Abstract
Turnip yellow mosaic virus (TYMV) RNA directs the translation of two overlapping open reading frames. Competing models have been previously published to explain ribosome access to the downstream polyprotein cistron. The Trojan horse model, based on cell-free experiments, proposes noncanonical cap-independent initiation in which the 3′-terminal tRNA-like structure (TLS) functionally replaces initiator tRNA, and the valine bound to the TLS becomes cis-incorporated into viral protein. The initiation coupling model, based on in vivo expression and ribosome toe-printing studies, proposes a variation of canonical leaky scanning. Here, we have re-examined the wheat germ extract experiments that led to the Trojan horse model, incorporating a variety of controls. We report that (1) translation in vitro from the polyprotein AUG of TYMV RNA is unchanged after removal of the 3′ TLS but is stimulated by the presence of a 5′-cap; (2) the presence of free cap analog or edeine (which interferes with initiation at the ribosomal P site and its tRNAi Met involvement) inhibits translation from the polyprotein AUG; (3) the toe-prints of immediately post-initiation ribosomes on TYMV RNA are similar with and without an intact TLS; and (4) significant deacylation of valyl-TYMV RNA in wheat germ extract can complicate the detection of cis-incorporation. These results favor the initiation coupling model.
Keywords: translation initiation, ribosome toe-printing, edeine inhibition, viral tRNA-like structure, valylation, in vitro translation, wheat germ extract
INTRODUCTION
Turnip yellow mosaic virus (TYMV) genomic RNA is 5′-capped and possesses two extensively overlapping open reading frames (ORFs) that are translated to yield p69, which serves as the viral movement protein and RNAi suppressor, and p206, the replication polyprotein (Fig. 1A; Dreher, 2004). The initiation codons of these two ORFs are only 7 nucleotides (nt) apart. Given that eukaryotic mRNAs are typically monocistronic, it has been of interest to understand how both overlapping ORFs are robustly expressed from TYMV RNA. The fact that the initiation context of the upstream AUG (AUG69) is suboptimal would suggest that expression could occur by cap-dependent leaky scanning (Kozak 2002). Indeed, using luciferase reporter mRNAs in a cowpea protoplast transient expression system (Matsuda and Dreher 2006), we have described a special variation of leaky scanning (termed “initiation coupling”) that allows dicistronic expression if initiation codons are closely spaced. Our experiments with TYMV-derived reporter RNAs in cell-based experiments revealed that (1) both ORFs are translated in a cap-dependent manner; (2) both ORFs are reached by ribosomes that have scanned the 5′-UTR; (3) the relative initiations from AUG69 and AUG206 are responsive to the context surrounding each initiation codon; (4) the close spacing of two initiation codons allows direct competition for initiation, rather than sequential 5′-dominant recognition as in typical leaky scanning; (5) expression from both ORFs is enhanced by the 3′-UTR containing a tRNA-like structure (TLS) that can be aminoacylated with valine (Matsuda and Dreher 2004); and (6) initiation behavior is similar on the reporter RNAs and authentic TYMV RNA (Matsuda and Dreher 2006).
FIGURE 1.
The 3′-TLS has no influence on translation of TYMV RNA in wheat germ extract. (A) Diagram of TYMV genomic and subgenomic RNAs. The genomic RNA (6318 nt) encodes overlapping ORFs (boxed) encoding p69 and p206 from AUG initiation codons only 7 nt apart. The p206 replication polyprotein is proteolytically cleaved to yield p141 and p66. Coat protein (CP) expression is silenced in the genomic RNA (CP ORF depicted with dashed box), and occurs from subgenomic RNA (693 nt). Both RNAs possess a 5′-cap and an 83-nt-long tRNA-like structure (TLS) at the 3′-terminus that can be aminoacylated with valine. (B) Quality control of RNA transcripts used for translation studies. Genome-length transcripts produced by T7 transcription were analyzed by electrophoresis in 1% agarose and stained with ethidium bromide. gRNA is full-length genomic RNA; gRNA-TLS257 is genomic RNA lacking the 3′-257 nt, while gRNA(AUG69KO) and gRNA(AUG206KO) are gRNAs with knock-out mutations of AUG69 and AUG206, respectively. (C) Translation products directed by TYMV RNA separated by 12% SDS-PAGE. TYMV virion RNA (vRNA) and gRNA and gRNA-TLS257 RNA transcripts were translated at final concentrations of 2.5 nM in wheat germ extract for 90 min in the presence of [35S]methionine. The identities of virus-encoded proteins indicated at left were confirmed by immunoprecipitation of vRNA translation products with antisera N206, C69, and anti-CP. (D) Analysis of TYMV translation products by 6% SDS-PAGE for clearer separation of p206, p141, and p69. (E) Translation of TYMV RNA in the presence of TY-266 RNA, which represents the 3′-266 nt of TYMV RNA. vRNA, gRNA, and gRNA-TLS257 were translated at 1.25 nM. Products were separated by 12% SDS-PAGE. All experiments were replicated twice.
This view of the translation of TYMV RNA differs radically from the Trojan horse expression mechanism previously proposed (Barends et al. 2003). Based on experiments conducted in unmodified commercially available wheat germ extracts, it was concluded that initiation at the two ORFs occurs by fundamentally different mechanisms. While initiation at the upstream AUG69 was proposed to occur via conventional cap-dependent recruitment of ribosomes with initiation codon decoding by methionyl-tRNAi Met, events at AUG206 were likened to initiation supported by the Cricket paralysis virus (CrPV) intergenic region internal ribosome entry site (IRES) (Jan 2005). Thus, neither the 5′-cap nor methionyl-tRNAi Met was postulated to be involved, but initiation was reported to depend on the presence of the 3′-TLS. Further, it was proposed that the TLS is involved in initiation at AUG206 by functioning directly at the decoding sites of the ribosome, with the potential to donate its 3′-esterified valine to the N terminus of the p206 polyprotein. Underlining the proposed contrast between ORF-69 and ORF-206 expression, the TLS was not reported to be necessary for ORF-69 expression in vitro.
In this report, we have addressed these conflicting views of TYMV RNA translation by reproducing key experiments conducted by Barends et al. (2003). Our experiments do not support the Trojan horse model postulating distinct initiation mechanisms at AUG69 and AUG206, but are consistent with the initiation coupling model proposed from our previous in vivo studies (Matsuda and Dreher 2006).
RESULTS
Robust translation of TYMV RNA lacking the 3′ TLS in wheat germ extract
To re-examine the translational behavior of TYMV RNA in vitro, we used the same commercially available (Promega) wheat germ extracts used by Barends et al. (2003), and high-quality in vitro transcribed RNAs (Fig. 1B). A key requirement in assessing the influence of the 3′-TLS on expression of ORF-69 and ORF-206 is the reliable identification of translation products. To verify product identification, we used ORF-specific antisera in immunoprecipitation experiments, and we translated RNA variants with mutated initiation codons. The translation of virion RNA (vRNA) in the presence of [35S]methionine produced two high-molecular-weight products derived from ORF-206, the unprocessed polyprotein (p206), and the N-terminal product released by proteolytic cleavage (p141) (Fig. 1C, lane 1; Fig. 1D, lane 3). As described previously (Bransom et al. 1991), most of the p206 is cleaved, giving rise to a prominent p141 band, while the C-terminal product, p66, is apparently unstable and lost. The identifications of p206 and p141 were verified by their immunoprecipitation with N206 antiserum (Fig. 1C, lane 4; Fig. 1D, lane 6) and by their near-absence among products translated from AUG206KO (knock-out) RNA (Fig. 1D, lane 2).
vRNA also supports the translation of p69 expressed from ORF-69. This product was identified by immunoprecipitation with C69 antiserum (Fig. 1C, lane 5; Fig. 1D, lane 7) and by its absence among products translated from AUG69KO RNA (Fig. 1D, lane 1). The 20-kDa coat protein (CP) was translated only from vRNA (Fig. 1C, lanes 1,6), an approximately equimolar mixture of genomic and subgenomic RNAs.
Apart from the production of CP, the expression profiles programmed by infectious genomic RNA produced by in vitro transcription (gRNA) (Weiland and Dreher, 1989) and by vRNA were similar (Fig. 1C, lanes 1,2; Fig. 1D, lanes 3,4), validating the use of transcript RNAs in these studies. As discussed below, we believe that the preferred method for producing genomic RNA lacking the TLS is by transcription from a defined template rather than via cleavage by ribonuclease H directed by an annealed DNA oligo, the method used by Barends et al. (2003). We produced gRNA-TLS257, which lacks the 3′-257 nt, by transcription of pTYMC linearized at the SmaI restriction site (Fig. 1B, lane 2).
Absence of the 3′-257 nt including the TLS did not alter the translation capacity of gRNA in wheat germ extracts (Fig. 1C, cf. lanes 2 and 3; Fig. 1D, cf. lanes 4 and 5). Similar observations were made with gRNA lacking the 3′-92 nt, produced from pTYMC linearized at the DraI site just upstream of the TLS (data not shown). We note that it is a common observation that 3′-UTRs exhibiting strong translation enhancing effects in cell-based assays (as in the case of the TYMV 3′-UTR) (Matsuda et al. 2004a; Matsuda and Dreher 2004) are not stimulatory in standard cell lysates (Leathers et al. 1993; Tanguay and Gallie 1996; Gallie et al. 2000; Michel et al. 2000).
Our results contrast with the observation that removal of the TLS from TYMV vRNA by oligomer-directed RNase H digestion led to a complete loss of ORF-206 translation, while p69 translation was not affected (Barends et al. 2003; Rudinger-Thirion 2005). We suggest two factors that may explain the differing results. First, RNase H treatment could lead to unplanned scission events, driven by transient oligomer annealing at nontarget sites, that compromise the translation of high MW proteins. The presence of unplanned cleavages is suggested by the appearance of a major new translation product (∼55 kDa) in Figure 6C of Barends et al. (2003). Second, Barends et al. (2003) employed no positive identifications in their assignments of ORF-69 or ORF-206 products and instead relied on 12% SDS-PAGE gels that do not clearly resolve high-molecular-weight products. These considerations cast doubt on the conclusion that TLS removal differentially affected the accumulations of p69 and p141/p206.
Barends et al. (2003) reported that the loss of high-molecular-weight translation products resulting from removal of the TLS could be rescued by addition in trans of the RNA fragment representing the 3′-terminal 260 nt of TYMV RNA, although the resolution of the PAGE system used did not allow a clear indication of which products were rescued. In similarly conducted experiments providing TY-266 RNA (representing the 3′-266 nt of TYMV RNA) in trans, we observed no change in the translation profile directed by full-length TYMV or TYMV RNA lacking the 3′-257 nt including the TLS (Fig. 1E).
Equivalent initiation behavior from AUG69 and AUG206 in the presence of initiation inhibitors
Unable to confirm the reported influence of the TLS on ORF-206 translation in wheat germ extracts, we next repeated experiments using edeine and m7GpppG cap analog as probes for initiation behavior. Barends et al. (2003) reported ORF-differential effects of edeine and cap analog on translation of TYMV vRNA and on valine donation from valyl-TYMV RNA, the latter being taken as an indirect measure of ORF-206 expression. Since edeine impairs tRNA binding to the ribosomal P site, where initiator tRNAi Met binds during initiation, resistance to this inhibitor indicates a noncanonical form of initiation that is likely independent of initiator tRNAi Met, as is translation dependent on the CrPV IRES (Sasaki and Nakashima 2000; Wilson et al. 2000a). This IRES is active in wheat germ extracts (Wilson et al. 2000b) and provided a convenient control for our experiments designed to test whether initiation at AUG206, but not at AUG69, is resistant to edeine and cap analog.
We placed the CrPV IRES between the ORFs of a dicistronic reporter mRNA (Fig. 2A) encoding Renilla luciferase (rLUC) and firefly luciferase (fLUC), downstream of a deletion-bearing EMCV IRES known to suppress reinitiation by ribosomes that have translated the upstream ORF. When produced in 5′-capped form, this RNA represented cap-dependent expression (rLUC) and cap- and tRNAi Met-independent expression (fLUC).
FIGURE 2.
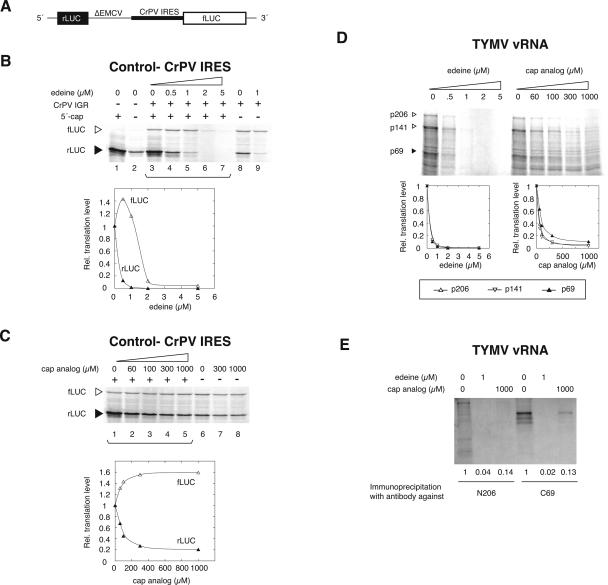
Effect of edeine and free cap analog on translation of TYMV RNA. (A) Diagram of dicistronic control RNA containing the CrPV intergenic region IRES positioned between Renilla (rLUC) and firefly luciferase (fLUC) ORFs. In order to repress reinitiation events, a nonfunctional deleted form of the EMCV IRES (ΔEMCV) was placed after the rLUC ORF. The translation of fLUC is under the control of the CrPV IRES, absent in an additional control RNA (construct not shown; analyzed in panel B, lanes 1,2). (B) Calibration of edeine inhibition using the dicistronic mRNA shown in A. Increasing amounts of edeine (0.5–5 μM) were added to translation reactions programmed with capped or uncapped RNA (20 nM) in the presence of [35S]methionine. Migration positions of fLUC and rLUC in the 12% SDS-PAGE are noted at left. (C) Calibration of cap analog inhibition using the dicistronic mRNA shown in A. Free cap analog (60–1000 μM) was present during the translation of capped or uncapped RNA. (D) Effect of edeine and cap analog on the translation of TYMV virion RNA. Wheat germ extracts were programmed with 20 nM TYMV RNA and products were resolved by 6% SDS-PAGE for quantitation of p206, p141, and p69 by phosphorimagery. The translation reactions in B to D were conducted in parallel. (E) Immunoprecipitation of viral proteins with N206 and C69 antisera (as in Fig. 1) after translation in the presence or absence of edeine and cap analog.
At 0.5 and 1 μM, edeine inhibited the production of rLUC from both uncapped and capped dicistronic control RNA (Fig. 2B, lanes 4,5,9). fLUC expression was dependent on the presence of the CrPV IRES (Fig. 2B, cf. lanes 3,8 and 1,2) and was not inhibited by 0.5 or 1 μM of edeine (Fig. 2B, lanes 4,5). Higher levels of edeine (lanes 6,7) inhibited all translation, consistent with its known ability to repress translational elongation at elevated concentrations (Kozak and Shatkin 1978; Odom et al. 1978). The presence of free m7GpppG cap analog during translation of the capped dicistronic control RNA strongly inhibited expression of rLUC, with no inhibition of fLUC expression (Fig. 2C). Translation of fLUC from the CrPV IRES in fact increased in the presence of cap analog, perhaps because of reduced competition from initiation via the 5′-cap. These experiments established conditions appropriate for distinguishing between conventional initiation (dependent on a cap and initiation factors) and initiation independent of cap and P-site loading (likely involving tRNAi Met).
When TYMV vRNA was translated in the presence of edeine or cap analog, equivalent inhibitory effects on the production of p206, p141, and p69 were observed (Fig. 2D). Protein identifications were once again confirmed by immunoprecipitation with N206 and C69 antisera (Fig. 2E). Strong inhibition of all products by 0.5 and 1 μM of edeine and by 100–1000 μM of cap analog mirrored the expression behavior of rLUC in the dicistronic control RNA (Fig. 2B,C), indicating that both ORF-69 and ORF-206 are expressed by conventional cap-dependent initiation. No indications of noncanonical initiation were observed. These results are consistent with our observations of the 5′-cap-dependency of TYMV RNA translation made from in vivo experiments (Matsuda et al. 2004a).
An additional experiment confirmed the cap-dependency of both ORF-69 and ORF-206 expression in wheat germ extracts. Capped gRNAs, whether full-length or lacking the TLS, supported three- to fivefold higher levels of translation than corresponding uncapped RNAs (data not shown). ORF-69 and ORF-206 expression responded similarly to the presence of a cap.
Disruption of ribosome toe-prints at AUG69 and AUG206 by edeine and cap analog, but not by the absence of the TLS
Since the question surrounding the mechanism of ORF-206 expression concerns the initiation step, we studied this step more directly by using toe-printing analysis, which assays the position of pre-elongation ribosomes stalled in the presence of cycloheximide. As reported previously (Matsuda and Dreher 2006), ribosomes produce very similar toe-prints on vRNA and gRNA (Fig. 3A, lanes 1,2). We now show that the same is true for RNA lacking the 3′-UTR, including the TLS (gRNA-TLS257) (Fig. 3A, lane 3). This result is consistent with our finding that the 3′-TLS is dispensable in the wheat germ translation system (Fig. 1B,C). In all three variants of genomic TYMV RNA, ribosomes are staged at both AUG69 and AUG206, with slightly more ribosomes (1.3- to 1.4-fold) located over AUG69 in each case (Fig. 3A, lanes 1–3). The two major toe-prints are dependent on the presence of AUG codons, as indicated by the loss of toe-prints upon mutation of AUG69 to AAG (gRNA[AUG69KO]) (Fig. 3B, lane 4) or mutation of AUG206 to ACG (gRNA[AUG206KO]) (Fig. 3B, lane 5).
FIGURE 3.
Toe-print analysis tracking translation initiation at AUG69 and AUG 206. (A) Effect of TLS removal. In the presence of cycloheximide, wheat germ extract was programmed with vRNA or with gRNA with and without the TLS (gRNA-TLS257). Radiolabeled primer, which anneals at 194 nt from the 5′-terminus of TYMV RNA, was extended by reverse transcriptase. The position of ribosomes stalled at AUG69 and AUG206 (presumably in the P-site) was confirmed with the loss of the corresponding band by mutation of each start site in gRNA(AUG69KO) and gRNA(AUG206KO). The positions of AUG69 and AUG206 on the sequencing ladder are noted by filled and open stars, respectively. The toe-prints (front edge of the ribosome) are located ~15 nt downstream of each AUG. As observed previously (Matsuda and Dreher 2006), mutation of AUG206 to ACG (lane 5) resulted in increased ribosome recognition of AUG69, a hallmark of modified leaky scanning at two closely spaced AUGs (termed “initiation coupling”). (B) Effect of edeine and cap analog on translation initiation on TYMV virion RNA. Wheat germ extract was first incubated with 2 μM of edeine or 1 mM of cap analog before being programmed with vRNA. Toe-prints were assayed as in panel A. (C) Effect of edeine and cap analog on translation initiation from CrPV IRES. Uncapped dicistronic RNA containing the CrPV IRES was programmed with wheat germ extract with the same treatments used in panel B. The radiolabeled primer (ZW4) was used to detect ribosomes occupying CCU (noted by filled stars in sequencing ladder) and ACA (open stars) in the P-site (Wilson et al. 2000a; Pestova and Hellen 2003). Nucleotide numbers refer to CrPV RNA. Note that the downstream band (arrowhead) is produced in the absence of ribosomes (lane 5) and is not a toe-print.
The addition of 2 μM of edeine to translation reactions (prior to addition of cycloheximide) drastically reduced the ribosomal toe-prints at both AUG69 and AUG206 on TYMV vRNA to 15% and 9%, respectively (Fig. 3B, cf. lanes 2 and 1). In contrast, the toe-print signal representing ribosomes staged after initiation on the control CrPV IRES was retained at a much higher level in the presence of edeine (48%) (Fig. 3C, cf. lanes 2 and 1). The 6-nt upstream shift of the toe-print on CrPV IRES RNA in the presence of edeine was previously reported in rabbit reticulocyte lysate (Wilson et al. 2000a; Pestova and Hellen 2003).
Addition of free cap analog also reduced the toe-print at both AUGs of TYMV vRNA, to about one-fourth the normal signal (Fig. 3B, cf. lanes 3 and 1). This result again contrasts with the behavior of ribosomes on the CrPV IRES, where toe-prints are little affected by the presence of cap analog (Fig. 3C, cf. lanes 3 and 1). These toe-printing studies thus support the experiments of Figures 1 and 2 in showing that initiation at both AUG69 and AUG206 is mechanistically equivalent and is conventional in relying on a 5′-cap and being sensitive to edeine inhibition. We observe no evidence for an alternative form of initiation at AUG206 that is similar to initiation at the CrPV IRES. In summary, our observations have not confirmed the report of Barends et al. (2003) that the 3′-TLS is involved in initiation at AUG206 in wheat germ extracts.
Detection of valine incorporated into translation products from valyl-TYMV TLS RNA by trans-esterification but not by direct donation
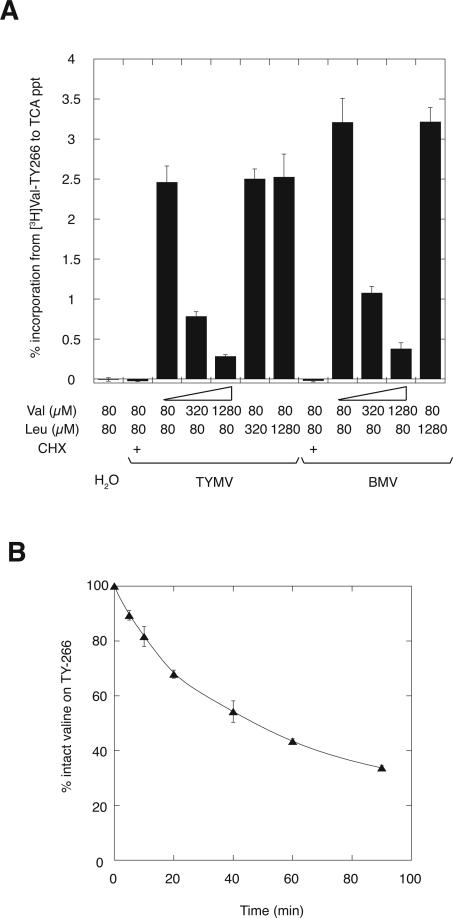
Another core observation underlying the Trojan horse model was the reported incorporation of [3H]valine from valylated TYMV RNA into TYMV translation products. The incorporation rate of [3H]valine into TCA-precipitable material was 4%–6% in cis from valylated TYMV vRNA and ∼3% in trans from valylated TYMV 3′-RNA fragment (Barends et al. 2003). Similarly, we observed that ∼2.5% of input [3H]valine, which was initially aminoacylated to TY-266 RNA, was detected in acid-precipitable substances (measured after RNA removal to exclude detection of valylated RNA) upon incubation in wheat germ extract programmed with TYMV vRNA (Fig. 4A, column 3). Such donation was undetectable in translation reactions with valylated TY-266 RNA as the sole exogenous RNA species (Fig. 4A, column 1). The incorporation of valine was abolished by the inclusion of cycloheximide in the translation reaction (Fig. 4A, column 2) and was progressively suppressed as the concentration of free valine in the incubation was increased (Fig. 4A, columns 3–5), but not as the concentration of free leucine was increased (Fig. 4A, columns 6,7). Similar incorporation properties were observed when BMV RNA was present in the translation extract (Fig. 4A).
FIGURE 4.
Incorporation of valine from valyl-TY266 RNA into translation products. (A) Incorporation of valine from [3H]valyl-TY266 RNA (120 nM) into base-resistant, acid-precipitable form in the presence of TYMV vRNA (40 nM), BMV RNA (0.25 μg/12.5 μL reaction), or neither (H20), in wheat germ extract. Cycloheximide (CHX) was present in control reactions, and free valine and leucine were present at the indicated concentrations. A linear relationship was observed between the amount of [3H]valine incorporated into acid-precipitable material and the specific activity of [3H]valine present in the TYMV and BMV translation reactions. (B) Deacylation of valyl-TY-266 RNA in the course of a translation reaction in wheat germ extract. [3H]valyl-TY266 RNA (120 nM) was incubated with wheat germ extract in the presence of 1280 μM unlabeled valine, and the integrity of the ester bond was monitored over the 90-min incubation.
These results are most consistent with the trans-incorporation of valine subsequent to its deacylation from TY-266 RNA (Fig. 4B) and reacylation to tRNAVal. Our results differ from those of Barends et al. (2003), who reported that incorporation was resistant to the presence of excess free valine and that no significant incorporation into acid-precipitable material occurred in the presence of BMV RNA. The reasons for these differences in experiments carried out with wheat germ extracts from the same source are unclear. However, our experiments do point out that significant deacylation and trans-incorporation can occur during standard incubations with wheat germ extracts, complicating the detection of exotic forms of incorporation like that suggested in the Trojan horse model. The difficulty in accounting for phenomena such as this has previously led to mistaken interpretations of direct translational incorporation of valine from valyl-TYMV RNA (Haenni et al. 1973, 1982).
DISCUSSION
With the present experiments and a previous report (Matsuda and Dreher 2006), we have now presented congruent results from both in vitro and vivo experiments showing that initiation from both AUG69 and AUG206 of TYMV RNA occurs by a conventional mechanism that depends on a 5′-cap and initiator tRNAi Met. We have obtained no evidence that radically different initiation schemes occur at these initiation sites, as proposed by the Trojan horse model. Direct attempts to repeat the key experiments of Barends et al. (2003) have been unable to reproduce the reported observations, in spite of care taken to include additional controls. Our use of positive identification of ORF-69 and ORF-206 products with antibodies and of an alternative way to prepare 3′-truncated genomic RNA lacking the TLS may underlie the conflicting results. With regard to the intriguing observation reporting transfer of valine from valyl-TYMV RNA into protein (Barends et al. 2003), a plausible explanation seems to have been offered by experiments with BMV RNA by the same investigators (Barends et al. 2004). Direct transfer of tyrosine to protein from tyrosyl-BMV RNA, but not from tyrosyl-tRNATyr, occurred in the presence of tyrosyl-tRNA synthetase. This reaction appears to be similar to one previously characterized for aspartyl-tRNA synthetase (Kern et al. 1985), in which the aspartyl moiety became ligated to an amino group in various settings, including lysine end-groups of proteins. Such a reaction catalyzed by valyl-tRNA synthetase could explain the dansyl chloride-sensitive valine incorporation in the presence of TYMV RNA that was interpreted as the positioning of valine at protein N termini (Barends et al. 2003).
Beyond our inability to garner experimental evidence supportive of the Trojan horse model, certain virological arguments make this expression mode unlikely. First, TYMV genome variants with GAC or AAC anticodons that should preclude base-pairing to the AUG206 initiation codon are highly infectious (Tsai and Dreher 1991). Second, a chimeric genome in which the TYMV TLS was replaced with the 3′-UTR from Erysimum latent tymovirus, which contains a vestigial TLS with no anticodon domain (Dreher and Goodwin 1998), is also infectious (Goodwin et al. 1997). Loss of the anticodon would prevent expression of the essential ORF-206 via the Trojan horse model of internal initiation. Third, if the Trojan horse mechanism applies to the expression of TYMV ORF-206 and explains ribosome access to the downstream AUG206, it should equally apply to the expression of the downstream ORFs of other tymoviral RNAs. Tymoviral RNAs all express their replication proteins from an ORF that initiates exactly 7 nt downstream of the first AUG, although the sequence surrounding the AUG initiation codons is not especially conserved (Fig. 5). It reveals no common features that would be expected in organizing the proposed interaction with the TLS anticodon. In addition, tymoviruses such as Erysimum latent virus and Dulcamara mottle virus (I. Tzanetakis and T. Dreher, unpubl.) lack a TLS with an anticodon domain, and Nemesia ring necrosis virus has a distinct, tobamoviral-like TLS (Koenig et al. 2005). There is an obvious lack of conservation in elements that are proposed to be involved in key steps of the Trojan horse model.
FIGURE 5.
Alignment of tymoviral RNA sequences surrounding the closely spaced AUG initiation codons.
MATERIALS AND METHODS
Preparation of RNAs
RNAs for in vitro translation were prepared by transcription with T7 RNA polymerase, in most cases in the presence of trace amounts of 32P-labeled CTP for quantification as described previously (Matsuda et al. 2004a). Capped RNAs were transcribed with mMessage mMachine (Ambion) according to the manufacturer’s recommendations. Uncapped RNAs were produced as described (Matsuda et al. 2004a). TYMV RNA transcripts were generated from pTYMC, which allows production of infectious genomic RNA (Weiland and Dreher 1989). Full-length RNA was produced from HindIII-linearized plasmid, and genomic RNA lacking the TLS was produced from pTYMC linearized with SmaI or DraI.
The dicistronic luciferase construct containing a portion of EMCV sequence that inhibits reinitiation or read-through of a termination codon was a kind gift from Drs. Peter Sarnow and Eric Jan. The dicistronic luciferase construct containing CrPV IRES was constructed as described by Wilson et al. (2000b) except that the CrPV IRES sequence was taken from pEJ6 (a gift from Peter Sarnow and Eric Jan). PCR amplification was used to generate a template for in vitro transcription of the 3′-terminal 266 nt of TYMV RNA (TY-266 RNA). The amplified DNA was purified by separation on a 6% denaturing polyacrylamide gel (Matsuda et al. 2004b). Radioactivity was omitted for the preparation of TY-266 RNA. The integrity of all RNAs was verified by gel electrophoresis.
Wheat germ in vitro translation
Wheat germ extract was purchased from Promega (no. L4380), and used according to the manufacturer’s directions in the presence of 100 mM potassium acetate for 90 min at 25°C. Products were labeled with [35S]methionine (0.86 μM, 43.5 TBq/mmol; Perkin Elmer), resolved by 6% or 12% SDS-PAGE, and detected by phosphorimagery. TYMV RNA present at 2.5 nM (as in Fig. 1) was in the linearly responsive range; higher RNA concentrations were used to detect CrPV IRES activity (Fig. 2) and for toe-prints. Where indicated, the given amounts of edeine and m7GpppG cap analog (Epicentre) were incubated with the wheat germ extract for 5 min at 25°C prior to the addition of mRNA.
Antibodies and immunoprecipitation
Translation products made from virion TYMV RNA were immunoprecipitated as described (Bransom et al. 1991) after incubation with the primary rabbit antisera TY-N206 (raised against the N-terminal domain of p206), TY-C69 (raised against the C-terminal domain of p69) (Bozarth et al. 1992), and anti-CP (raised against TYMV virions).
Ribosome toe-print assay
Toe-print assays (primer extension-inhibition assay) were performed as described (Matsuda and Dreher 2006) using the Promega wheat germ extract, with cycloheximide added at time 0. The signal ratio for toe-prints at AUG69 to AUG206 (1.3–1.4) (Fig. 3A) differed from the ratio of 0.43 previously observed (Matsuda and Dreher 2006), presumably because of the different source and amounts of wheat germ extract used (50% of the reaction volume used in this study, rather than 30%). The primer used with the dicistronic luciferase RNA that contains the CrPV IRES was ZW4 (5′-TCCAGGAACCAGGGCGTA-3′), priming in the fLUC coding region (Wang and Sachs 1997). For toe-prints assayed in the presence of edeine or cap analog, these reagents were added as described for translation reactions.
Assessment of incorporation of valine from val-TYMV TLS into translation products
Gel-purified TY-266 RNA was aminoacylated with [3H]valine (24 Ci/mmol; NEN) as described previously (Matsuda et al. 2004b), followed by removal of unincorporated [3H]valine with a Micro Bio-Spin 6 column (Bio-Rad) pre-equilibrated with 5 mM of sodium acetate (pH 5.5) and ethanol precipitation. Integrity and quantity of RNA were monitored by 6% denaturing PAGE. Valylation extent of TY-266 RNA (∼60%) was confirmed by TCA precipitation onto paper filters and liquid scintillation counting.
To study valine trans-incorporation into protein, TYMV virion RNA (1 μg; ∼0.5 pmol) or Brome mosaic virus (BMV) virion RNA (Promega, 0.25 μg) was incubated with [3H]valyl-TY-266 RNA (1.5 pmol) in 12.5 μL translation reactions containing 1000 pmol of each amino acid. Some translation reactions contained excess amounts of either unlabeled valine or leucine (4 or 16 nmol). Translation reactions were carried out in the presence or absence of 1 mg/mL cycloheximide for 90 min at 25°C. Reactions were terminated by the addition of 1.25 μL of 770 mM NaOH followed by further incubation for 15 min at 25°C. This step ensures deacylation of >99% [3H]valine from RNA (data not shown). The [3H]valine incorporation was analyzed by TCA precipitation onto filters and liquid scintillation counting (Dreher et al. 1999).
Deacylation of val-TYMV TLS during translation reaction
[3H]valyl-TY-266 RNA (120 nM) was incubated with wheat germ extract in the absence of other exogenous RNA up to 90 min at 25°C. At each specified time, a portion of the reaction was TCA precipitated on a filter, and analyzed by liquid scintillation counting.
ACKNOWLEDGMENTS
We thank Peter Sarnow and Eric Jan (Stanford University) for providing the dicistronic plasmid and pEJ6; Matthew Sachs and Anthony Gaba for instruction and advice in the toe-printing procedure; and George Rohrmann, Matthew Sachs, and members of the Dreher laboratory for help with the manuscript. This work was supported by NSF grant MCB0235563.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.244407.
REFERENCES
- Barends, S., Bink, H.H., van den Worm, S.H., Pleij, C.W., Kraal, B. Entrapping ribosomes for viral translation: tRNA mimicry as a molecular Trojan horse. Cell. 2003;112:123–129. doi: 10.1016/s0092-8674(02)01256-4. [DOI] [PubMed] [Google Scholar]
- Barends, S., Rudinger-Thirion, J., Florentz, C., Giegé, R., Pleij, C.W., Kraal, B. tRNA-like structure regulates translation of Brome mosaic virus RNA. J. Virol. 2004;78:4003–4010. doi: 10.1128/JVI.78.8.4003-4010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozarth, C.S., Weiland, J.J., Dreher, T.W. Expression of ORF-69 of turnip yellow mosaic virus is necessary for viral spread in plants. Virology. 1992;187:124–130. doi: 10.1016/0042-6822(92)90301-5. [DOI] [PubMed] [Google Scholar]
- Bransom, K.L., Weiland, J.J., Dreher, T.W. Proteolytic maturation of the 206-kDa nonstructural protein encoded by turnip yellow mosaic virus RNA. Virology. 1991;184:351–358. doi: 10.1016/0042-6822(91)90851-2. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W. Pathogen profile. Turnip yellow mosaic virus: Transfer RNA mimicry, chloroplasts and a C-rich genome. Mol. Plant Pathol. 2004;5:367–375. doi: 10.1111/j.1364-3703.2004.00236.x. [DOI] [PubMed] [Google Scholar]
- Dreher, T.W., Goodwin, J.B. Transfer RNA mimicry among tymoviral genomic RNAs ranges from highly efficient to vestigial. Nucleic Acids Res. 1998;26:4356–4364. doi: 10.1093/nar/26.19.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher, T.W., Uhlenbeck, O.C., Browning, K.S. Quantitative assessment of EF-1α.GTP binding to aminoacyl-tRNAs, aminoacyl-viral RNA, and tRNA shows close correspondence to the RNA binding properties of EF-Tu. J. Biol. Chem. 1999;274:666–672. doi: 10.1074/jbc.274.2.666. [DOI] [PubMed] [Google Scholar]
- Gallie, D.R., Ling, J., Niepel, M., Morley, S.J., Pain, V.M. The role of 5′-leader length, secondary structure and PABP concentration on cap and poly(A) tail function during translation in Xenopus oocytes. Nucleic Acids Res. 2000;28:2943–2953. doi: 10.1093/nar/28.15.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin, J.B., Skuzeski, J.M., Dreher, T.W. Characterization of chimeric turnip yellow mosaic virus genomes that are infectious in the absence of aminoacylation. Virology. 1997;230:113–124. doi: 10.1006/viro.1997.8475. [DOI] [PubMed] [Google Scholar]
- Haenni, A.L., Prochiantz, A., Bernard, O., Chapeville, F. TYMV valyl-RNA as an amino-acid donor in protein biosynthesis. Nat. New Biol. 1973;241:166–168. doi: 10.1038/newbio241166a0. [DOI] [PubMed] [Google Scholar]
- Haenni, A.L., Joshi, S., Chapeville, F. tRNA-like structures in the genomes of RNA viruses. Prog. Nucleic Acid Res. Mol. Biol. 1982;27:85–104. doi: 10.1016/s0079-6603(08)60598-x. [DOI] [PubMed] [Google Scholar]
- Jan, E. Divergent IRES elements in invertebrates. Virus Res. 2005;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Kern, D., Lorber, B., Boulanger, Y., Giegé, R. A peculiar property of aspartyl-tRNA synthetase from bakers’ yeast: Chemical modification of the protein by the enzymatically synthesized aminoacyl adenylate. Biochemistry. 1985;24:1321–1332. [Google Scholar]
- Koenig, R., Barends, S., Gultyaev, A.P., Lesemann, D.E., Vetten, H.J., Loss, S., Pleij, C.W. Nemesia ring necrosis virus: A new tymovirus with a genomic RNA having a histidylatable tobamovirus-like 3′ end. J. Gen. Virol. 2005;86:1827–1833. doi: 10.1099/vir.0.80916-0. [DOI] [PubMed] [Google Scholar]
- Kozak, M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M., Shatkin, A.J. Migration of 40 S ribosomal subunits on messenger RNA in the presence of edeine. J. Biol. Chem. 1978;253:6568–6577. [PubMed] [Google Scholar]
- Leathers, V., Tanguay, R., Kobayashi, M., Gallie, D.R. A phylogenetically conserved sequence within viral 3′ untranslated RNA pseudoknots regulates translation. Mol. Cell. Biol. 1993;13:5331–5347. doi: 10.1128/mcb.13.9.5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, D., Dreher, T.W. The tRNA-like structure of Turnip yellow mosaic virus RNA is a 3′-translational enhancer. Virology. 2004;321:36–46. doi: 10.1016/j.virol.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Matsuda, D., Dreher, T.W. Close spacing of AUG initiation codons confers dicistronic character on a eukaryotic mRNA. RNA. 2006;12:1338–1349. doi: 10.1261/rna.67906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, D., Bauer, L., Tinnesand, K., Dreher, T.W. Expression of the two nested overlapping reading frames of turnip yellow mosaic virus RNA is enhanced by a 5′ cap and by 5′ and 3′ viral sequences. J. Virol. 2004a;78:9325–9335. doi: 10.1128/JVI.78.17.9325-9335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda, D., Yoshinari, S., Dreher, T.W. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004b;321:47–56. doi: 10.1016/j.virol.2003.10.028. [DOI] [PubMed] [Google Scholar]
- Michel, Y.M., Poncet, D., Piron, M., Kean, K.M., Borman, A.M. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- Odom, O.W., Kramer, G., Henderson, A.B., Pinphanichakarn, P., Hardesty, B. GTP hydrolysis during methionyl-tRNAf binding to 40 S ribosomal subunits and the site of edeine inhibition. J. Biol. Chem. 1978;253:1807–1816. [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudinger-Thirion, J., Olsthoorn, R.C.L., Giegé, R., Barends, S. Idiosynchratic behaviour of tRNA-like structures in translation of plant viral RNA genomes. J. Mol. Biol. 2005;355:873–878. doi: 10.1016/j.jmb.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Sasaki, J., Nakashima, N. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc. Natl. Acad. Sci. 2000;97:1512–1515. doi: 10.1073/pnas.010426997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, C.-H., Dreher, T.W. Turnip yellow mosaic virus RNAs with anticodon loop substitutions that result in decreased valylation fail to replicate efficiently. J Virol. 1991;65:3060–3067. doi: 10.1128/jvi.65.6.3060-3067.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay, R.L., Gallie, D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Sachs, M.S. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa . Mol. Cell. Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiland, J.J., Dreher, T.W. Infectious TYMV RNA from cloned cDNA: Effects in vitro and in vivo of point substitutions in the initiation codons of two extensively overlapping ORFs. Nucleic Acids Res. 1989;17:4675–4687. doi: 10.1093/nar/17.12.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000a;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- Wilson, J.E., Powell, M.J., Hoover, S.E., Sarnow, P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000b;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]