Abstract
Rpp20 and Rpp25 are subunits of the human RNase MRP and RNase P endoribonucleases belonging to the Alba superfamily of nucleic acid binding proteins. These proteins, which bind very strongly to each other, transiently associate with RNase MRP. Here, we show that the Rpp20-Rpp25 heterodimer is resistant to both high concentrations of salt and a nonionic detergent. The interaction of Rpp20 and Rpp25 with the P3 domain of the RNase MRP RNA appeared to be strongly enhanced by their heterodimerization. Coimmunoprecipitation experiments demonstrated that only a single copy of each of these proteins is associated with the RNase MRP and RNase P particles in HEp-2 cells. Both proteins accumulate in the nucleoli, which in case of Rpp20 is strongly dependent on its interaction with Rpp25. Finally, the results of overexpression and knock-down experiments indicate that their expression levels are codependent. Taken together, these data indicate that the Rpp20-Rpp25 heterodimerization regulates their RNA-binding activity, subcellular localization, and expression, which suggests that their interaction is also crucial for their role in RNase MRP/P function.
Keywords: heterodimer, RNA processing, RNase MRP, RNase P, snoRNP
INTRODUCTION
The RNase MRP complex is a small ribonucleoprotein particle composed of an essential RNA subunit (normally ∼300 nucleotides) (Sbisa et al. 1996; Piccinelli et al. 2005) and ∼10 protein subunits. This macromolecular complex and its constituents have been identified in several vertebrates, invertebrates, plants, and lower eukaryotes (Ziehler et al. 2001), but not in eubacteria or archaebacteria (Zhu et al. 2006). RNase P, a structurally and functionally related ribonucleoprotein complex, has been reported to occur in all three domains of life (Walker and Engelke 2006). Both complexes catalyze essential cellular endoribonucleolytic RNA processing events that are directly or indirectly linked to the cellular translation machinery. RNase P is an essential factor in tRNA maturation as it removes the 5′ leader sequence of pre-tRNA (Jarrous 2002). RNase MRP is involved in the processing of pre-rRNA, more specifically the generation of the 5′ end of 5.8S rRNA by cleaving the internal transcribed spacer 1 (ITS1) (Schmitt and Clayton 1993; Lygerou et al. 1996). Additional substrates for RNase MRP have been identified in both mammals and yeast. In human, murine, and bovine cells, RNase MRP has been reported to function in mitochondrial DNA replication by its involvement in the endoribonucleolytic generation of an RNA species that is required for the priming of this process (Chang and Clayton 1987). Recently, a role for RNase MRP in yeast cell cycle regulation was reported by Schmitt and collaborators, who showed that RNase MRP plays a role in the degradation of the mRNA encoding the mitosis specific cyclin Clb2 (Gill et al. 2004).
The RNA subunit of human RNase MRP, which has been shown to be essential for its enzymatic activity in a yeast system (Schmitt and Clayton 1992), can adopt a typical structure that is also found or predicted for the RNase MRP and RNase P RNAs from other species (Reddy and Shimba 1995; Frank et al. 2000; Walker et al. 2005; Walker and Avis 2005). Because of the structural similarities of the RNA subunits, an evolutionary relation between RNase MRP and RNase P was proposed (Reddy and Shimba 1995; Collins et al. 2000; Zhu et al. 2006). This is further supported by the fact that both complexes share several protein subunits (Walker and Engelke 2006).
Distinct structural elements of the RNase MRP and RNase P RNAs have been demonstrated to be required for protein subunit interactions (Pluk et al. 1999; van Eenennaam et al. 2002; Welting et al. 2004) and to be involved in particle assembly (Li and Altman 2002), subcellular localization (Jacobson et al. 1995, 1997), and enzymatic activity (Li et al. 2004). The P3 domain of RNase MRP RNA not only is a structural moiety where several protein subunits interact with the RNA (Yuan et al. 1991; Pluk et al. 1999) but also is important for the accumulation of the RNase MRP RNA in the nucleoli, where pre-rRNA processing occurs (Jacobson et al. 1995). The human P3 domain has also been reported to be involved in the recognition of RNase MRP by so-called anti-Th/To autoantibodies found in the serum of systemic sclerosis, polymyositis, Raynaud's phenomenon, and systemic lupus erythematosus patients (Welting et al. 2003). Although originally a 40-kDa protein subunit was believed to contain the major Th/To determinants, we showed that the 25- and 20-kDa protein subunits Rpp25 and Rpp20, previously referred to as MRP25 and MRP20, represent the subunits most frequently targeted by such autoimmune sera (Pluk et al. 1999; van Eenennaam et al. 2002). Rpp25 and Rpp20 both interact with the P3 domain of the RNase MRP RNA and are also associated with human RNase P, which explains why most anti-Th/To-positive patient sera coprecipitate RNase MRP and RNase P (Liu et al. 1994; Jarrous et al. 1998; van Eenennaam et al. 2002; Guerrier-Takada et al. 2002). Recently, we showed that the recombinant Rpp20 and Rpp25 proteins very efficiently bind to each other in vitro (Welting et al. 2004), suggesting that they may associate as a heterodimer with the RNase MRP and RNase P complexes. Rpp20 was reported to exhibit ATPase activity and to interact with the proteins SMN, Hsp27 and KIAA0065 (Jiang and Altman 2001; Hua and Zhou 2004). Interestingly, in contrast to the other RNase MRP protein subunits, both Rpp20 and Rpp25 do not sediment at 60–80S in glycerol gradients and thus might dissociate from RNase MRP before or during its association with pre-rRNA containing particles, which sediment at this region of the gradients (Welting et al. 2006). Moreover, both Rpp20 and Rpp25 share sequence homology with archaeal proteins from the Alba family (Aravind et al. 2003), further supporting a putative functional relationship between Rpp20 and Rpp25. To obtain more insight into the role of the Rpp20 and Rpp25 proteins in the function of RNase MRP and RNase P, we studied their dimerization in more detail, their association with RNase MRP/RNase P particles, their subcellular localization, and the interdependence of their expression levels.
RESULTS
Rpp20-Rpp25 heterodimerization
To further characterize the strong interaction between Rpp20 and Rpp25 (Welting et al. 2004), bacterially expressed GST-Rpp25 was incubated with in vitro translated, radiolabeled Rpp20 and precipitated with glutathione-Sepharose. In agreement with previous observations, Rpp20 was almost quantitatively coprecipitated with GST-Rpp25, whereas no detectable coprecipitation was observed with the control proteins GST and GST-Rrp41p (Rrp41p is a subunit of the exosome complex) (Fig. 1A). Note that in vitro translated Rpp20 migrates as a doublet, which is most likely due to translation initiation at both the start codon at the 5′ end of the VSV-tag encoding sequence and at the start codon of the Rpp20 cDNA sequence. Similar results were observed for the reciprocal pull-down assay with GST-Rpp20 and in vitro translated Rpp25 (data not shown). In the GST pull-down assay, the GST-tagged recombinant proteins were used in a large excess (∼1000-fold) in comparison with the in vitro translated proteins. To investigate whether similar interaction efficiencies are observed when both interacting proteins are used at relatively low and approximately equal concentrations, in vitro translated Rpp20 and Rpp25 were incubated and immunoprecipitated with polyclonal rabbit sera against Rpp20 or Rpp25. The specificity of these antibodies was substantiated by the specific precipitation of the cognate in vitro translated proteins (Fig. 1B, lanes 1–3, 5–7). When both proteins were mixed prior to the immunoprecipitations, Rpp25 was efficiently coprecipitated with the anti-Rpp20 antibodies and vice versa (Fig. 1B, lanes 4,8, respectively). When the number of radiolabeled residues per molecule is taken into account (three for VSV-Rpp20 and five for VSV-Rpp25), it can be concluded that about the same number of Rpp20 and Rpp25 molecules is precipitated by both antibodies. Thus also under these conditions, Rpp20 and Rpp25 efficiently interact with each other.
FIGURE 1.
Rpp20-Rpp25 heterodimerization. Rpp20 and Rpp25 were translated in vitro using a rabbit reticulocyte lysate system. The 35S-labeled proteins were separated by SDS-PAGE and visualized by autoradiography. (A) In vitro translated Rpp20 was incubated with GST-Rpp25 and GST and GST-Rrp41p as controls. GST-tagged and associated proteins were pulled down by glutathione-Sepharose. (Lane 1) In vitro translated Rpp20 (input, 100% of the amount incubated with the GST-tagged proteins). (Lanes 2–4) Radiolabeled protein bound to GST, GST-Rrp41p, and GST-Rpp25, respectively. (B ) In vitro translated Rpp20, Rpp25, or a mixture of these proteins was immunoprecipitated with polyclonal anti-Rpp20 and anti-Rpp25 rabbit sera. (Lanes 1,5) Rpp20 and Rpp25, 10% input. (Lanes 2,3) Immunoprecipitates of Rpp20 with anti-Rpp20 and anti-Rpp25 antibodies, respectively. (Lanes 6,7) Immunoprecipitates of Rpp25 with anti-Rpp25 and anti-Rpp20 antibodies, respectively. (Lanes 4,8) Immunoprecipitates of a Rpp20/Rpp25 mixture with anti-Rpp20 and anti-Rpp25 antibodies, respectively. (C) GST-Rpp25 and GST as a control were incubated with in vitro translated Rpp20 under standard conditions (100 mM KCl, no Triton X-100, lanes 2,3) and in the presence of increasing KCl (upper panel) or Triton X-100 (lower panel) concentrations. (Lane 1) One hundred percent of input material. (Lanes 2–13) GST (even lanes) and GST-Rpp25 (odd lanes) bound material precipitated under the conditions indicated above the lanes. (D) GST pull-down analysis of in vitro translated Rpp25 with equimolar amounts of GST-Rpp20 and GST-Rpp20 amino acid substitution mutants. (Lane 1) In vitro translated Rpp25 (input). (Lanes 2–6) Material bound to GST, GST-Rpp20, GST-Rpp20D36E, GST-Rpp20N40Q, and GST-Rpp20G59S, respectively.
To gain more insight into the nature of the Rpp20-Rpp25 interaction, GST pull-down experiments were performed in the presence of increasing concentrations of salt or a nonionic detergent to interfere with hydrophilic and hydrophobic bonds, respectively. The results in Figure 1C show that the interaction between Rpp20 and Rpp25 is resistant to both high salt (up to 4 M KCl) and high detergent (up to 20% Triton X-100) concentrations. A role for intermolecular disulfide bridge formation was excluded by the addition of increasing concentrations (up to 700 mM) of β-mercaptoethanol (data not shown). Taken together, these data substantiate the very strong, noncovalent interaction between Rpp20 and Rpp25 and suggest that this heterodimerization is not mainly determined by either ionic or hydrophobic bonds.
Rpp20 was previously reported to exhibit ATPase and GTPase activity, and substitution of amino acids in regions that show homology with the ABC ATPase signature motif or the DIxxN sequence element, which is homologous to the ATPase motif of the Upf1p subfamily of DEAD box RNA helicases, led to the inhibition of ATP hydrolysis by recombinant, bacterially expressed Rpp20 (Li and Altman 2001). Although the presence or absence of ATP did not affect the interaction between Rpp20 and Rpp25 (data not shown), we investigated whether the Rpp20 mutations in the ABC ATPase signature motif and DIxxN element interfere with Rpp20-Rpp25 dimerization. Three different Rpp20 substitution mutants were expressed as GST-fusion proteins, and equal amounts of GST-Rpp20, GST-Rpp20D36E, GST-Rpp20N40Q, and GST-Rpp20G59S were used in a GST pull-down assay with in vitro translated Rpp25 in the absence of ATP (Fig. 1D). As observed before, wild-type GST-Rpp20 efficiently interacted with Rpp25 (lane 3). Similar levels of Rpp25 coprecipitation were observed with mutants Rpp20D36E and Rpp20G59S. In contrast, substitution of asparagine-40 by a glutamine (Rpp20N40Q) strongly diminished the interaction with Rpp25 (lane 5). Similar results were obtained when ATP was included in this experiment. These results show that asparagine-40 of Rpp20 plays a crucial role in the interaction with Rpp25. The efficient interactions between the ATPase-deficient Rpp20 mutants D36E and G59S (Li and Altman 2001) and Rpp25 substantiate that heterodimerization of these two proteins is independent of ATP.
The Rpp20-Rpp25 heterodimer efficiently interacts with the P3 domain of human RNase MRP RNA
Previously, UV-crosslinking and GST pull-down analyses showed that both Rpp20 and Rpp25 interact with or are in close proximity to the RNA subunit of human RNase MRP, in particular the P3 domain of this RNA molecule (Yuan et al. 1991; Pluk et al. 1999; Welting et al. 2004). To investigate the binding of Rpp20, Rpp25 and/or the heterodimer of these proteins to the P3 RNA, streptavidin pull-down experiments were performed using in vitro transcribed, biotinylated P3 RNA and radiolabeled, in vitro translated Rpp20 and Rpp25. Rpp20 alone did not detectably interact with the P3 RNA, whereas Rpp25 was reproducibly coprecipitated, albeit at relatively low levels (Fig. 2A). The coprecipitation of Rpp25 was specific, because in the absence of the biotinylated P3 RNA no coprecipitation was observed. The efficient heterodimerization of in vitro translated Rpp20 and Rpp25 (Fig. 1B) allowed us to investigate the interaction of the dimer with the P3 RNA. The results in Figure 2A show that this dimer indeed efficiently associated with the P3 domain. Significant amounts of Rpp20 were now coprecipitated with the P3 RNA, and the efficiency by which Rpp25 interacted with P3 RNA was strongly enhanced (note that equal amounts of Rpp25 were added to the incubations in the absence and in the presence of Rpp20). These results not only indicate that, in contrast to Rpp20, Rpp25 is able to interact directly and independently with the P3 domain but also demonstrate that the Rpp20-Rpp25 heterodimer binds more strongly to the P3 domain than Rpp25 alone.
FIGURE 2.
Interaction of Rpp20 and Rpp25 with the P3 domain of human RNase MRP RNA. Biotin-labeled P3 RNA was incubated with in vitro translated Rpp20 and/or Rpp25. Subsequently, P3 RNA-bound material was isolated by precipitation with streptavidin-agarose and analyzed by SDS-PAGE and autoradiography. (A, lanes 1,2) In vitro translated Rpp20 and Rpp25, respectively (100% of the amount used for P3 binding). (Lane 3) Control without P3 RNA. (Lanes 4,5) Precipitated material after incubation with only Rpp20 or only Rpp25. (Lane 6) Precipitated material after incubation with a mixture of Rpp20 and Rpp25. (Lanes 7–11) Precipitated Rpp25 after incubation with P3 RNA in the presence of purified, bacterially expressed GST-Rpp20, GST-Rpp20D36E, GST-Rpp20N40Q, GST-Rpp20G59S, and GST, respectively. (B) Binding of in vitro translated Rpp20 and Rpp25 to biotinylated P3 RNA after incubations with decreasing amounts of Rpp25 (upper panel) or Rpp20 (lower panel) and constant amounts of Rpp20 (upper panel) or Rpp25 (lower panel). (Lanes 1–5) One hundred percent of (diluted) input material (threefold dilution series of the respective proteins in lanes 2–5). (Lanes 6–9) Precipitated material of mixtures of Rpp20 and Rpp25 corresponding to the amounts in lane 1 in combination with that in lanes 2–5.
To substantiate the importance of Rpp20-Rpp25 dimerization for efficient interactions with the P3 domain, the effect of the Rpp20 amino acid substitution mutations was studied using the GST-Rpp20 mutants in the streptavidin pull-down assay. Indeed, the presence of mutant Rpp20N40Q, which did not efficiently dimerize with Rpp25 (see Fig. 1D), did not lead to an enhanced interaction between Rpp25 and the P3 domain (Fig. 2A, lane 9). In contrast, the Rpp20 mutations that did not affect dimerization, resulted in wild-type levels of Rpp25–P3 RNA interaction (lanes 7,8,10). Thus, the dimerization of Rpp20 and Rpp25 is necessary for the efficient interaction of these proteins with the P3 domain of human RNase MRP RNA.
To further investigate the importance of Rpp20-Rpp25 heterodimerization for their interaction with the P3 domain, a pull down experiment was performed with decreasing amounts of either Rpp20 or Rpp25. Decreasing the amount of Rpp25 in the presence of a constant amount of Rpp20 led to a concomitant decrease in the coprecipitation of Rpp20 (Fig. 2B, upper panel). Similarly, decreasing the amount of Rpp20 in the presence of a constant amount of Rpp25 gradually decreased the coprecipitation of Rpp25 (Fig. 2B, lower panel). These results are fully consistent with the binding of Rpp20 and Rpp25 as a dimer to the P3 domain of RNase MRP RNA.
Stoichiometry of Rpp20 and Rpp25 interaction with RNase MRP/RNase P
The results of the experiments described above indicated that Rpp20 and Rpp25 bind in a one-to-one ratio to the P3 domain of RNase MRP RNA in vitro, which may reflect the situation in the RNase MRP/RNase P particles. To investigate the stoichiometry of the Rpp20 and Rpp25 association with RNase MRP and RNase P particles, we transfected HEp-2 cells with constructs encoding VSV-tagged versions of both proteins or an “empty” VSV-tag vector as a control. Subsequently, extracts from cells expressing the tagged proteins were subjected to immunoprecipitation with anti-VSV-tag antibodies. The immunoprecipitated material was separated by SDS-PAGE and analyzed by Western blotting. Staining with anti-Rpp20 and anti-Rpp25 antisera showed that the expression levels of VSV-Rpp20 and VSV-Rpp25 were similar to those of the corresponding endogeneous proteins (Fig. 3, input lanes). The VSV-tags of Rpp20 and Rpp25 did not interfere with their incorporation in ribonucleoprotein complexes, as evidenced by the coprecipitation of endogenous Rpp30 and Rpp40 (Fig. 3) with anti-VSV-tag antibodies, and by the coprecipitation of the RNase MRP and RNase P RNAs (Welting et al. 2006; data not shown). The lack of coprecipitation of the endogeneous Rpp20 and Rpp25 proteins with their respective VSV-tagged counterparts indicated that in these cells the RNase MRP and RNase P ribonucleoprotein complexes contain only a single copy of these proteins.
FIGURE 3.
Stoichiometry of Rpp20 and Rpp25 binding to the RNase MRP and RNase P complexes. HEp-2 cells were transiently transfected with constructs encoding VSV-tagged variants of both Rpp20 and Rpp25. As a control, the “empty” VSV-tag vector was used in parallel. Cell lysates were subjected to immunoprecipitation with anti-VSV-tag antibodies, and immunoprecipitated material was analyzed by SDS-PAGE and Western blotting using polyclonal antisera against Rpp30, Rpp40, Rpp20, and Rpp25 as indicated on the left of each panel. Two percent of the input material was loaded in lanes 1–3, and the immunoprecipitated material was loaded in lanes 4–6. (Lanes 1,4) Lysate from cells transfected with the control vector; (lanes 2,5) lysate from cells expressing VSV-Rpp20; (lanes 3,6) lysate from cells expressing VSV-Rpp25. Note that the VSV-tagged proteins have a somewhat higher molecular weight than their endogenous counterparts.
Subcellular distribution of Rpp20 and Rpp25
The results described above raised the question whether heterodimerization of Rpp20 and Rpp25 is important for their proper subcellular localization. To be able to study the Rpp20 localization by immunofluorescence, a newly developed mouse monoclonal antibody (mAb) against Rpp20 (1F11) was characterized. mAb 1F11 is specific for Rpp20 and is reactive with Rpp20 in both immunoblotting and immunoprecipitation (Supp. Fig. 1; supplemental material can be found at http://www.modiquest.com or can be requested by sending an e-mail message to G.Pruijn@ncmls.ru.nl.).
Immunofluorescent staining of HEp-2 cells with 1F11 showed that the highest concentrations of Rpp20 are found in the nucleoli (Fig. 4A,B). The subcellular localization of Rpp25 was visualized by the polyclonal anti-Rpp25 antibodies. As can be seen in Figure 4, C and D, also Rpp25 accumulates in the nucleoli. Both Rpp20 and Rpp25 seemed to be enriched in specific subnucleolar compartments. To investigate whether these subnucleolar staining patterns are identical for both proteins, which would be expected based upon their efficient interaction, cells were stained with both antibodies in combination with Alexa Fluor 488 and Alexa Fluor 555 conjugated secondary antibodies. The overlay of both staining patterns obtained by confocal laser scanning microscopy demonstrated that Rpp20 and Rpp25 almost completely colocalize in these cells (Fig. 4E–H). To investigate whether the subnucleolar regions showing the strongest staining with anti-Rpp20 and anti-Rpp25 antibodies corresponded to either the dense fibrillar component (DFC) or the granular component (GC) of the nucleoli, double immunofluorescence experiments were performed with antibodies to fibrillarin as a marker for the DFC and with antibodies to nucleophosmin/B23 as a marker for the GC. The results (Fig. 4I–P) showed that the regions containing the highest concentrations of Rpp20 and Rpp25 correspond to the GC.
FIGURE 4.
Subcellular localization of Rpp20 and Rpp25. Immunofluorescence staining of HEp-2 cells with antibodies to Rpp20 (B; mAb 1F11) and Rpp25 (D; polyclonal rabbit serum). A and C show the phase contrast images corresponding to B and D, respectively. The bars in A and C correspond to 10 μm. (E–H) Double immunofluorescence staining of Rpp20 (E, G, H) and Rpp25 (F, G, H). (I–L) Double immunofluorescence staining of Rpp25 (I, K, L) and fibrillarin (J, K, L). (M–P) Double immunofluorescence staining of Rpp25 (M, O, P) and nucleophosmin/B23 (N–P). The images in panels E–P were recorded by confocal microscopy. Bars in panels E–P correspond to 1 μm.
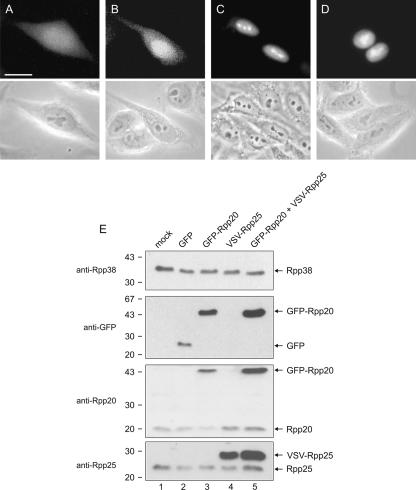
The specific (sub)nucleolar localization of Rpp20 does not reflect the previously reported subcellular localization of Rpp20 fused to three Flag-tags overexpressed in transfected HeLa cells (Hua and Zhou 2004). In that study the overexpressed Rpp20 protein diffusely distributed throughout the cytoplasm and nucleus, with higher concentrations in the nucleus. Since this might be due to overexpression of the human Rpp20 protein, we generated an expression construct encoding a GFP-Rpp20 fusion protein to study whether overexpression of the Rpp20 fusion protein results in a different subcellular distribution pattern. Indeed, in transiently transfected HEp-2 cells, GFP-Rpp20 diffusely distributed throughout the cell with a somewhat higher concentration in the nucleus, very similar to the previously reported localization of overexpressed Rpp20 (Fig. 5B). The expression level of GFP-Rpp20 was determined by Western blotting using both anti-GFP and anti-Rpp20 antibodies. These results confirmed that the GFP-Rpp20 fusion protein, migrating at 45 kDa in agreement with its predicted molecular mass, was indeed overexpressed in comparison with the endogenous Rpp20 protein (Fig. 5E). Importantly, it should be noted that only a subset of the cells will express the GFP-tagged protein. Similarly, the localization of overexpressed Rpp25 was determined using a construct encoding a VSV-tagged Rpp25 protein. In contrast to Rpp20, VSV-Rpp25, which like GFP-Rpp20 was expressed to higher levels than its endogenous counterpart, showed the strongest staining in the nucleoli (Fig. 5C). In addition, a diffuse nucleoplasmic staining was observed for VSV-Rpp25. Because the levels of endogenous Rpp25 were not detectably affected in the cells overexpressing GFP-Rpp20 (Fig. 5E), the differences between the distributions of the endogenous and overexpressed Rpp20 might be due to the limiting amounts of Rpp25, which do not allow efficient heterodimerization of the overexpressed Rpp20 protein. To investigate this possibility, Rpp20 and Rpp25 were simultaneously expressed to higher levels in the same cells. HEp-2 cells were cotransfected with constructs encoding GFP-Rpp20 and VSV-Rpp25, and the localization of GFP-Rpp20 was studied by fluorescence microscopy. The high expression level of VSV-Rpp25 in GFP-Rpp20 expressing cells, which was confirmed by Western blotting (Fig. 5E), indeed affected the subcellular distribution of GFP-Rpp20. In these cells not only did GFP-Rpp20 accumulate in the nuclei, but an enrichment in nucleolus-like regions was observed as well (Fig. 5D). These results strongly suggest that the interaction with Rpp25 is required for the efficient nuclear entry of Rpp20 as well as for its accumulation in the nucleoli.
FIGURE 5.
Overexpression of Rpp20 and Rpp25. HEp-2 cells were transfected with constructs encoding GFP (A), GFP-Rpp20 (B), GFP-Rpp25 (C), or both GFP-Rpp20 and VSV-Rpp25 (D) and after 24 h the localization of GFP(-tagged) proteins was visualized by fluorescence microscopy (upper panels). The corresponding phase contrast images are shown in the lower panels. The bar in A corresponds to 10 μm. (E ) The expression of GFP-Rpp20 and VSV-Rpp25 in the transfected cells was analyzed by Western blotting using antibodies to Rpp38, GFP, Rpp20, and Rpp25 (from top to bottom). (Lane 1) Extract from mock-transfected cells; (lanes 2–4) extracts from cells transfected with GFP, GFP-Rpp20, and VSV-Rpp25, respectively; (lane 5) extract from cells cotransfected with GFP-Rpp20 and VSV-Rpp25. The positions of molecular mass markers is indicated on the left (kDa), and the positions of the endogenous and ectopically expressed proteins are marked by arrows on the right.
Rpp20 and Rpp25 expression levels are dependent on each other
The observation that heterodimerization of Rpp20 and Rpp25 is important for their correct subcellular distribution raised the question whether their expression levels are interdependent. The results in Figure 5E already suggested that coexpression of VSV-Rpp25 led to increased GFP-Rpp20 expression levels and vice versa. This may be explained by stabilization of these proteins by their heterodimerization, thereby decreasing their turnover rates. To further investigate this issue, the expression levels of the endogenous Rpp20 and Rpp25 proteins were knocked-down by RNA interference. Transfection of HEp-2 cells with siRNAs specific for the Rpp20 and Rpp25 mRNAs resulted in efficient knock-down of the target proteins within 48 h. Re-transfection of these cells at 48 h with the same siRNAs resulted in nearly complete knock-down of both proteins, as demonstrated by Western blotting of cell lysates obtained after 96 h (Fig. 6). Strikingly, an almost equally efficient codepletion of Rpp25 with the siRNA for Rpp20 and of Rpp20 with the siRNA for Rpp25 was observed. In contrast, the expression levels of other RNase MRP/RNase P proteins, Rpp30, Rpp38, and Rpp40, and of an unrelated protein, Rrp4p were not detectably affected by Rpp20 or Rpp25 knock-down. These results indeed indicate that the expression levels of Rpp20 and Rpp25 are codependent.
FIGURE 6.
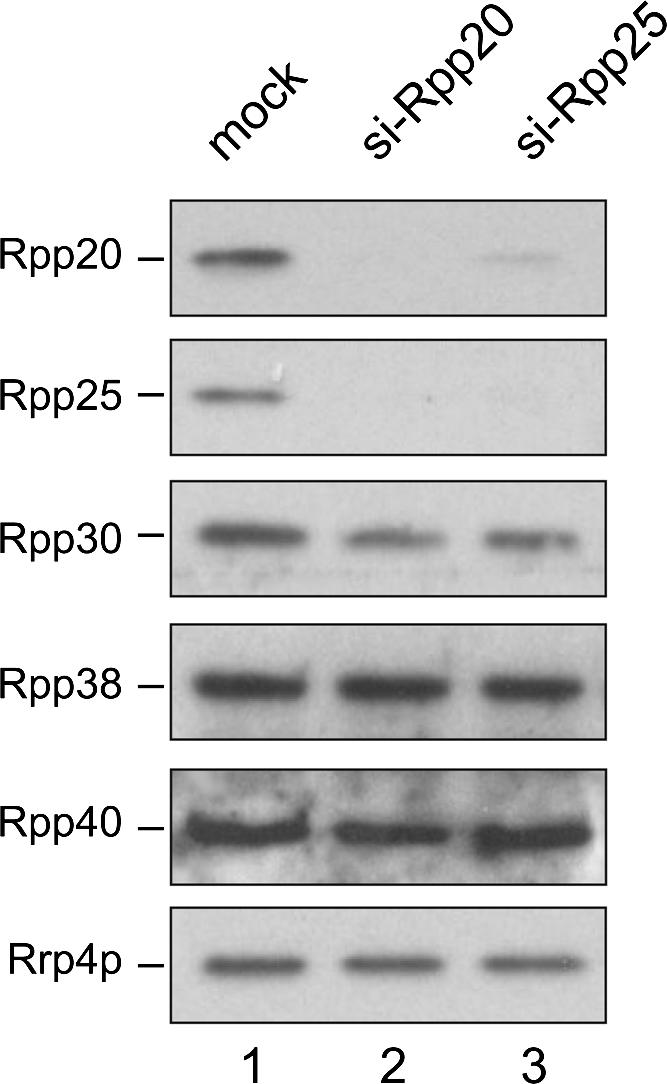
Down-regulation of Rpp20 and Rpp25 expression by RNAi. The expression of either Rpp20 or Rpp25 was inhibited by transfection of HEp-2 cells with corresponding siRNAs. After a second transfection with the same siRNAs at 48 h, cells were lysed at 96 h and the lysates analyzed by Western blotting. An extract from mock transfected cells was generated in parallel as a control. Rpp20 was detected using mAb 1F11, and Rpp25 was detected with a polyclonal rabbit serum against Rpp25. The expression of other RNase MRP/RNase P subunits (Rpp30, Rpp38, and Rpp40) and of an exosome subunit (Rrp4p) in these cells was analyzed using polyclonal rabbit sera specific for each of these proteins.
DISCUSSION
In this study we present evidence that the heterodimerization of Rpp20 and Rpp25 is not only extremely stable but also important for various aspects of their biological function as subunits of RNase MRP and RNase P. The intimate relationship between these proteins is further substantiated by the fact that their expression levels seem to be highly codependent.
The Rpp20-Rpp25 heterodimer is associated with both RNase MRP and RNase P. Recent evidence suggests that the association with RNase MRP is dynamic, because these proteins were not found in 60–80S complexes isolated from HEp-2 cells, in contrast to other RNase MRP subunits. Because these high-molecular-weight complexes most likely represent preribosomal complexes, we proposed that the Rpp20-Rpp25 dimer dissociates from RNase MRP either before or during its assembly with preribosomes and reassociates after the RNase MRP catalyzed processing event of pre-rRNA (Welting et al. 2006). Currently, it is unknown whether a similar transient dissociation of the Rpp20-Rpp25 dimer from RNase P complex may occur during pre-tRNA processing by this enzyme.
The association of Rpp25 with the human RNase MRP and RNase P complexes is most likely mediated by a combination of protein–RNA and protein–protein interactions. In agreement with its RNA-binding capacity, Rpp25 directly contacts the P3 domain of the RNase MRP and RNase P RNAs (Pluk et al. 1999; Guerrier-Takada et al. 2002; van Eenennaam et al. 2002; Welting et al. 2004) and has been shown to bind to several protein subunits (Welting et al. 2004). Yuan and coworkers showed that the Rpp25 protein most likely interacts with the distal stem-loop of the P3 domain (Yuan et al. 1991). Also for Rpp20 direct contacts with the P3 domain have been reported (Pluk et al. 1999), but in addition, Rpp20 binds only to Rpp25 and none of the other protein subunits in vitro (Welting et al. 2004). Taken together, these data strongly suggest that the association of the Rpp20-Rpp25 heterodimer with the RNase MRP and RNase P RNAs is mediated by multiple interactions, most of which involve Rpp25. Although the association of Rpp20 with RNase MRP seems to be mainly mediated by Rpp25, additional protein–protein interactions between Rpp20 and other subunits may contribute to the association of Rpp20, which is supported by the results of yeast two-hybrid experiments (Jiang and Altman 2001). The results in Figure 2 indicate that the binding affinity of Rpp25 for the P3 domain is much higher than that of Rpp20. In agreement with these observations, Guerrier-Takada and coworkers (2002) showed that Rpp20 is not able to interact directly with the P3 domain of RNase P RNA in vitro, and Jiang and colleagues did not detect a direct interaction of Rpp20 with full-length RNase P RNA in a yeast-three-hybrid screening (Jiang et al. 2001). Nevertheless, Rpp20 is predicted to contribute to the binding of the Rpp20-Rpp25 dimer to the P3 domain, because (1) close contacts have been detected by UV-crosslinking (Pluk et al. 1999; van Eenennaam et al. 2002), and (2) Rpp20 enhances the efficiency of Rpp25 binding to the P3 domain. Alternatively, the enhanced binding efficiencies may be explained by conformational changes in the proteins induced by their heterodimerization or by structural changes in the P3 RNA resulting from their binding.
Attempts to obtain more insight in the mode of the interaction between Rpp20 and Rpp25 failed, because under conditions where either ionic or hydrophobic or both types of interactions were destabilized, heterodimerization was still observed at levels very similar to those observed under milder conditions. The previously reported ATPase activity associated with Rpp20 did not appear to be important for its interaction with Rpp25, because amino acid substitutions that abrogated the ATPase activity did not affect the binding to Rpp25. Moreover, the addition of a nonhydrolyzable analog of ATP (AMP-PNP) did not affect Rpp20-Rpp25 heterodimerization (data not shown). However, substitution of asparagine-40 by glutamine, which led to a moderate reduction of the ATPase activity (Li and Altman 2001), severely reduced the affinity of Rpp20 for binding to Rpp25. Since the conservative amino acid mutation at position 40 is not expected to cause major structural changes in the Rpp20 protein, this residue is predicted to play a crucial role in the interaction with Rpp25. Our data do not exclude the possibility that the binding of Rpp25 to Rpp20 blocks the binding of NTP. Further studies will be required to investigate this issue.
Despite extensive knowledge on their protein composition, the stoichiometry of protein subunit binding to the human RNase MRP and RNase P complexes has not been documented so far. The results of the experiments in which VSV-tagged versions of Rpp20 and Rpp25 were expressed next to the endogenous nontagged counterparts (Fig. 3) indicated that the vast majority, if not all, of these ribonucleoprotein complexes contains only a single copy of each of these proteins. This is consistent with the binding of a single Rpp20-Rpp25 heterodimer to RNase MRP and RNase P. Further experiments will be required to investigate whether the same is true for the other protein subunits. The results shown in Figure 3 also show that under these conditions RNase MRP and RNase P are not physically associated with each other and that both complexes exist as monomers.
In agreement with their efficient heterodimerization, the expression levels of Rpp20 and Rpp25 appeared to be codependent, as indicated by the results of overexpression and knock-down experiments. Regulation of their expression levels most likely occurs on the protein level. Since similar amounts of plasmid DNA were transfected and the overexpressed proteins are under transcriptional control of constitutively active CMV promoters, the increased signals of coexpressed GFP-Rpp20 and VSV-Rpp25 can be explained by a reduced turnover rate, which is most likely resulting from an increased stability due to their heterodimerization.
The functional relevance of Rpp20-Rpp25 heterodimerization is further emphasized by the Rpp25-dependent subcellular localization of Rpp20. This phenomenon may also explain the localization observed for the Flag-tagged human Rpp20 protein reported by Hua and colleagues (Hua and Zhou 2004). They observed a very similar subcellular distribution of overexpressed Flag-tagged Rpp20 protein as we observed for GFP-Rpp20. In both cases the limited amount of Rpp25 present in the transfected cells is likely to be insufficient for the efficient accumulation of tagged Rpp20 in the nucleoli. The colocalization of Rpp25 with nucleophosmin/B23 demonstrated that the highest concentrations of the Rpp20-Rpp25 dimer are found in the granular component of the nucleolus. The GC is believed to be the area of the nucleolus where the late processing of pre-rRNA and ribosome assembly takes place. RNase MRP has been demonstrated to play a role in late steps of ITS1 processing (Cohen et al. 2003; Thiel et al. 2005), consistent with its accumulation in the GC (Reimer et al. 1988). Paradoxically, we have recently observed that the Rpp20-Rpp25 dimer is not present in 60–80S complexes, which are most likely preribosomal complexes containing processing factors such as RNase MRP. The transient association of the Rpp20-Rpp25 heterodimer with RNase MRP and possibly RNase P may play a crucial role in the regulation of their endonucleolytic activities. One possibility is that the binding of Rpp20 and Rpp25 to the P3 domain of the RNase MRP and RNase P RNAs target these complexes to the GC. Indeed, the P3 domain was previously demonstrated to be required for their nucleolar accumulation (Jacobson et al. 1995). Another possibility is that the association of the Rpp20-Rpp25 dimer blocks the endonucleolytic activity of these enzymes, and that their dissociation leads to their activation. Interestingly, the P3 domain is located close to the region of the RNA that is believed to be directly involved in substrate binding and cleavage. Alternatively, the Rpp20-Rpp25 heterodimer may also be involved in the correct assembly of the RNase MRP and RNase P complexes by recruiting the RNAs via their binding to the P3 domain. In this regard, Li and Altman (2002) reported that the human RNase P RNA, when lacking the P3 region, is not able to assemble into intact ribonucleoprotein complexes. A final possibility is that the main function of Rpp25 is to mediate and stabilize the association of Rpp20 with RNase MRP and RNase P and that Rpp20 is acting as a functional bridge (Li and Altman 2001), which mediates the recruitment of additional factors that are temporarily, and possibly only under certain conditions, required for optimal functioning of these enzymes. Candidates for such factors are SMN, Hsp27, and KIAA065, which all have been reported as interacting partners for Rpp20 (Jiang and Altman 2001; Hua and Zhou 2004).
MATERIALS AND METHODS
GST-fusion proteins
The cDNAs of Rpp20 (wild type and mutants) and Rpp25 were cloned into vector pGEX2T'G (Welting et al. 2004). The cDNAs encoding the Rpp20 amino acid substitution mutants (D36E, N40Q, and G59S) were kindly provided by Cecilia Guerrier-Takada (Yale University, New Haven, CT). GST-fusion proteins were expressed in Escherichia coli BL21(DE3)pLysS and purified by standard procedures. The purity and concentrations of the GST-fusion proteins were determined by SDS-PAGE and Coomassie brilliant blue staining. The purified proteins were supplemented with 10% glycerol (final concentration) and stored at −70°C.
In vitro translation of RNase MRP proteins
The coding sequences of Rpp20 and Rpp25 were cloned into the pCI-Neo vector in-frame with a sequence encoding a vesicular stomatitis virus glycoprotein-epitope (VSV-tag) (Welting et al. 2004). For in vitro transcription, these constructs were linearized with NotI, isolated by gel electrophoresis, and transcribed using T7 RNA polymerase in the presence of the GpppG cap analog. Subsequently, the transcripts were used to produce N-terminally VSV-tagged Rpp20 and Rpp25 in a rabbit reticulocyte lysate translation system (Promega) in the presence of 35S-labeled methionine.
In vitro transcription and biotinylation of P3 RNA
To generate in vitro transcribed and biotinylated P3 RNA, a previously described transcription construct was used which encodes nucleotides 22–67 of the human RNase MRP RNA (Pluk et al. 1999). After linearization with HindIII, in vitro transcription was performed according to standard procedures using T7 RNA polymerase. The P3 RNA was cotranscriptionally biotinylated by the addition of biotin-11-UTP (PerkinElmer Life Sciences) in the transcription reaction. The transcription reaction was stopped by phenol/chloroform/isoamylalcohol extraction, and the remaining mononucleotides were removed by a Sephadex G-50 spin column. Finally, the RNA was precipitated with isopropanol, and after washing, the pellet was dissolved in water to a final concentration of 0.1 μg/μL and stored at −70°C.
GST pull-down experiments
GST pull-down experiments were performed essentially as described before (Welting et al. 2004). Glutathione-Sepharose beads were incubated with GST (fusion) protein in a buffer containing 20 mM HEPES/KOH (pH 7.6), 100 mM KCl, 0.5 mM EDTA, 0.05% Nonidet P-40 (NP-40), 1 mM dithiotreitol, 5 mM MgCl2, 0.02% BSA, and complete protease inhibitor (Roche) for 30 min at room temperature. Subsequently, the coated beads were incubated with in vitro translated, 35S-labeled protein for 2 h at 4°C under continuous agitation in the presence or absence of 1 mM ATP. Finally, the bound proteins were analyzed by SDS-PAGE and autoradiography.
Expression of tagged Rpp20 and Rpp25 in transfected HEp-2 cells
The coding sequences of Rpp20 and Rpp25 were cloned in the mammalian expression vectors pCI-Neo and pEGFP-C3 in-frame with the VSV-tag (see above) and the coding sequence for green fluorescent protein (GFP), respectively. For transient expression of N-terminally tagged proteins, 4 × 106 HEp-2 cells were transfected with 10 μg purified plasmid DNA by electroporation using the Biorad GenePulser II at 260 V; 950 μF. After transfection, the cells were cultured in a CO2 incubator at 37°C in Dulbecco's Minimal Essential Medium (GIBCO BRL) supplemented with 10% fetal calf serum on glass slides or in T75 culture flasks. After 24 h the cells were processed for fluorescent microscopy or harvested for immunoprecipitation experiments.
Immunoprecipitation
Transiently transfected HEp-2 cells were homogenized by sonication in a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM KCl, 2 mM EDTA, 0.05% NP-40, and 1 mM dithioerytritol (DTE). For anti-VSV-tag immunoprecipitations, 100 μg rabbit-anti-mouse immunoglobulin antibodies (Dako) were coupled to 20 μL Protein-A–agarose beads (Kem-En-Tec) at room temperature for 1 h in 500 μL of buffer containing 500 mM NaCl, 20 mM Tris-HCl (pH 8.0), and 0.05% NP-40 (IPP500). After coupling, the beads were washed three times with IPP500, and subsequently, a monoclonal anti-VSV-tag antibody was coupled to the rabbit-anti-mouse coated Protein-A–agarose beads under identical conditions as described above. For immunoprecipitations with polyclonal antisera, 5 μL of rabbit serum was coupled to 10 μL of Protein-A–agarose beads at room temperature for 1 h in 500 μL IPP500. The beads were washed three times with IPP500 and once with IPP150 (identical to IPP500, but with 150 mM NaCl). The clarified cell lysates or in vitro translated proteins were added to the beads, and the immunoprecipitation was performed under continuous agitation at 4°C in a total volume of 750 μL. After 2 h, the beads were washed three times with IPP150, and the immunoprecipitated material was either dissolved in SDS-PAGE sample buffer or subjected to RNA isolation using Trizol (Invitrogen).
Streptavidin-biotin pull-down
Biotinylated P3 RNA (100 ng) was mixed with 1 μL 10-fold concentrated pull-down buffer 100 (PB100: 100 mM KCl, 20 mM HEPES/KOH at pH 7.6, 0.5 mM EDTA, 0.05% NP-40, 1 mM DTE), 1 μg calf thymus competitor tRNA, and appropriate amounts of in vitro translated, 35S-labeled Rpp20 and/or Rpp25. Water was added to a final reaction volume of 10 μL. The mixture was incubated for 1 h at 0°C. Subsequently, 10 μL of streptavidin-Sepharose (Amersham Biosciences) was added together with 100 μL PB100 and incubated under continuous agitation for 1 h at 4°C. The beads were washed three times with PB100, and the precipitated proteins were dissolved in SDS-PAGE sample buffer, separated by SDS-PAGE, and visualized by autoradiography.
Indirect immunofluorescence
HEp-2 cells were cultured to 70% confluency on glass slides and fixed with methanol. The fixed cells were rehydrated in PBS for 5 min. Protein-A–purified mouse monoclonal antibody against Rpp20 (1F11, ModiQuest) was used in a 200-fold dilution; polyclonal rabbit serum against Rpp25 was 100-fold diluted and culture supernatants containing mouse monoclonal antibodies against fibrillarin (ASWU1) (Monestier et al. 1994), and nucleophosmin/B23 (37/5.1) (Spector et al. 1984) were 10-fold diluted in PBS. The cells were incubated with the primary antibodies for 1 h at room temperature. Subsequently, the slides were washed five times with PBS and incubated with 100-fold diluted Alexa Fluor 488-conjugated goat-anti-rabbit antibodies or Alexa Fluor 555-conjugated goat-anti-mouse antibodies (Molecular Probes). For double stainings, the procedure was repeated as described above. Stained cells were mounted in fluorescent mounting medium (Dako) and visualized by fluorescence microscopy using an Olympus BH-2 microscope or a Leica DM IRBE confocal laser scanning microscope.
RNA interference
For knock-down of Rpp20 by RNAi, an siRNA duplex targeting Rpp20, si-Rpp20–1, was designed using standard guidelines (Elbashir et al. 2002): 5′-CCAUCAACCGCGCCAUCAATT-3′. The siRNA duplex sequence targeting Rpp25, si-Rpp25–1, was adapted from the siRNA sequence described by Zhang and Altman (2004): 5′-GUGCGCCGAGAUCCUCAAGTT-3′. Both duplexes were purchased from Eurogentec and contain 3′ dTdT overhangs. For siRNA transfection, 5 × 104 HEp-2 cells were transfected with 20 pmol of siRNA duplex using oligofectamine transfection reagent (Invitrogen) according to the protocol provided by the manufacturer. After 48 h the cells were harvested, lysed, and processed for Western analysis or re-transfected with siRNA using the protocol described above.
Western blotting
Cell extracts and recombinant or immunoprecipitated proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes by electroblotting. Immunodetection of Rrp4p, GFP, Rpp20, Rpp25, Rpp30, Rpp38, and Rpp40 was performed using polyclonal rabbit antisera against these proteins (Eder et al. 1997; Jarrous et al. 1998; Allmang et al. 1999; Jarrous et al. 1999; Guerrier-Takada et al. 2002) and a monoclonal mouse antibody against GST and Rpp20 (ModiQuest). Horseradish peroxidase-conjugated polyclonal swine anti-rabbit and rabbit anti-mouse immunoglobulin antibodies were purchased from Dako and applied as secondary antibody. Bound antibodies were visualized by enhanced chemiluminescence detection procedures.
ACKNOWLEDGMENTS
We thank Cecilia Guerrier-Takada and Sidney Altman (Yale University, New Haven, CT) for their kind gifts of antibodies and cDNAs, and Wiljan Hendriks (Radboud University Nijmegen, Nijmegen, The Netherlands), David Tollervey (University of Edinburgh, Scotland, UK), Pui-Kwong Chan (Baylor College of Medicine, Houston, TX), and Marc Monestier (Temple University School of Medicine, PA) for providing various antibodies.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.237807.
REFERENCES
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., Mitchell, P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L., Iyer, L.M., Anantharaman, V. The two faces of Alba: The evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, D.D., Clayton, D.A. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, A., Reiner, R., Jarrous, N. Alterations in the intracellular level of a protein subunit of human RNase P affect processing of tRNA precursors. Nucleic Acids Res. 2003;31:4836–4846. doi: 10.1093/nar/gkg691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, L.J., Moulton, V., Penny, D. Use of RNA secondary structure for studying the evolution of RNase P and RNase MRP. J. Mol. Evol. 2000;51:194–204. doi: 10.1007/s002390010081. [DOI] [PubMed] [Google Scholar]
- Eder, P.S., Kekuda, R., Stolc, V., Altman, S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc. Natl. Acad. Sci. 1997;94:1101–1106. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., Tuschl, T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Frank, D.N., Adamidi, C., Ehringer, M.A., Pitulle, C., Pace, N.R. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill, T., Cai, T., Aulds, J., Wierzbicki, S., Schmitt, M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Eder, P.S., Gopalan, V., Altman, S. Purification and characterization of Rpp25, an RNA-binding protein subunit of human ribonuclease P. RNA. 2002;8:290–295. doi: 10.1017/s1355838202027954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, Y., Zhou, J. Rpp20 interacts with SMN and is re-distributed into SMN granules in response to stress. Biochem. Biophys. Res. Commun. 2004;314:268–276. doi: 10.1016/j.bbrc.2003.12.084. [DOI] [PubMed] [Google Scholar]
- Jacobson, M.R., Cao, L.G., Wang, Y.L., Pederson, T. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J. Cell Biol. 1995;131:1649–1658. doi: 10.1083/jcb.131.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, M.R., Cao, L.G., Taneja, K., Singer, R.H., Wang, Y.L., Pederson, T. Nuclear domains of the RNA subunit of RNase P. J. Cell Sci. 1997;110:829–837. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jarrous, N. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous, N., Eder, P.S., Guerrier-Takada, C., Hoog, C., Altman, S. Autoantigenic properties of some protein subunits of catalytically active complexes of human ribonuclease P. RNA. 1998;4:407–417. [PMC free article] [PubMed] [Google Scholar]
- Jarrous, N., Eder, P.S., Wesolowski, D., Altman, S. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA. 1999;5:153–157. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T., Altman, S. Protein-protein interactions with subunits of human nuclear RNase P. Proc. Natl. Acad. Sci. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, T., Guerrier-Takada, C., Altman, S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Altman, S. A subunit of human nuclear RNase P has ATPase activity. Proc. Natl. Acad. Sci. 2001;98:441–444. doi: 10.1073/pnas.021555498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Altman, S. Partial reconstitution of human RNase P in HeLa cells between its RNA subunit with an affinity tag and the intact protein components. Nucleic Acids Res. 2002;30:3706–3711. doi: 10.1093/nar/gkf499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Zaman, S., Langdon, Y., Zengel, J.M., Lindahl, L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Res. 2004;32:3703–3711. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.H., Yuan, Y., Reddy, R. Human RNaseP RNA and nucleolar 7-2 RNA share conserved “To” antigen-binding domains. Mol. Cell. Biochem. 1994;130:75–82. doi: 10.1007/BF01084270. [DOI] [PubMed] [Google Scholar]
- Lygerou, Z., Allmang, C., Tollervey, D., Seraphin, B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Monestier, M., Losman, M.J., Novick, K.E., Aris, J.P. Molecular analysis of mercury-induced antinucleolar antibodies in H-2S mice. J. Immunol. 1994;152:667–675. [PubMed] [Google Scholar]
- Piccinelli, P., Rosenblad, M.A., Samuelsson, T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk, H., van Eenennaam, H., Rutjes, S.A., Pruijn, G.J., van Venrooij, W.J. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, R., Shimba, S. Structural and functional similarities between MRP and RNase P. Mol. Biol. Rep. 1995;22:81–85. doi: 10.1007/BF00988710. [DOI] [PubMed] [Google Scholar]
- Reimer, G., Raska, I., Scheer, U., Tan, E.M. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp. Cell Res. 1988;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Sbisa, E., Pesole, G., Tullo, A., Saccone, C. The evolution of the RNase P- and RNase MRP-associated RNAs: Phylogenetic analysis and nucleotide substitution rate. J. Mol. Evol. 1996;43:46–57. doi: 10.1007/BF02352299. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.E., Clayton, D.A. Yeast site-specific ribonucleoprotein endoribonuclease MRP contains an RNA component homologous to mammalian RNase MRP RNA and essential for cell viability. Genes & Dev. 1992;6:1975–1985. doi: 10.1101/gad.6.10.1975. [DOI] [PubMed] [Google Scholar]
- Schmitt, M.E., Clayton, D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae . Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector, D.L., Ochs, R.L., Busch, H. Silver staining, immunofluorescence, and immunoelectron microscopic localization of nucleolar phosphoproteins B23 and C23. Chromosoma. 1984;90:139–148. doi: 10.1007/BF00292451. [DOI] [PubMed] [Google Scholar]
- Thiel, C.T., Horn, D., Zabel, B., Ekici, A.B., Salinas, K., Gebhart, E., Ruschendorf, F., Sticht, H., Spranger, J., Muller, D., et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 2005;77:795–806. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eenennaam, H., Vogelzangs, J.H., Lugtenberg, D., Van Den Hoogen, F.H., van Venrooij, W.J., Pruijn, G.J. Identity of the RNase MRP- and RNase P-associated Th/To autoantigen. Arthritis Rheum. 2002;46:3266–3272. doi: 10.1002/art.10673. [DOI] [PubMed] [Google Scholar]
- Walker, S.C., Avis, J.M. Secondary structure probing of the human RNase MRP RNA reveals the potential for MRP RNA subsets. Biochem. Biophys. Res. Commun. 2005;335:314–321. doi: 10.1016/j.bbrc.2005.07.074. [DOI] [PubMed] [Google Scholar]
- Walker, S.C., Engelke, D.R. Ribonuclease p: The evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, S.C., Aspinall, T.V., Gordon, J.M., Avis, J.M. Probing the structure of Saccharomyces cerevisiae RNase MRP. Biochem. Soc. Trans. 2005;33:479–481. doi: 10.1042/BST0330479. [DOI] [PubMed] [Google Scholar]
- Welting, T.J., Raijmakers, R., Pruijn, G.J. Autoantigenicity of nucleolar complexes. Autoimmun. Rev. 2003;2:313–321. doi: 10.1016/s1568-9972(03)00029-6. [DOI] [PubMed] [Google Scholar]
- Welting, T.J., van Venrooij, W.J., Pruijn, G.J. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welting, T.J., Kikkert, B.J., van Venrooij, W.J., Pruijn, G.J. Differential association of protein subunits with the human RNase MRP and RNase P complexes. RNA. 2006;12:1373–1382. doi: 10.1261/rna.2293906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y., Tan, E., Reddy, R. The 40-kilodalton to autoantigen associates with nucleotides 21 to 64 of human mitochondrial RNA processing/7-2 RNA in vitro. Mol. Cell. Biol. 1991;11:5266–5274. doi: 10.1128/mcb.11.10.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Altman, S. Inhibition of the expression of the human RNase P protein subunits Rpp21, Rpp25, Rpp29 by external guide sequences (EGSs) and siRNA. J. Mol. Biol. 2004;342:1077–1083. doi: 10.1016/j.jmb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Stribinskis, V., Ramos, K.S., Li, Y. Sequence analysis of RNase MRP RNA reveals its origination from eukaryotic RNase P RNA. RNA. 2006;12:699–706. doi: 10.1261/rna.2284906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler, W.A., Morris, J., Scott, F.H., Millikin, C., Engelke, D.R. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]