Abstract
Kinetoplastid mitochondrial mRNA editing is a post-transcriptional process of uridine insertion and deletion. Editing is mediated by small RNA molecules termed guide RNAs (gRNAs). Most gRNAs are encoded by numerous small circular DNA minicircles, while the protein coding mitochondrial genes are encoded on a separate, larger genome called the maxicircle. In order to provide a workbench for the analysis of RNA editing in kinetoplastids and a well-annotated set of guide RNAs for Trypanosoma brucei, we generated the kinetoplastid RNA editing sequence search tool (KISS) (http://gmod.mbl.edu/kiss/). KISS is a pipeline and database that uses BLAST comparisons and minicircle sequence motifs to annotate potential gRNAs and cognate mRNA editing sequence. KISS 1.0 contains all previously known minicircle and maxicircle data from Trypanosoma brucei plus >400 new minicircle sequences. Using an online format, KISS 1.0 allows the mapping and visualization of all known T. brucei gRNAs to minicircle genes and to potential mRNA substrates for RNA editing.
Keywords: Trypanosoma brucei, RNA editing, guide RNA, minicircle
INTRODUCTION
The mitochondrial genome of Trypanosoma brucei is composed of two classes of circular DNA molecules, maxicircles and minicircles (Simpson 1997; Stuart et al. 2005). Each mitochondrion contains ∼50 maxicircles (∼20 kb) and 5000–10,000 minicircles (∼1 kb). Maxicircles encode 18 protein-coding genes, two ribosomal RNAs, and two guide RNAs (gRNAs). Transcripts from 12 of the protein-coding genes are edited. The extent of editing ranges from four uridines inserted in the cytochrome c oxidase II (COXII) mRNA to hundreds of uridines added and dozens of uridines deleted in the cytochrome c oxidase III (COXIII) mRNA. Each minicircle encodes 3–5 small RNA molecules, called guide RNAs (gRNAs) that contain the information needed for the editing of mRNAs (Blum and Simpson 1990; Pollard et al. 1990). Guide RNAs and their cognate mRNAs specifically interact by the formation of a short anchor duplex of 5–10 base pairs (bp) that forms between the 5′ end of the gRNAs and sequences in the mRNA immediately 3′ to editing sites (Blum and Simpson 1990; Decker and Sollner-Webb 1990; Pollard et al. 1990). Adjacent to the anchor sequence of the gRNA is the “guiding region,” which directs the precise insertion or deletion of uridine residues in the mRNA. Finally, all mature gRNAs have a 3′ poly(U) tail that is added post-transcriptionally to stabilize the gRNA pre-mRNA interaction (Blum and Simpson 1990).
It has been estimated that a minimum of 200 gRNAs are necessary to direct the editing of all of the mitochondrial mRNAs of T. brucei (Corell et al. 1993). The minicircle genome, however, is considerably more complex. It contains >200 minicircle sequence classes, each encoding 3–5 gRNA genes. Several redundant gRNA genes, with different sequences but the identical anchor regions, have been identified (Koslowsky et al. 1992; Corell et al. 1993; Riley et al. 1994). In addition, several gRNA genes have been proposed that have extensive mismatches to their potential cognate mRNA (Corell et al. 1993; Hong and Simpson 2003).
We have developed a comprehensive online database that allows precise annotation of gRNA genes as well as the target region on the mitochondrial mRNAs. This greatly extends and integrates the existing gRNA and RNA editing databases (Souza et al. 1997; Simpson et al. 1998). We have added 439 minicircle sequences and thereby increased the previous number of known minicircles and annotated gRNAs by >10-fold. Furthermore, we added 300 cDNA sequences from gRNAs of T. brucei, verifying expression of many of the predicted gRNA genes.
MINICIRCLE AND cDNA LIBRARIES
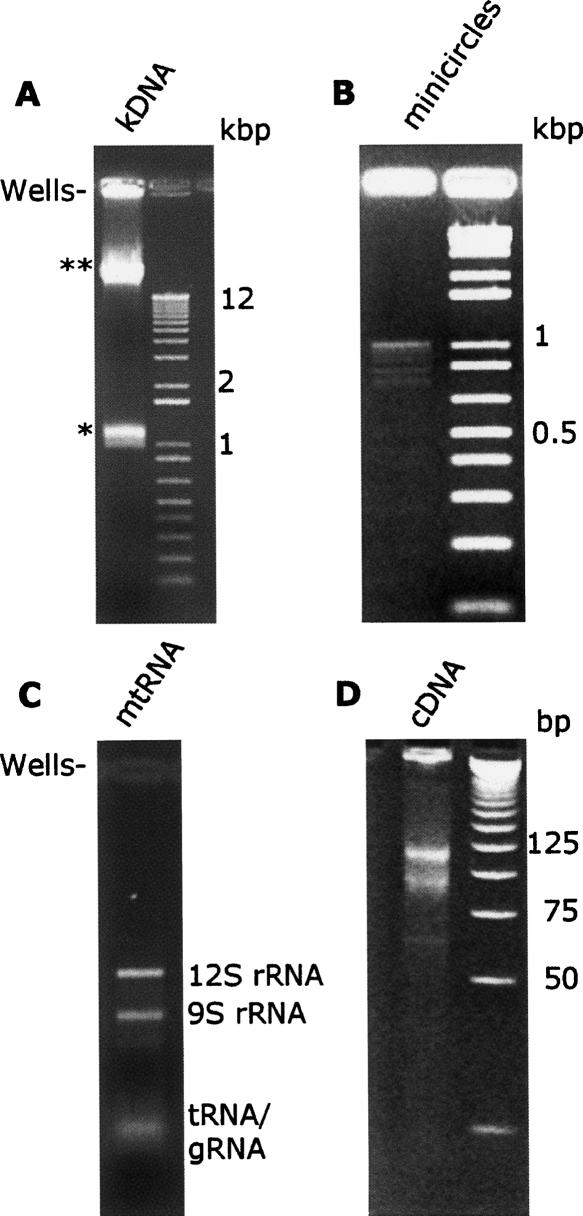
We cloned and sequenced >400 minicircles from the procyclic, insect developmental stage of T. brucei [Trypanosome Research Edinburg University 667 (TREU667)]. After cell lysis, kDNA was isolated using a sucrose cushion and ultracentrifugation. Purified kDNA was treated with topoisomerase II to liberate minicircles from the kDNA network (Fig. 1A). After gel purification, the minicircles were linearized using the restriction enzyme TaqI (NEB; Fig. 1B). A minor portion of the minicircles was cleaved multiple times by TaqI, resulting in restriction fragments less than the full-length 1 kilobase pair (kbp) minicircle. Digested minicircles were cloned and sequenced without any further size fractionation, resulting in a library with partial and full-length minicircles. The mean size of the minicircles sequenced was 767 bp, with a size range from 300 bp to 2.2 kbp. All full-length minicircles contained a conserved region with the origin for minicircle replication as well as at least two predicted gRNA genes. Minicircles with no predicted gRNA genes were not identified. Minicircles were annotated as redundant if they shared >95% sequence similarity over 90% of their sequence length. By these standards, we calculated the range of distribution of redundant minicircles to be 58 with a median distribution of 12, while the mode of distribution was still 1. RNA was isolated from purified mitochondria of bloodstream and procyclic developomental stages of T. brucei. Mitochondrial RNA was used to clone and sequence >300 cDNAs prepared from gRNAs isolated from procyclic T. brucei. cDNA libraries were constructed using the Creator SMART cDNA library construction kit with some modifications to the manufacturer's recommendations (Clontech). We used a poly dA primer (5′-ATTCTAGAGCGGCCGCGACATGAAAAAAAAAAAAAAAN-3′) to prime reverse transcription from the poly(U) tail of the gRNAs. After amplification with internal primers (see Creator SMART instructions), we sized selected cDNAs from 75 bp to 100 bp and from 100 bp to 125 bp on polyacrylamide gels (Fig. 1D). Subsequently cDNAs were cloned into pCR 2.1 (Invitrogen). The average size of cDNAs from both libraries was 45 bp with a size range from 21 bp to 60 bp. The average number of each sequence recovered in our library was 1.5 with the mode of distribution being 1.
FIGURE 1.
Preparation of minicircle and gRNA libaries. (A) Kinetoplast DNA preparation from procyclic T. brucei. Minicircles (*) were released from kDNA network by treatment with topoisomerase II and were resolved on a 1% agarose gel. Contaminating nuclear DNA (**) run as an unresolved band above the 12-kbp marker. Remnants of kDNA network were retained in the wells. (B) Linearized minicircle DNA for construction of a minicircle DNA library. Decatenated minicircles (from panel A) were gel purified, linearized by digestion with TaqI, and resolved on a 2% agarose gel. (C) Mitochondrial RNA (mtRNA) for construction of a cDNA library. Mitochondrial RNA from purified mitochondria was resolved on a 1.2% methyl mercury gel. (D) cDNA was prepared from gRNAs (cDNA). The guide-cDNA was amplified from total mitochondrial RNA (from panel C) using poly(A) oligonucleotides and a stringent size selection. Two cDNA populations of the sizes 75–100 nt and 100–125 nt were purified on a 6% polyacrylamide gel and used for cloning.
DATABASE
In silico identification of gRNA genes from minicircle sequences
All minicircle sequences were aligned to edited mRNA sequences using the WUBLAST program (Lopez et al. 2003). A modified similarity matrix was created in order to allow for G-U base pairing (http://gmod.mbl.edu/kiss/). Hits were considered valid if the length was >20 bp with no gaps, allowing for G-U base pairing. In order to screen out false positives, we disregarded hits with more 80% T or more then 80% C in the match sequence. This criterion is empirical and does not reflect any experimental evidence. Yet we assumed that long stretches of a single nucleotide are unlikely to be functional molecules. Furthermore, this criterion will also help in the future versions where cDNA sequences from polyadenylated mRNAs will be included.
Additional features analyzed
The inverted repeats on the minicircles were predicted using the EMBOSS program PALINDROME (Rice et al. 2000), using the constraints of a minimum length of 14 and a maximum length of 30 while allowing three mismatches. These settings lead in most cases to multiple inverted repeats surrounding a certain gRNA gene on one minicircle. However, more stringent conditions lead to loss of many inverted repeats surrounding potential gRNA genes. In order not to exclude data, the more relaxed settings were used in this version. The origin of replication and the surrounding conserved region was annotated using BLAST.
Description of the database
In order to create a workbench for the analysis of RNA editing in kinetoplastids, we designed the relational database, KISS, which can be accessed from http://gmod.mbl.edu/kiss. KISS is a pipeline that allows the annotation and visualization of sequence data from minicircles and maxicircles as well as cDNA data from gRNAs. KISS 1.0 contains all previously known T. brucei minicircle and maxicircle sequence data plus 439 new minicircle sequences with the annotated corresponding gRNA genes. In addition, KISS contains 300 cDNA sequences from gRNAs, verifying the expression of a significant portion of the predicted gRNA genes.
Web interface
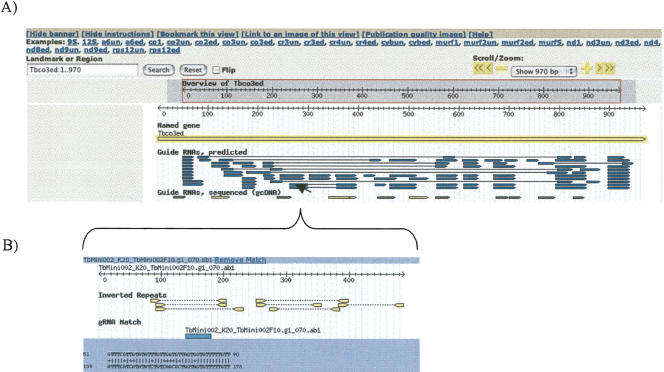
KISS contains four different interfaces: (1) “Home” displays an introduction into RNA editing and some statistics of the underlying sequence data. (2) “Gbrowse” is the main interface that allows the comparison of maxicircle and minicircle sequences (Stein et al. 2002). The maxicircle gene names are displayed in the top part and can be used to search for gRNA genes predicted from minicircles and guide cDNAs that match a particular edited (ed) or pre-edited (un) maxicircle sequence (Fig. 2). The database can also be searched using a sequence name, gene name, locus, or other landmark. The maxicircle gene is displayed as a long yellow bar along with several different tracks, which can be selected in the bottom part of the page. The “gRNAs gene” track, for example, shows all gRNA genes that have been predicted from minicircles for the selected maxicircle sequence as blue bars. The alignment between gRNA and maxicircle sequence can be examined by hovering over the gRNA feature. The hover window contains the alignment, the positioning of the predicted gRNA gene on its minicircle, as well as the inverted repeats on the minicircle. Upon selection of the gRNA gene feature, a new window opens and displays the corresponding minicircle along with two additional tracks, the inverted repeats and the known conserved region. Similarly, the “gcDNA” track shows cDNA sequences from gRNAs that match the selected maxicircle sequence. The track definitions can be reviewed in a separate window by selecting any of the track features. (3) “Gblast” is a BLAST interface that allows the user to search the sequenced minicircles, predicted gRNAs, and maxicircle genes. Gblast uses BLAST in conjunction with standard matrices or the modified similarity matrix, which is permissive of G-U base pairings. (4) “Downloads” provides information on and results of various automated analyses as a bulk download. The user can download minicircle and predicted gRNA gene sequences.
FIGURE 2.
Screen shot from the GBROWSE menu. (A) Displayed is the edited sequence of COXIII (long yellow bar) together with the binding sites of gRNAs as predicted from minicircles (short blue bars), gcDNAs (short green bars) from this study, or previously predicted gRNAs (short yellow bars). Lines between two blue bars indicate that the gRNAs reside on the same minicircle. gRNAs with overlapping target regions are displayed in stacks. (B) Hovering over a displayed gRNA (short blue bar) opens a small window, which shows the alignment between the maxicircle transcript (top strand; 5′ to 3′) and the gRNA (bottom strand; 3′ to 5′). Furthermore, it displays the positioning of the gRNA gene on the corresponding minicircle together with the surrounding inverted repeats.
Availability
The database KISS is accessible via http://gmod.mbl.edu/kiss/. Sequence data can be downloaded individually as well as in a flat file format for most of the features provided. All available sequence data have also been deposited to one of the major sequence databases if not done previously. Users of the database should cite this publication. Corrections, new entries, errors, and omissions or other materials for inclusion in the database are welcome. Submission of new sequence data will be accepted in FASTA format.
Future directions
The next release of the database will include cDNA sequence data of mitochondrial mRNA from different life stages of the parasite. We are also in the process of sequencing additional minicircles and gRNA/cDNAs in order to achieve full gRNA coverage of all edited mitochondrial transcripts from T. brucei. These sequence data will also be included in the next release.
ACKNOWLEDGMENTS
We thank the members of the GID laboratory for extensive and productive discussions. In particular, Matt Beverly, Robert Sabatini, Rudo Kieft, Daniel Golden, and Justin Widener. We also acknowledge the “The U-insertion/deletion Edited Sequence Database” and “the gRNA Database,” which were very helpful for this work. We are grateful to the National Institutes of Health, the Ellison Medical Foundation, the W.M. Keck Foundation, and the G. Unger Vetlesen Foundation for funding this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.232907.
REFERENCES
- Blum, B., Simpson, L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Corell, R.A., Feagin, J.E., Riley, G.R., Strickland, T., Guderian, J.A., Myler, P.J., Stuart, K. Trypanosoma brucei minicircles encode multiple guide RNAs which can direct editing of extensively overlapping sequences. Nucleic Acids Res. 1993;21:4313–4320. doi: 10.1093/nar/21.18.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Sollner-Webb, B. RNA editing involves indiscriminate U changes throughout precisely defined editing domains. Cell. 1990;61:1001–1011. doi: 10.1016/0092-8674(90)90065-m. [DOI] [PubMed] [Google Scholar]
- Hong, M., Simpson, L. Genomic organization of Trypanosoma brucei kinetoplast DNA minicircles. Protist. 2003;154:265–279. doi: 10.1078/143446103322166554. [DOI] [PubMed] [Google Scholar]
- Koslowsky, D.J., Goringer, H.U., Morales, T.H., Stuart, K. In vitro guide RNA/mRNA chimaera formation in Trypanosoma brucei RNA editing. Nature. 1992;356:807–809. doi: 10.1038/356807a0. [DOI] [PubMed] [Google Scholar]
- Lopez, R., Silventoinen, V., Robinson, S., Kibria, A., Gish, W. WU-Blast2 server at the European Bioinformatics Institute. Nucleic Acids Res. 2003;31:3795–3798. doi: 10.1093/nar/gkg573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, V.W., Rohrer, S.P., Michelotti, E.F., Hancock, K., Hajduk, S.L. Organization of minicircle genes for guide RNAs in Trypanosoma brucei . Cell. 1990;63:783–790. doi: 10.1016/0092-8674(90)90144-4. [DOI] [PubMed] [Google Scholar]
- Rice, P., Longden, I., Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Riley, G.R., Corell, R.A., Stuart, K. Multiple guide RNAs for identical editing of Trypanosoma brucei apocytochrome b mRNA have an unusual minicircle location and are developmentally regulated. J. Biol. Chem. 1994;269:6101–6108. [PubMed] [Google Scholar]
- Simpson, L. The genomic organization of guide RNA genes in kinetoplastid protozoa: Several conundrums and their solutions. Mol. Biochem. Parasitol. 1997;86:133–141. doi: 10.1016/s0166-6851(97)00037-6. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Wang, S.H., Thiemann, O.H., Alfonzo, J.D., Maslov, D.A., Avila, H.A. U-insertion/deletion Edited Sequence Database. Nucleic Acids Res. 1998;26:170–176. doi: 10.1093/nar/26.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, A.E., Hermann, T., Goringer, H.U. The guide RNA database. Nucleic Acids Res. 1997;25:104–106. doi: 10.1093/nar/25.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L.D., Mungall, C., Shu, S., Caudy, M., Mangone, M., Day, A., Nickerson, E., Stajich, J.E., Harris, T.W., Arva, A., et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, K.D., Schnaufer, A., Ernst, N.L., Panigrahi, A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]