribution of MLN51 via Western blotting. Significantly, gradient fractions enriched in mRNPs (i.e., 13–17), did not contain detectable levels of MLN51, whereas CBP80, Aly/REF, and Magoh were clearly present (Fig. 3B). Taken together, these results confirm that our MS2 affinity-purified, in vitro spliced mRNPs lack MLN51.
FIGURE 3.
MLN51 is not associated with purified, spliced mRNPs. (A) Western blot of proteins recovered from HeLa nuclear extract (lane 1) or MS2-affinity selected AdML-M3 mRNPs (lane 2). Proteins were separated on a 12% polyacrylamide-SDS gel, transferred to nitrocellulose and visualized by staining with antibodies against CBP80, MLN51, Aly/REF, and Magoh as indicated. (B) MLN51 does not co-sediment with partially purified mRNPs. Splicing reactions containing AdML-M3 mRNPs were fractionated on a glycerol gradient. RNA and proteins were recovered and analyzed by denaturing PAGE or SDS-PAGE as described in Figures 1 and 2. 32P-labeled mRNA was detected by autoradiography (upper panel) and the distribution of MLN51, CBP80, Aly/REF, and Magoh across the gradient (lower panels) was determined by Western blot as described in (A).
Protein composition of heparin-treated, spliced mRNPs
To investigate which proteins are stably associated with the spliced mRNA, we affinity-purified AdML mRNPs as described above, except that heparin (which dissociates loosely associated proteins) was added during the last 5 min of the RNase H digestion. Heparin treatment led to a shift in the mRNP peak from the 30S to 25S region of the glycerol gradient, indicating that some protein factors had indeed dissociated (data not shown). MS analyses of this “core-mRNP” (Table 1) revealed that significantly fewer proteins bind mRNA in this complex than in mRNPs isolated in the absence of heparin. Several, but not all, of the SR and hnRNP proteins detected in the native mRNPs, as well as all THO proteins and the general RNA-binding proteins NFAR1, NF45, and YB-1, were no longer detected. Likewise, snRNP-associated proteins were no longer found. A subset of the EJC proteins (i.e., RNPS1 and Pinin) were also lost; UAP56 was not detected in the core AdML-M3 mRNP, but found in heparin-treated β-globin mRNPs (Table 1). The association of the remaining EJC proteins (Y14, Magoh, Aly/REF, Acinus, and eIF4A3) and the CBC proteins was heparin-resistant. Similarily, while the DEAD-box helicases DDX5 and DDX9 were lost, DDX3 was still present. Finally, with the exception of TRAP150, the novel mRNP proteins BCLAF1, ELG, and hNHN1 remained associated with the mRNA in the presence of heparin, indicating that they are stably associated.
Identification of proteins binding unspliced AdML mRNA
To distinguish between proteins that are deposited on mRNAs solely as a consequence of splicing, versus those that can bind in a splicing-independent manner, we affinity-selected unspliced mRNPs generated after incubating m7G-capped, intronless AdML RNA (AdMLΔI-M3) under splicing conditions. Essentially only AdMLΔI-M3 mRNA was detected in the eluate by PhosphorImager analysis or silver staining (Supplemental Fig. S-2; http://edoc.mpg.de/288481/fulltext/30456/0), indicating that highly pure mRNPs had been isolated. A comparison of the protein contents of spliced and unspliced AdML mRNPs revealed that the majority of the mRNP proteins identified above (i.e., excluding known general RNA binding proteins) associate with mRNAs as a consequence of splicing. As expected, the unspliced mRNA did not acquire any members of the canonical EJC (Table 1). The absence of EJC proteins was also confirmed by Western blot analysis with antibodies against RNPS1, Y14, and Magoh (Fig. 4, lane 2). Interestingly, the DEAD-box helicases DDX3 and DDX5, a subset of the THO proteins and the novel proteins ELG and hNHN1, were also not detected, suggesting that their association with mRNPs is dependent on splicing. In contrast, three other TREX proteins (UAP56, Aly/REF, and THOC1) were detected in unspliced AdM3ΔI mRNPs by MS (Table 1) and by Western blotting (Fig. 4, lane 2). Of the newly identified mRNP proteins, BCLAF1 and TRAP150 were also detected, indicating that their association with mRNA can occur independent of splicing.
FIGURE 4.
Western analyses of the protein composition of purified mRNPs. mRNPs generated from intronless control mRNA (AdMLΔI-M3, lane 2) and pre-mRNAs with a truncated 5′ exon (AdML15-M3, lane 3) or wild-type 5′ exon (AdML-M3, lane 4) were purified under standard conditions, or after addition of m7GppG to the splicing reaction (AdML-M3ΔCBC, lane 5). A mock selection was performed with MBP-MS2 fusion protein (lane 1). Westerns were performed with ∼200 fmol of each mRNP (according to cpm of RNA) and the indicated antibodies as described in Figure 3.
Protein composition of spliced mRNPs lacking an EJC
Unspliced mRNPs lacked not only EJC components but also a number of additional proteins, suggesting that EJC formation is required for the association of the latter with the mRNA. To determine which proteins associate with spliced mRNPs independently of the EJC, we affinity-selected mRNPs generated from the AdML15-M3 pre-mRNA, which contains a truncated, 15-nt long exon 1. This pre-mRNA undergoes splicing, but the mRNA product is defective in EJC assembly due to the lack of an EJC-binding platform 20–24 nts upstream of its exon–exon boundary (Le Hir et al. 2001). Analysis of the RNA composition of the affinity-selected AdML15-M3 mRNPs (Supplemental Fig. S-3; http://edoc.mpg.de/288481/fulltext/30456/0) indicated that they were purified to near homogeneity. MS (Table 1) and Western blotting (Fig. 4, lane 3) confirmed that EJC proteins were not present in purified AdML15-M3 mRNPs. Intriguingly, we also did not find any TREX proteins, including those that had been previously detected in both spliced and unspliced mRNPs (Table 1 and Fig. 4, cf. lanes 2,4,5). Moreover, DDX5 and DDX3 were also not detected, suggesting that they, together with the TREX proteins, associate with the mRNA via the EJC. In contrast, all of the novel mRNP proteins, including those whose association is dependent on splicing (i.e., ELG, hNHN1), were detected in AdML15-M3 mRNPs; thus, their association with mRNA does not require EJC formation.
RNA sequence requirements for recruitment of EJC proteins to the spliceosome
Most EJC proteins appear to be recruited to the pre-mRNA after B-complex formation, but prior to/during the formation of the C complex (Jurica et al. 2002; Reichert et al. 2002; Deckert et al. 2006). Although stable deposition of the EJC on spliced mRNA requires the presence of >17 nt upstream of the exon/exon junction (Le Hir et al. 2001), it is presently not known whether this EJC-binding platform is required for the recruitment of EJC proteins to the spliceosome. We thus affinity-purified a mixture of B*/C spliceosomal complexes formed on wild-type or truncated exon 1 MINX pre-mRNA containing an intronic MS2-tag and a 3′ splice site mutation (AG→GG) that abolishes the second step of splicing. Analysis of the RNA content of purified complexes revealed that they contained both unspliced pre-mRNA and splicing intermediates, but no mRNA (Fig. 5A, lanes 1–4). Quantification of the radioactively labeled RNA species showed that in both cases ∼85% of the pre-mRNA had undergone the first step of splicing and thus was present in the form of C complexes. Purified spliceosomes contained only small amounts of the U1 and U4 snRNAs (Fig. 5A), further indicating that predominantly a mixture of C and catalytically activated B* complexes had been isolated in both cases. As evidenced by Western blot analyses, the EJC proteins Y14, Magoh, RNPS1, Aly/REF, and UAP56 were present in purified B*/C spliceosomes, regardless of whether an EJC-assembly platform in the –20/24 region of exon 1 was present or not (Fig. 5B, lanes 1,2). In contrast, consistent with results shown above, spliced mRNPs containing a truncated exon 1, did not retain these EJC-associated proteins (Fig. 5B, lane 4). These data suggest that the recruitment of at least a subset of EJC proteins to spliceosomes does not require an EJC-assembly platform on the pre-mRNA. Thus, their absence from purified mRNPs with a truncated 5′ exon does not appear to be due to inhibition of their initial recruitment to the pre-mRNA, but rather likely the result of impaired EJC assembly at the time of mRNA formation (see Discussion).
FIGURE 5.
EJC proteins associate with spliceosomal C complexes formed on pre-mRNA with a truncated 5′ exon. (A) RNA content of MS2 affinity-purified B*/C spliceosomes. Spliceosomes were allowed to form on wild-type MINX pre-mRNA (E1-WT, lanes 1 and 2) or MINX pre-mRNA with a truncated 15 nt long 5′ exon (E1–15, lanes 3 and 4). After affinity purification of B*/C complexes, RNA was recovered and visualized by PhosphorImager analysis (lanes 1 and 3) and silver staining (lanes 2 and 4). Pre-mRNA and splicing intermediates are indicated on the right, and products of RNase H digestion are marked with asterisks. (B) Western analyses (see Figure 3) were performed with purified B*/C complexes or mRNPs with a wild-type or truncated 5′ exon (as indicated above each lane) and those antibodies indicated at the right. CBP80 is a loading control.
The cap-binding complex is not required for EJC formation in vitro
A common structural feature of all RNAPII transcripts is their m7G-cap structure and the cap-binding protein dimer CBP80/20. The latter aids in the recruitment of various factors during pre-mRNA processing (see Discussion). We next analyzed the role of the CBC in the recruitment of EJC proteins to mRNAs. To prevent CBP80/20 association, we supplemented in vitro splicing reactions with the cap analog m7GpppG, and affinity-selected spliced AdML-M3 mRNPs from these reactions after incubating under splicing conditions. Addition of m7GpppG had no effect on spliceosome assembly (Fig. 6A) nor on the efficiency of splicing (Fig. 6B) of m7G-capped AdML-M3 pre-mRNA. Western analyses of wild-type AdML-M3 mRNPs or mRNPs isolated in the presence of m7GpppG (designated AdMLΔCBC) demonstrated that the latter were devoid of CBP80 (Fig. 4, cf. lanes 4,5), confirming that the addition of cap analog effectively inhibits CBC formation. Significantly, the EJC and/or TREX-associated proteins RNPS1, Y14, Magoh, Aly/REF, UAP56/DDX39, and THOC1, as well as DDX3, were detected both in wild-type, spliced mRNPs and in those lacking the CBC (Fig. 4, lanes 4,5). In contrast, none of these proteins were precipitated when a control affinity-selection was performed with free MS2-MBP fusion protein (Fig. 4, lane 1). To rule out that CBP80 associates prior to mRNP formation but is subsequently lost, we isolated A, B, and B*/C spliceosomal complexes formed in the presence of cap analog; in no case was CBP80 detectable by Western blotting (data not shown). Finally, we assayed for EJC assembly on the AdML and AdMLΔCBC mRNAs by performing RNase H digestions with DNA oligonucleotides complementary to the EJC-assembly platform and sequences immediately up- and downstream of it (Fig. 6C). Significantly, the –20/24 region of both spliced mRNAs was resistant to oligonucleotide-directed RNase H cleavage, while the flanking sequences were efficiently cleaved. Taken together, these results indicate that the recruitment of EJC proteins to the pre-mRNA and the subsequent assembly of a stable EJC at the –20/24 region of a spliced mRNA does not require the presence of the CBC.
FIGURE 6.
The CBC is dispensable for EJC formation in vitro. An excess of m7GpppG does not affect (A) splicing complex formation nor (B) splicing. Splicing was performed with AdML-M3 pre-mRNA ±m7GpppG (as indicated) for the indicated times. Splicing complex formation or splicing was analyzed on an agarose gel or by denaturing PAGE, respectively, followed by PhosphorImager analysis. The positions of the various spliceosomal complexes (A) or pre-mRNA/intermediates/products (B) are indicated at the right. (C) The absence of the CBC does not inhibit stable EJC formation. EJC formation at the −20/24 region was analyzed by RNase H digestion of the 5′ exon after allowing for the formation of spliced mRNP in the absence (lanes 1–5) or presence (lanes 6–10) of m7GpppG. −40, −22, and −8 indicate the central nucleotide of the complementary DNA oligo used, relative to the exon–exon junction. Arrows on the right mark actual/expected positions of RNase H cleavage products. RNAs were analyzed by denaturing PAGE and visualized with a PhosphorImager.
Several newly identified mRNP proteins require the CBC for mRNA association
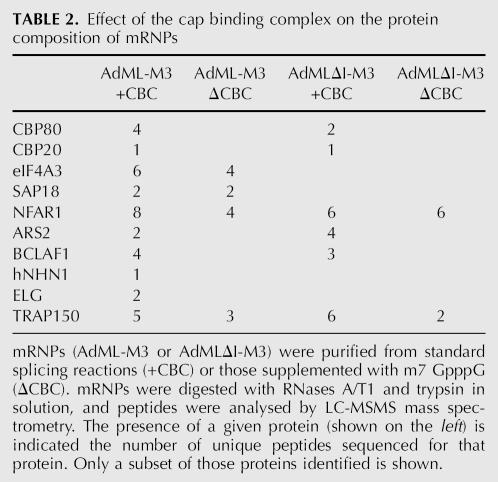
The newly identified mRNP proteins BCLAF1, ELG, hNHN1, and TRAP150, and also the previously identified ARS2, were detected in spliced mRNPs lacking an EJC. Moreover, with the exception of TRAP150, they were insensitive toward heparin treatment, indicating their tight association with the mRNP. Targeted RNAse H digestion indicated that regions of the mRNA flanking the –20/24 position are not bound by protein, suggesting that the aforementioned proteins might associate with other regions of the mRNA or via the CBC. To test the latter possibility, CBC-bound and ΔCBC AdML mRNPs were purified and, as antibodies against the newly identified proteins were not available, mRNPs were analyzed for the presence of these proteins by MS. In this instance, mRNPs were first digested with RNAses A/T1, and proteins were subsequently digested in-solution with trypsin (i.e., without prior SDS-PAGE fractionation as above) prior to LC-MS/MS mass spectrometry. The protein composition of the wild-type mRNPs determined via this MS method was very similar to that described above (Table 2; data not shown). Although the previously undetected EJC protein SAP18 could now be found, MLN51 remained undetectable. Consistent with our initial observations, ARS2, BCLAF1, ELG, hNHN1, and TRAP150 were detected in spliced AdML-M3 mRNPs containing the CBC (Table 2). In contrast, only TRAP150 was detected in spliced AdMLΔCBC mRNPs, indicating that the association of ARS2, BCLAF1, ELG, and hNHN1 is mediated, at least in part, by the CBC.
TABLE 2.
Effect of the cap binding complex on the protein composition of mRNPs
DISCUSSION
We have succeeded in purifying under native conditions a series of spliced and unspliced mRNPs from HeLa nuclear extract, and determining their protein composition by MS. These studies identified a number of proteins previously not known to be mRNP-associated. A comparison of the protein composition of wild-type mRNPs with those of unspliced mRNPs and spliced mRNPs lacking an EJC revealed that the association of several mRNP-associated proteins, while dependent on splicing, does not require formation of the EJC. Characterization of spliceosomal B*/C complexes formed on a pre-mRNA containing a truncated 5′ exon demonstrated that the EJC binding platform at the –20/24 region of the 5′ exon is not required for the initial recruitment of EJC proteins during splicing. Finally, we demonstrate a differential requirement of the CBC for the recruitment of EJC proteins versus several of the newly identified mRNP proteins.
Proteins associated with native affinity-purified, spliced mRNPs
Our affinity-purified mRNPs were essentially free of spliceosomes, as judged by the fact that (1) only faint amounts of unspliced pre-mRNA but no splicing intermediates were detected and (2) MS did not uncover splicing factors acting during activation, catalysis, or spliceosome disassembly (e.g., hPrp16, hSlu7, or hPrp43). However, trace amounts of spliceosomal snRNAs were detected by silver staining, and a number of snRNP-associated proteins were identified by MS. That the identified snRNPs are merely contaminants and do not represent mRNPs complexed with snRNPs (i.e., mRNPs not completely released from postspliceosomal snRNP-containing complexes) is supported by the fact that (1) mRNPs complexed with one or more snRNPs would be expected to sediment with a higher S value, and (2) several proteins identified have never been detected in late spliceosomal complexes (e.g., U1–70K or U4/U6-associated proteins), whereas snRNP proteins specifically associated with the 35S form of the U5 snRNP (i.e., Prp19 and associated proteins), which is formed late in the splicing pathway, were not found. Although the association of one or more individual snRNP proteins with the mRNP cannot be rigorously excluded, they do not appear to be bona fide mRNP proteins. First, several snRNP-associated proteins (but none of the EJC, TREX, or newly identified mRNP proteins) were also eluted from an amylose matrix in control pulldowns with nontagged AdML pre-mRNA, indicating that they bind nonspecifically. Second, the number of unique peptides identified for many of these proteins by MS is relatively low (especially if one takes their size into account). Finally, immunoprecipitation experiments indicated that the U4/U6–61K protein is not associated with mRNPs.
As expected, affinity-purified, spliced AdML and β-globin mRNPs contained most components known to associate with the EJC in the nucleus. Consistent with previous reports (Le Hir et al. 2001), their association was dependent on splicing (i.e., they were not found in AdMLΔI-M3 mRNPs) and also on the presence of a binding platform in the –20/24 region of the 5′ exon (i.e., they were absent from AdML15-M3 mRNPs). Several, loosely associated EJC proteins such as SRm160, TAP/NXF1, and Upf3a/b, were not detected, suggesting they were lost during purification. More surprisingly, we did not detect the EJC core component MLN51 either by different MS methods or by Western blotting in any of our purifed mRNPs. Interestingly, MLN51 was previously not detected by MS in AdML mRNPs affinity-purified in the presence of heparin (Tange et al. 2005); however, in contrast to our studies, subsequent Western blotting experiments confirmed its presence. Furthermore, it was detected by MS in double-affinity-purified EJC complexes subsequently purifed from the above-mentioned mRNPs (Tange et al. 2005). Whether the inability to detect MLN51 in our purified mRNPs is merely a detection problem, or reflects its true absence, is presently unclear. In vitro reconstitution experiments revealed that the association of Magoh, Y14, and eIF4A3 with mRNA is not strictly dependent upon the presence of MLN51 (Ballut et al. 2005). Thus, although stable EJCs form on our in vitro spliced mRNPs (Fig. 6C), it is conceivable that MLN51 might, nonetheless, be absent. MLN51 is a predominantly cytoplasmic protein, consistent with the idea that it might be stably incorporated into EJCs first in the cytoplasm. Although MLN51 was previously found in purified mRNPs and EJCs generated in nuclear extract (Tange et al. 2005), these experiments were performed in the presence of an excess of GST-Y14/Magoh, which may have driven the low amounts of nuclear MLN51 into EJC complexes. Likewise, TAP-tagged MLN51 is found in EJCs formed after in vitro splicing in Hela nuclear extract, when the latter is supplemented with an excess of the tagged protein (Degot et al. 2004; Ballut et al. 2005). Overexpressed EYFP-tagged MLN51 was recently shown to co-localize with Magoh in nuclear speckles (Degot et al. 2004); however, whether endogenous nuclear MLN51 behaves in a similar manner is not clear.
Recent immunoprecipitation experiments indicated that components of the human TREX complex are associated with spliced mRNPs, and also, to a much lesser extent with unspliced mRNPs (Masuda et al. 2005). Indeed, with the exception of THOC6, we detected nearly all known components of the human TREX complex in spliced AdML mRNPs. Based on the number of unique peptides identified (and also taking their molecular weight into consideration), some of the THO proteins identified appear to be underrepresented. Thus, they may be loosely-associated mRNP components that are readily lost during affinity selection. Consistent with this idea, only UAP56, Aly/REF and THOC2 were found in β-globin mRNPs, and only the association of UAP56 and Aly/REF withstood heparin-treatment, indicating that they, in contrast to other TREX proteins, are stably associated. Similarily, aside from Aly/REF, no TREX-associated proteins were previously found by MS in heparin-treated, affinity-purified, spliced mRNPs (Tange et al. 2005). The association of the THO proteins with purified mRNPs was dependent on both splicing (with the curious exception of THOC1) and EJC formation (Table 1; Figure 4), consistent with the idea that the export factors Aly/REF and/or UAP56—which are also EJC-associated—or potentially other EJC components, mediate their association with the mRNA.
Multiple members of the SR and hnRNP protein families were found in purified mRNPs. Native mRNPs isolated after in vitro splicing contained both shuttling hnRNP proteins (A1, A2/B1, K, and L), which accompany the mRNA to the cytoplasm, and nonshuttling proteins (e.g., hnRNP C), which can retain mRNAs in the nucleus. The latter are typically removed via interactions with matrix-associated proteins and other cofactors (e.g., the nuclear pore complex, Lamins, TAP/p15), prior to mRNA export. SRp20 and 9G8 are shuttling SR-proteins that are not retained in the nucleus, but rather accompany the mRNP to the cytoplasm (Huang et al. 2003). In good agreement with previous studies (Dreyfuss et al. 2002 and references therein), the shuttling proteins hnRNP A1, A2/B1 and L, and SRp20 and 9G8 remain stably bound to the mRNA even in the presence of heparin (Table 1). SRp20 and 9G8 were shown to interact with RNAs independent of splicing (Cavaloc et al. 1999). However, most SR proteins were not detected in AdMLΔI-M3 mRNPs (with the exception of SRp20), suggesting that their recruitment is splicing-dependent.
Identification of novel mRNP-associated proteins
Several proteins previously not known to be mRNP-associated were detected in purified mRNPs (but not in mock affinity-selections), including the DEAD-box helicases DDX3, DDX5/p68, and DDX9. DDX3 is a nucleo-cytoplasmic shuttling protein that was recently implicated in the nuclear export of unspliced HIV RNAs (Yedavalli et al. 2004). It is a predominantly cytoplasmic protein, but can be detected by immunofluorescence in a speckle-like pattern in HeLa cell nuclei (Owsianka and Patel 1999) and also has been found in a mixture of affinity-purified human spliceosomes and mRNPs (Zhou et al. 2002) and spliceosomal B complexes (Deckert et al. 2006). DDX3 was also detected in mRNA transport granules in murine neurons (Kanai et al. 2004). DDX5/p68 is an essential splicing factor that facilitates unwinding of the U1 snRNA/5′ splice site base-pairing interaction (Liu 2002). Consistent with its role in splicing, it has been detected in numerous spliceosomal complexes, including A, B*, and C (Hartmuth et al. 2002; Zhou et al. 2002; Deckert et al. 2006). Interestingly, DDX3 is stably associated with mRNPs after heparin treatment, and DDX3 and DDX5 are exclusively found in spliced mRNAs carrying an EJC (Table 1), implicating EJC components in their association with the mRNA. DDX9 (RNA helicase A) has also been detected in a mixture of affinity-purified spliceosomes (Rappsilber et al. 2002; Zhou et al. 2002), but its association with mRNPs is nonetheless not dependent on splicing nor on the presence of an EJC (Table 1). It plays a role in CREB-dependent transcription, but has also been implicated in the nuclear export of unspliced viral RNAs (Li et al. 1999). Whether it (as well as DDX3) also contributes to mRNA export is presently not clear.
ELG, hNHN1, BCLAF1, and TRAP150 were also identified for the first time in affinity-purified, spliced mRNPs. All of these proteins have no known function in splicing or mRNA export. Essentially nothing is known about hNHN1 or ELG and with the exception of a CCCH zinc finger in hNHN1, they do not contain motifs suggestive of a particular function. Nonetheless, hNHN1 was identified in affinity-selected pre-catalytic spliceosomal B complexes, whereas the human ELG protein was first found in C complexes isolated in the absence of heparin (S. Bessonov, H. Urlaub, and R. Lührmann, unpubl.). Thus they associate during splicing prior to mRNA formation. Interestingly, the association of ELG and hNHN1 (but not BCLAF1 or TRAP150) with mRNA was dependent on splicing, but did not require EJC formation. Rather, as discussed below, their mRNP association was dependent on the presence of CBP80/CBP20.
BCLAF1 is a Bcl-2-associated protein and acts as a death-promoting transcriptional repressor during apoptosis (Kasof et al. 1999). The thyroid-hormone receptor associated protein 3 (TRAP150 or Thrap3) has been described as a member of the multiprotein complex TRAP/Mediator, involved in transcriptional activation of various genes (for review, see Malik and Roeder 2000). However, TRAP150 lacks known conserved domains or sequence motifs connected with splicing or transcription. While TRAP150 dissociates from mRNA under stringent conditions—like general RNA-binding proteins (e.g., YB-1, NFAR1)—BCLAF1, ELG, and hNHN1 are stable components of heparin-treated mRNPs. However, in previous studies (Tange et al. 2005), they were not detected by MS in MS2 affinity-purified mRNPs isolated in the presence of heparin, presumably due to the higher concentration of heparin used in the latter studies.
An EJC binding platform at position −20/24 of exon 1 is not required for EJC protein recruitment
With the exception of Pinin and Acinus, which are found in complex A and B, respectively, the canonical EJC proteins appear to associate with the pre-mRNA first after catalytic activation of the spliceosome (i.e., in spliceosomal C complexes) (Jurica et al. 2002; Reichert et al. 2002; S. Bessonov, H. Urlaub, and R. Lührmann, unpubl.). By isolating spliceosomal B*/C complexes assembled on a pre-mRNA with a truncated 5′ exon and analyzing their composition by Western blotting, we demonstrate that the EJC proteins Aly/REF, UAP56, Magoh, Y14, and RNPS1 do not require the EJC binding platform at the –20/24 region of the 5′ exon for their association with the pre-mRNA. As only a handful of EJC proteins were analyzed, we cannot rule out that one or more EJC proteins may associate with spliceosomes in a manner dependent on this region. Nonetheless, our data indicate that there is a differential requirement for the −20/24 region for initial recruitment of EJC proteins and formation of a stable EJC on spliced mRNA. Interestingly, previous studies revealed that at least nine proteins contact the 5′ exon in a highly dynamic manner during splicing (Reichert et al. 2002) At the time of exon ligation, a new set of proteins associates with position −24. Although Aly/REF has been shown to crosslink to the –20/24 region in the spliceosomal C complex (Reichert et al. 2002), the absence of this region does not significantly affect its association with catalytically active spliceosomes (Fig. 5). Nonetheless, Aly could play a role in providing positional information for EJC deposition and as nucleation factor for EJC assembly in the –20/24 region. It will be interesting in the future to determine which spliceosomal proteins are required for the recruitment of EJC proteins and whether EJC proteins are recruited as preorganized subcomplexes.
Influence of the cap binding complex on the association of proteins with mRNA
The m7G-cap plays an essential role in several steps of RNA metabolism, including pre-mRNA splicing, 3′ end formation, and RNA transport (for review, see Lewis and Izaurralde, 1997), as well as translation and NMD (Hosoda et al. 2005) It is tightly bound by CBP80 and CBP20, which form the so-called cap binding complex (CBC). The CBC has been shown to aid in the recruitment of factors during splicing, including facilitating the interaction of the U1 snRNP with the cap-proximal 5′ splice site (Lewis et al. 1996). The CBC also physically interacts with pre-mRNA 3′ end processing factors, thereby aiding 3′ end formation (Flaherty et al. 1997). Here we have tested whether the CBC aids the association of components of the spliced mRNP. Western blot analysis and targeted RNase H digestion revealed that an EJC is assembled on both CBC-bound and ΔCBC mRNPs (Figs. 4, 6). Thus, EJC formation during in vitro splicing in HeLa nuclear extracts occurs independent of the cap binding complex. During the course of this work, in vitro reconstitution experiments with recombinant Magoh, Y14, eIF4A3, and MLN51/BTZ revealed that the stable interaction of these core EJC components with RNA does not require the presence of an m7G cap, nor CBP80/20 (Ballut et al. 2005). The newly identified proteins ELG, hNHN1, and BCLAF1, on the other hand, and also ARS2, were not detected in purified mRNPs lacking a CBC (Table 2). Whether these proteins directly contact the CBC, or their association is indirectly stabilized by it, is presently not clear.
MATERIALS AND METHODS
Construction of plasmids and in vitro transcription of RNAs
pAdML-M3 has been described (Zhou et al. 2002), and intronless AdMLΔI-M3 and AdML15-M3 were generated by PCR-based techniques. pGlo-M3 was produced by overlap-extension of a PCR product of the β-globin pre-mRNA. All in vitro transcriptions were performed with T7 RNA polymerase from PCR-generated DNA templates.
In vitro splicing and MS2 affinity purification of mRNPs
Recombinant MBP-MS2 fusion protein was purified essentially as described (Jurica et al. 2002). HeLa nuclear extract was prepared according to Dignam et al. (1983), except initial cell lysis was performed in MC buffer (10 mM Hepes, pH 7.6; 10 mM KOAc; 0.5 mM MgOAc; 5 mM DTT; 1× Complete Protease Inhibitor). In vitro splicing reactions contained 40% HeLa nuclear extract in Buffer D (20 mM Hepes-KOH, pH 7.9; 10% glycerol; 100 mM KCl; 1.5 mM MgCl2; 0.5 mM DTT), supplemented with 1.5 mM ATP and 15 mM creatine phosphate. The final concentrations of MgCl2 and KCl were adjusted to 3.5 mM and 80 mM, respectively. For splicing analysis, RNA was recovered and separated on a 7 M urea-12% polyacrylamide gel and detected by autoradiography. For analysis of spliceosome assembly, AdML-M3 pre-mRNA was incubated under splicing conditions for 0–20 min, 2 μL of heparin (4 mg/mL) were added to 10 μL of the splicing reaction and complexes were analyzed on a 1.5% native agarose gel.
To isolate mRNPs, 15 pmol/mL of pre-mRNA were preincubated with a 30–50-fold molar excess of purified MBP-MS2 fusion protein for 30 min on ice. After incubation under splicing conditions for 75 min, three DNA oligonucleotides complementary to intronic sequences of either the AdML or β-globin pre-mRNA were added and the reaction mixture was incubated for 20 min at 30°C. In some instances heparin was added (final concentration 0.25 mg/mL) after 15 min of RNAse H digestion, and reactions were incubated at 30°C for 5 min. For preparative isolation of mRNPs, 700 μL of an in vitro splicing reaction were loaded onto a 4.4 mL linear 10%–30% glycerol gradient containing 20 mM Hepes-KOH 7.9, 150 mM NaCl, 1.5 mM MgCl2. Gradients were centrifuged at 55,000 rpm in a Sorvall TH-660 rotor and harvested manually in 175 μL fractions from the top. RNA and protein were extracted from 100 μL of odd-numbered fractions and analyzed as described below. Gradient fractions containing mRNP were pooled and passed twice over a 150 μL amylose column. After washing with 30 column volumes of MBP150 buffer (20 mM Hepes-KOH 7.9, 150 mM NaCl, 1.5 mM MgCl2), mRNPs were eluted with MBP150 buffer containing 12 mM maltose. Isolation of ΔCBC mRNPs was performed as above after adding 1 mM m7GpppG to the nuclear extract and preincubating for 10 min on ice. Detection of a stable EJC at position −20/24 was determined by RNase H digestion as previously described (Le Hir et al. 2000).
Purification of activated spliceosomal B*/C complexes
To isolate a mixture of B* and C spliceosomal complexes, splicing was performed in vitro with MINX pre-mRNA containing two intronic MS2-RNA aptamers and a 3′ splice site mutation (i.e., AG→GG) that abolishes the second step of splicing. Pre-mRNAs containing either a wild-type exon 1 (WT) or a truncated exon 1 (E1–15, 15 nt in length) were incubated under splicing conditions for 70 min and RNase H digestions were performed with oligos directed against nt –24 to +6 (WT) or nt –6 to +6 (E1–15) relative to the 5′ss to remove pre-mRNAs not engaged in C complexes (Jurica et al. 2002). Spliceosomes assembled on both substrates were purified by MS2 affinity selection as described above.
Antibodies and immunoblotting
Mouse monoclonal anti-MBP antibody was obtained from Biozol. Rabbit polyclonal peptide antibodies raised against peptides of U4/U6–61K, CBP80, hSnu23, Aly/REF, and UAP56 were affinity-purified on a SulfoLink column (Pierce) containing the cognate peptide. All other antisera were used without prior affinity purification. For immunoblotting, proteins extracted from purifed mRNP complexes were separated on a 12% polyacrylamide gel and transferred to nitrocellulose. Immunostaining was performed with an ECL detection kit (Amersham).
Mass spectrometry
MS was performed with SDS-PAGE separated proteins as previously described (Deckert et al. 2006) or alternatively, mRNPs were digested in-solution with RNAses A and T1 for 30 min at 37°C, followed by overnight digestion with trypsin, and peptides were analyzed in an LC-coupled ESI Q-ToF (Waters) and/or a linear iontrap (4000 QTrap, Applied Biosystems) mass spectrometer under improved conditions (S. Bessonov, H. Urlaub, and R. Lührmann, unpublished).
ACKNOWLEDGMENTS
We acknowledge the excellent technical assistance of M. Raabe and U. Plessmann with all MS techniques. Y14, Magoh and RNPS1 antibodies were a kind gift from E. Izaurralde. Antibodies against DDX3, NFAR-1, pThoc1, and MLN51 were kindly provided by A. Patel, S.-E. Behrens, D. Goodrich, and C. Tomasetto, respectively. We thank R. Reed, J. Tazi, and J. Vilardell for providing pAdML-M3, pGlo-M3S1, and pMBP-MS2, respectively, and P. Woolley for helpful discussions. This work was supported by grants from the DFG, the BMBF, Fonds der Chemischen Industrie, and the Ernst Jung Stiftung to R.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.336807.
REFERENCES
- Applequist, S.E., Selg, M., Raman, C., Jack, H.M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballut, L., Marchadier, B., Baguet, A., Tomasetto, C., Seraphin, B., Le Hir, H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Cavaloc, Y., Bourgeois, C.F., Kister, L., Stevenin, J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckert, J., Hartmuth, K., Boehringer, D., Behzadnia, N., Will, C.L., Kastner, B., Stark, H., Urlaub, H., Lührmann, R. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 2006;26:5528–5543. doi: 10.1128/MCB.00582-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degot, S., Le Hir, H., Alpy, F., Kedinger, V., Stoll, I., Wendling, C., Seraphin, B., Rio, M.C., Tomasetto, C. Association of the breast cancer protein MLN51 with the exon junction complex via its speckle localizer and RNA binding module. J. Biol. Chem. 2004;279:33702–33715. doi: 10.1074/jbc.M402754200. [DOI] [PubMed] [Google Scholar]
- Dignam, J.D., Lebovitz, R.M., Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie, J., Dreyfuss, G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr. Biol. 2002;12:1060–1067. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- Dreyfuss, G., Kim, V.N., Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Flaherty, S.M., Fortes, P., Izaurralde, E., Mattaj, I.W., Gilmartin, G.M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl. Acad. Sci. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, N.H., Kunz, J.B., Neu-Yilik, G., Breit, S., Viegas, M.H., Hentze, M.W., Kulozik, A.E. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol. Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Hachet, O., Ephrussi, A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., Urlaub, H., Vornlocher, H.P., Will, C.L., Gentzel, M., Wilm, M., Lührmann, R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda, N., Kim, Y.K., Lejeune, F., Maquat, L.E. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- Huang, Y., Gattoni, R., Stevenin, J., Steitz, J.A. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Jurica, M.S., Licklider, L.J., Gygi, S.R., Grigorieff, N., Moore, M.J. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA. 2002;8:426–439. doi: 10.1017/s1355838202021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai, Y., Dohmae, N., Hirokawa, N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kasof, G.M., Goyal, L., White, E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 1999;19:4390–4404. doi: 10.1128/mcb.19.6.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka, N., Dreyfuss, G. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon–exon junction complex. J. Biol. Chem. 2004;279:7009–7013. doi: 10.1074/jbc.M307692200. [DOI] [PubMed] [Google Scholar]
- Lau, C.K., Diem, M.D., Dreyfuss, G., Van Duyne, G.D. Structure of the Y14-Magoh core of the exon junction complex. Curr. Biol. 2003;13:933–941. doi: 10.1016/s0960-9822(03)00328-2. [DOI] [PubMed] [Google Scholar]
- Le Hir, H., Izaurralde, E., Maquat, L.E., Moore, M.J. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir, H., Gatfield, D., Izaurralde, E., Moore, M.J. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.D., Izaurralde, E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Lewis, J.D., Izaurralde, E., Jarmolowski, A., McGuigan, C., Mattaj, I.W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes & Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- Li, J., Tang, H., Mullen, T.M., Westberg, C., Reddy, T.R., Rose, D.W., Wong-Staal, F. A role for RNA helicase A in post-transcriptional regulation of HIV type 1. Proc. Natl. Acad. Sci. 1999;96:709–714. doi: 10.1073/pnas.96.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Lin, R.I., Lai, M.C., Ouyang, P., Tarn, W.Y. Nuclear Pnn/DRS protein binds to spliced mRNPs and participates in mRNA processing and export via interaction with RNPS1. Mol. Cell. Biol. 2003;23:7363–7376. doi: 10.1128/MCB.23.20.7363-7376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z.R. p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol. Cell. Biol. 2002;22:5443–5450. doi: 10.1128/MCB.22.15.5443-5450.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M.L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M., Reed, R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., Lührmann, R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- Malik, S., Roeder, R.G. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Masuda, S., Das, R., Cheng, H., Hurt, E., Dorman, N., Reed, R. Recruitment of the human TREX complex to mRNA during splicing. Genes & Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., Badolato, J., Kobayashi, R., Zhang, M.Q., Gardiner, E.M., Krainer, A.R. Purification and characterization of human RNPS1: A general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nott, A., Le Hir, H., Moore, M.J. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owsianka, A.M., Patel, A.H. Hepatitis C virus core protein interacts with a human DEAD box protein DDX3. Virology. 1999;257:330–340. doi: 10.1006/viro.1999.9659. [DOI] [PubMed] [Google Scholar]
- Rappsilber, J., Ryder, U., Lamond, A.I., Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel, J., Herold, A., Gari, K., Kocher, T., Rode, M., Ciccarelli, F.L., Wilm, M., Izaurralde, E. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster . Nat. Struct. Mol. Biol. 2004;11:558–566. doi: 10.1038/nsmb759. [DOI] [PubMed] [Google Scholar]
- Reichert, V.L., Le Hir, H., Jurica, M.S., Moore, M.J. 5′ Exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes & Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe, M.E., Tange, T.O., Thomas, D.R., Moore, M.J., Grigorieff, N. The three-dimensional architecture of the EJC core. J. Mol. Biol. 2006;360:743–749. doi: 10.1016/j.jmb.2006.05.049. [DOI] [PubMed] [Google Scholar]
- Tange, T.O., Nott, A., Moore, M.J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tange, T.O., Shibuya, T., Jurica, M.S., Moore, M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Lou, P.J., Leu, S., Ouyang, P. Modulation of alternative pre-mRNA splicing in vivo by pinin. Biochem. Biophys. Res. Commun. 2002;294:448–455. doi: 10.1016/S0006-291X(02)00495-3. [DOI] [PubMed] [Google Scholar]
- Wiegand, H.L., Lu, S., Cullen, B.R. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. Sci. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will, C.L., Lührmann, R. Spliceosome structure and function. In: Cech T.R., Atkins J.F., editors. The RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; NY: 2006. pp. 369–400. [Google Scholar]
- Yedavalli, V.S., Neuveut, C., Chi, Y.H., Kleiman, L., Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., Licklider, L.J., Gygi, S.P., Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]