Abstract
Estimates of missense error rates (misreading) during protein synthesis vary from 10−3 to 10−4 per codon. The experiments reporting these rates have measured several distinct errors using several methods and reporter systems. Variation in reported rates may reflect real differences in rates among the errors tested or in sensitivity of the reporter systems. To develop a more accurate understanding of the range of error rates, we developed a system to quantify the frequency of every possible misreading error at a defined codon in Escherichia coli. This system uses an essential lysine in the active site of firefly luciferase. Mutations in Lys529 result in up to a 1600-fold reduction in activity, but the phenotype varies with amino acid. We hypothesized that residual activity of some of the mutant genes might result from misreading of the mutant codons by tRNALys UUUU, the cognate tRNA for the lysine codons, AAA and AAG. Our data validate this hypothesis and reveal details about relative missense error rates of near-cognate codons. The error rates in E. coli do, in fact, vary widely. One source of variation is the effect of competition by cognate tRNAs for the mutant codons; higher error frequencies result from lower competition from low-abundance tRNAs. We also used the system to study the effect of ribosomal protein mutations known to affect error rates and the effect of error-inducing antibiotics, finding that they affect misreading on only a subset of near-cognate codons and that their effect may be less general than previously thought.
Keywords: protein synthesis, accuracy, misreading errors, paromomycin, streptomycin
INTRODUCTION
To insure survival, cells must accurately and efficiently translate information encoded in messenger RNA (mRNA) sequences into proteins. Maximizing accuracy and efficiency puts conflicting demands on the translational machinery (for review, see Parker 1989). The resolution of these conflicting demands has involved the evolution of multiple accuracy mechanisms. These mechanisms include substrate recognition and proofreading steps. For example, during aminoacylation of tRNAs, aminoacyl-tRNA synthases accept correct amino acids that fit into a pocket in the substrate-binding site but reject poorly fitting incorrect substrates (Sankaranarayanan and Moras 2001). Discrimination based on size and shape of similar amino acids (e.g., valine and isoleucine) is insufficient to explain the observed accuracy, as originally suggested by Pauling (1957). A proofreading step, in which incorrectly acylated amino acids are hydrolyzed, further increases accuracy at the cost of energy (Sankaranarayanan and Moras 2001). Aminoacylation errors occur approximately once in 106 events (Söll 1990; Schulman 1991). Similar two-stage error correction schemes involving a proofreading step insure accuracy in all other steps of genetic information transfer, including DNA replication, RNA transcription, and translation of mRNA sequences into proteins (for review, see Galas et al. 1986).
Aminoacylation is much more accurate than other steps in protein synthesis, however, and most errors occur during the process of mRNA decoding, which occurs on the ribosome by the recruitment of aminoacyl-tRNAs (aa-tRNAs) to the aa-tRNA-binding site, the A site. Estimates of the frequency of missense errors range from about 10−3 to 10−4 per codon (Loftfield and Vanderjagt 1972; Edelmann and Gallant 1977; Parker et al. 1980, 1983; Ellis and Gallant 1982; Bouadloun et al. 1983; Khazaie et al. 1984; Parker and Holtz 1984; Rice et al. 1984; Laughrea et al. 1987; Toth et al. 1988; Ulrich et al. 1991; Weickert and Apostol 1998). Based on these data, Parker (1989) estimated an average frequency no higher than 4 × 10−4 per codon.
For 30 years the prevailing model considered kinetic proofreading (Hopfield 1974; Ninio 1975) the major determinant of translational accuracy. Under this model, ribosomes would use the greater intrinsic instability of incorrect aminoacyl-tRNA complexes on the ribosome to select against them in a two-stage discrimination reaction (for review, see Thompson 1988; Rodnina et al. 2005). Aminoacyl-tRNAs bind to the ribosomal A site in a ternary complex with elongation factor 1A (EF-1A) and GTP. The model proposed that incorrect ternary complexes dissociated much more rapidly than correct complexes in a first stage of discrimination (initial binding). Ribosome binding of the ternary complex stimulates GTP hydrolysis, after which, in a second discrimination stage (proofreading), incorrect aminoacyl-tRNAs again tend to dissociate from the ribosome•EF1A•GDP complex, while correct ones remain bound. Under this model, a 100-fold preference for correct tRNAs at each stage compound to provide the observed ∼10,000-fold preference (Thompson 1988). These effects result from a difference in the equilibrium binding constants for correct (cognate) and incorrect (near and noncognate) complexes. More recent work shows that these thermodynamic effects cannot explain the observed accuracy (for review, see Rodnina et al. 2005). Instead, the most critical parameter in discrimination is the rate of activation of the intrinsic GTPase of EF-1A, which is almost three orders of magnitude faster for correct complexes. The ribosome cannot maximize discrimination using these differences in tRNA stability because it proceeds to the next step in the reaction sequence too rapidly to allow the full thermodynamic difference to be achieved (Gromadski and Rodnina 2004a).
The preference for cognate tRNA results from changes in the ribosome structure created by an induced fit mechanism. Cognate codon–anticodon complexes form a web of interactions with nucleotides in the rRNA and portions of the ribosomal protein rpS12 (Ogle et al. 2001, 2002). These contacts require the geometry of Watson–Crick base pairs (bp), which do not form when a near-cognate codon binds the A site (Ogle et al. 2002). Our current model of discrimination argues that interactions with the cognate codon–anticodon complex both stabilize correct complexes and cause the large-scale ribosomal rearrangement that ultimately increases the rate of GTPase activation (for review, see Rodnina et al. 2005).
Why do the estimates of missense errors vary so widely? The source of variation appears to be that different studies have measured different errors involving mismatches at different positions of the 3-bp codon•anticodon complex. In addition, the studies used different methods and reporter systems in detecting these errors, making it difficult to compare the results. For example, several investigators relied on the knowledge that some Escherichia coli proteins naturally do not contain particular amino acids, allowing them to estimate misincorporation by measuring the amounts of these amino acids incorporated in vivo. Measurements of misincorporation of cysteine into flagellin provided an estimate, a frequency of about 10−4 per codon for the first position misreading of the CGU/C arginine codons by Cys GCA (Edelmann and Gallant 1977). Subsequently, researchers have measured misincorporation of cysteine into ribosomal proteins (Bouadloun et al. 1983; Laughrea et al. 1987) or the bacteriophage T7 0.3 protein (Rice et al. 1984), and misincorporation of histidine, tryptophan, and lysine into bacteriophage Qβ proteins (Khazaie et al. 1984). Misreading in these cases involved errors at each of the three codon positions; the misincorporation rates varied from 2 × 10−2 (Khazaie et al. 1984) to 2.5 × 10−4 (Bouadloun et al. 1983; Laughrea et al. 1987).
A second method to estimate error frequency relies on changes in charge caused by the substitution of an amino acid resulting in an altered isoelectric point. The charge difference allows separation of mistranslated forms of a protein from correctly translated species during two-dimensional gel electrophoresis. A series of studies on translation of the MS2 coat protein in E. coli used this method to detect lysine for asparagine substitutions (Parker and Friesen 1980; Parker et al. 1980, 1983; Johnston et al. 1984; Parker and Holtz 1984). They estimated that tRNALys UUU misreads the third position of AAU at a rate of 2–6 × 10−3 per codon and AAC at about 2 × 10−4 per codon. In a second study, the frequency of lysine for asparagine substitutions in the Qβ coat protein was estimated at 5–6 × 10−3 (Khazaie et al. 1984).
A third approach uses a reporter gene bearing a mutation altering an essential amino acid (Toth et al. 1988; Cornut and Willson 1991). Misincorporation of the wild-type amino acid at the mutant codon would restore enzymatic activity. The misincorporation frequency is approximately the ratio of the enzyme activity expressed from the mutant to that of the wild-type control. Toth et al. (1988) estimated the frequency that tRNASer GCU misreads a GGC glycine codon as about 10−3 per codon using a mutant form of β-lactamase.
Each of these studies considered at most a few of the possible errors for a particular tRNA. Thus, the reported estimated error frequency averages a fraction of all potential errors. The average includes rates of many distinct events, differing in the type of mispaired nucleotides, the codon position affected, the identity of the codon, and the reporter gene used. It is therefore unclear how each of these parameters affects the variability in reported error rates. In particular, no study has estimated error frequency variability among the possible errors at a single codon in E. coli. In this report, we describe a single, positive reporter system that measures the frequency of all near-cognate misreading errors by tRNALys UUU at a single position in the firefly luciferase gene. We show that the misreading error frequency does, in fact, vary widely. In addition, we show that misreading frequency reflects not simply the details of codon•anticodon pairing, but depends critically on how cognate and near-cognate aa-tRNAs compete for the codon. We have used this system to study the effect on misreading of error-inducing antibiotics and ribosomal protein mutations that either increase or decrease errors. We found significant misreading of only a subset of the near-cognate codons and no significant effect on any noncognate. This suggests that these perturbations have a far less general effect than was previously thought.
RESULTS
The frequency of translational misreading errors varies widely in E. coli
In order to better understand the range of translational missense error rates in vivo in E. coli, we developed a reporter system to quantify the frequency of each type of misreading error at a defined codon. Our approach was to measure activity of mutant forms of the firefly luciferase (F-luc) enzyme with inactivating mutations in an essential active-site lysine residue. The chosen codon, Lysine 529 (K529) of F-luc, highly conserved within a superfamily of adenylate-forming enzymes, orients the substrates ATP and luciferin in the active site (Branchini et al. 2000). The substrates make multiple contacts with the lysine side chain. No other side chain could interact equivalently, suggesting that any protein lacking this residue should be inactive. As expected, missense mutations at K529 reduce enzymatic activity up to 1600-fold; even a conservative arginine for lysine substitution has ∼625-fold lower activity. These proteins, however, retain significant activity, which we hypothesized might result from mistranslation of the mutant transcripts to incorporate lysine at position 529, producing a small proportion of wild-type enzyme. This activity would be a measure of misreading by tRNALys UUUU, the cognate tRNA for the lysine codons AAA and AAG.
To accurately measure the residual activity of mutant forms of F-luc we used a dual reporter system consisting of a translational fusion of F-luc to a second luciferase from Renilla, R-luc. Our system is derived from one created by Grentzmann et al. (1998) as described in Materials and Methods. Because the two proteins are expressed as a single polypeptide, their relative concentrations are always identical. Any difference in the activity of F-luc relative to R-luc, therefore, must reflect a change in the activity of the protein. Also, because the reaction conditions of the two luciferases are so different, it is possible in a single tube to assay each enzyme in succession in a sample using an automated luminometer.
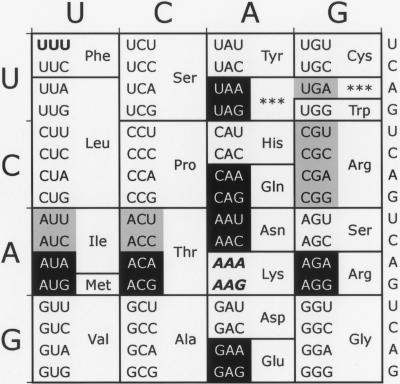
To test all possible near-cognate misreading errors by tRNALys UUUU, we replaced the K529 codon with all codons that differ by a single nucleotide from the lysine codons AAA or AAG (Fig. 1). In each case, tRNALys UUUU can recognize the mutant codon as a near-cognate substrate with only one base mismatch. We also replaced the AAA lysine codon with a UUU phenylalanine codon. Because tRNALys UUUU can form no base pairs with UUU, the codon serves as the negative control for misreading; any activity observed for this mutant must have a different cause. We have expressed these mutant forms of F-luc in E. coli and have measured their activity to estimate the frequency with which tRNALys UUUU misreads each near-cognate codon during normal translation and in the presence of various error-inducing antibiotics and ribosomal mutations that either increase or decrease translational accuracy.
FIGURE 1.
Near- and noncognate mutations for Lysine-529. The two Lys codons are shown in italics in this standard genetic code table. Near-cognate codons, differing at one codon position, are shown in reverse on black. Noncognate synonymous codons, differing in more than one codon position, are shown in black on gray. The control UUU codon is shown in boldface.
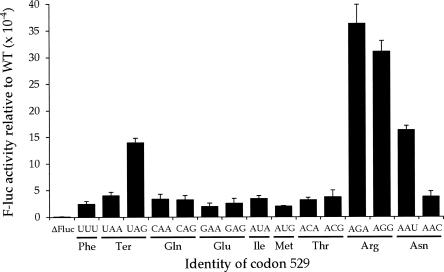
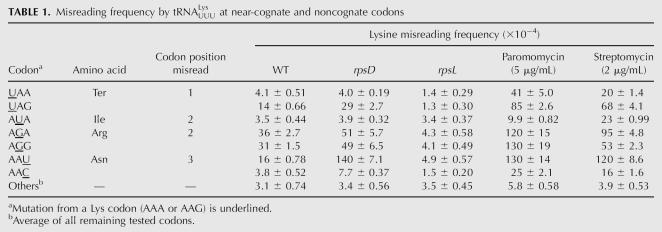
Among the 14 near-cognate codon 529 mutants, the F-luc activity varies by a factor of >10 (Fig. 2). Ten of the mutants had activities 1.6-fold higher than the UUU control. These mutants had an average apparent error frequency, determined as the ratio of mutant to the wild-type activities, of 3.7 × 10−4. Four codons (UAG, AGG, AGA, and AAU) had significantly higher activity (P < 0.05, determined by a homoscedastic two-tailed Student's t-Test, which assumes equal variance), up to 15-fold higher than the control. The apparent error frequencies for these mutants ranged from a low of 1.4 × 10−3, for UAG, to 3.6 × 10−3 per codon, for AGA (Table 1). These highly error-prone codons include a termination codon (UAG), two arginine codons (AGA, AGG), and an asparagine codon (AAU).
FIGURE 2.
Variation in F-luc activity of K5290 mutants. The graph shows the F-luc expression of the indicated constructs expressed as a fraction of the expression of wild-type F-luc. The mutants include a deletion of the entire firefly luc gene (Fluc) and those replacing the K529 codon (AAA) with the indicated codon. Indicated below each codon is the amino acid it encodes. Error bars are standard errors of the mean.
TABLE 1.
Misreading frequency by tRNALys UUUU at near-cognate and noncognate codons
Two mechanisms could explain the higher activity of some near-cognate mutants: partial activity of the mutant protein (functional replacement) or production of small amounts of wild-type protein (misreading). Three of the mutations introduce conservative substitutions, from lysine to arginine (K529R) or asparagine (K529N). These amino acids could provide partial enzymatic function. This model fails to explain the UAG nonsense mutant, however, because a truncated protein resulting from premature termination should have no activity. In addition, the two mutations introducing arginine (encoded by AGA and AGG) and the two introducing asparagine (encoded by AAU and AAC) under this hypothesis should produce proteins with identical sequences and, therefore, identical activities. The error frequencies for AGA (3.6 × 10−3) and AGG (3.1 × 10−3) are quite similar, but those for AAU (1.6 × 10−3) and AAC (3.8 × 10−4) differ significantly, which is inconsistent with the functional replacement model.
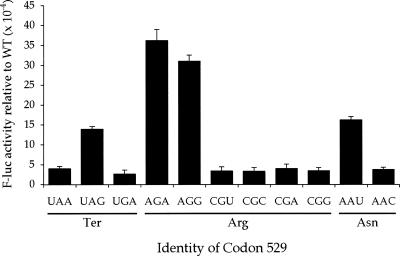
Under the misreading model, the residual activity would result from a small amount of active (incorrectly translated) molecules in a background of an excess of inactive (correctly translated) molecules. The misreading model makes a strong prediction about the phenotype of mutations that replace lysine codons with noncognate codons. Because these codons make two or more mismatches with the anticodon of tRNALys UUUU, they should allow much lower rates of misreading and therefore have much lower activity. The functional replacement model, in contrast, predicts that all synonymous codon mutations, those introducing the same amino acid, should have equivalent activity. To test these predictions we measured the activity of all mutations involving codons synonymous with the four near-cognate mutants with the highest activity. AAU, UAG, AGA, and AGG have a total of six synonymous codons that are noncognate codons for tRNALys UUUU: the termination codons UAA, and UGA and the arginine codons CGU, CGC, CGA, and CGG (Fig. 1). As shown in Figure 3, the activities of the six mutants involving noncognate synonymous codons were all near that of the noncognate UUU mutant and much lower than the synonymous near-cognate codon mutations. These data are inconsistent with the functional replacement model and consistent with the misreading model, suggesting that the unusually high activities of the UAG, AGA, AGG, and AAU mutants result from near-cognate reading of the mutant codons by tRNALys UUUU.
FIGURE 3.
High residual F-luc activity of some K529 codons results from near-cognate decoding. A comparison of the F-luc activity relative to wild type of synonymous near- and noncognate codon mutants. Codon identity is shown as in Figure 2.
Translational misreading errors are affected by competition between cognate and near-cognate tRNA species
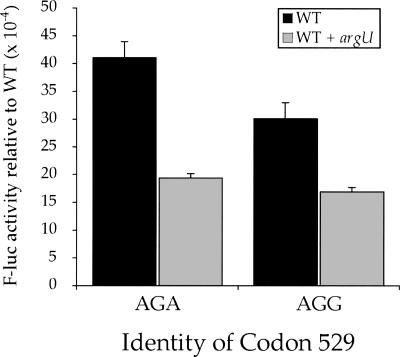
The two codons misread most frequently by tRNALys UUUU, AGA and AGG, are among the most rare codons in the E. coli genome and are normally decoded by the minor tRNAArg UCU (Ikemura 1981). The kinetic rate constant for codon recognition measured in vitro is the same for all tRNAs, 100–140 (μM aa-tRNA)−1 sec−1 (Gromadski et al. 2006), but because the rate constant is first-order with respect to the tRNA concentration, a low-abundance tRNA should recognize its cognate codon less rapidly than an abundant tRNA. We suspect that the slow recognition by their cognate tRNA explains the extremely high error frequency at AGA and AGG codons.
If the low abundance of tRNAArg UCU is responsible for frequent errors on AGA and AGG, then overexpressing tRNAArg UCU should reduce the frequency that they are decoded as lysine. The argU gene encodes tRNAArg UCU. To overexpress this tRNA species, we transformed the low-copy plasmid pAlter-Ex2/argU (Ruzicka et al. 2000) along with the reporter plasmid bearing the K529R(AGA) and K529R(AGG) mutant forms of F-luc. As shown in Figure 4, misreading of AGA and AGG by near-cognate tRNALys UUUU decreased almost twofold in the presence of excess cognate tRNAArg UCU. This strongly suggests that competition for decoding between cognate and near-cognate tRNAs present at different cellular concentrations can control the misreading error frequencies.
FIGURE 4.
Overexpression of tRNAArg UCU reduces misreading errors at Arg codons. The F-luc activity relative to wild type is shown for mutants replacing the K529 codon with the Arg codons AGA or AGG. Black bars indicate expression in a wild-type genetic background (WT) and gray bars indicate expression in the presence of overexpressed tRNAArg UCU.
The aminoglycoside antibiotics streptomycin and paromomycin increase misreading of some near-cognate codons
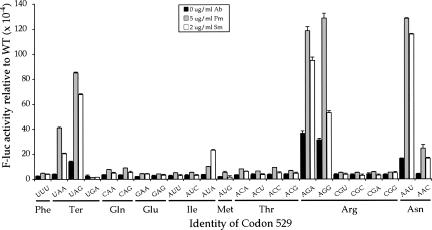
As a further test of whether the excess activities observed with some of the near-cognate codon mutants results from misreading, we measured the effect of error-inducing antibiotics on the activity of near-cognate and noncognate mutant forms of F-luc. At sublethal concentrations, the aminoglycoside antibiotic streptomycin can cause phenotypic reversion, suggesting that it reduces translational accuracy (for review, see Kurland et al. 1996). Mutations resistant to streptomycin map to the rpsL gene, which encodes ribosomal protein S12 (rpS12) and causes translational hyperaccuracy. Very strongly hyperaccurate mutants are streptomycin dependent (StrD) because their viability requires the accuracy-reducing effect of streptomycin. Aminoglycosides like streptomycin or paromomycin increase misreading by accelerating the rate of acceptance of near-cognate aa-tRNAs relative to cognates (Pape et al. 2000). We were interested to see whether the accuracy effects of these antibiotics varied with identity of the codon being misread.
Surprisingly, paromomycin strongly increased the error frequency of only a subset of the 14 near-cognate K529 mutants and had little or no effect on any of the noncognate mutants (Fig. 5). For each of the four especially error prone near-cognate mutants, UAG, AGA, AGG, and AAU, misreading increased from 3.3- to 7.9-fold in the presence of paromomycin (Table 1). The estimated misreading frequencies were extremely high at three codons, AGA, AGG, and AAU, with the frequencies rising to >10−2 per codon. In addition, misreading on two other codons, UAA (stop) and AAC (Asn), was very significantly above background, increasing by a factor of 10-fold for UAA and 6.4-fold for AAC. The effect of paromomycin on the remaining 18 mutants tested averaged less than twofold with an average error frequency of 5.8 × 10−4 per codon.
FIGURE 5.
Aminoglycoside antibiotics increase misreading on a subset of near-cognate codons. The F-luc activities relative to wild type are shown for mutants carrying the indicated K529 codon replacements in the presence of no antibiotic (black bars), 5 μg/mL paromomycin (gray bars), or 2 μg/mL streptomycin (white bars).
The increases in misreading caused by streptomycin were qualitatively similar. The drug caused large increases in misreading at the same six codons affected most strongly by paromomycin: UAA, UAG, AGA, AGG, AAU, and AAC as well as at a seventh codon, AUA (Fig. 5; Table 1). The average increase in misreading for these codons was 4.6-fold, to an average misreading frequency of 5.6 × 10−3. The remaining 17 codons tested showed no significant increase in misreading (P > 0.05). These codons showed an average 1.3-fold increase in misreading to an average frequency of 3.9 × 10−4.
Streptomycin and paromomycin significantly increased misreading of only a subset of codons near cognate for tRNALys UUU. Among those affected were all but one of the codons requiring a U•U mismatch in the first or second codon positions; UAA, UAG, and AUA showed an increase, but AUG did not. The other four codons affected included the two requiring mispairing at the wobble position (AAU and AAC) and the Arg codons, AGA and AGG. Grosjean et al. (1978) noted that mismatches involving uridines had the least destabilizing effect on codon–anticodon complexes, that U•U mismatches were especially stable as were U•G mismatches in the middle codon position. They also showed that U•U and U•C wobble mismatches could readily be formed (Grosjean et al. 1978). Their results are completely consistent with our observations.
An rpsD mutation increases misreading of several near-cognate substrates
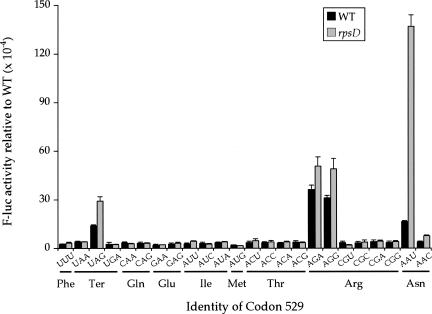
Mutations in the rpsD gene, encoding ribosomal protein S4 (rpS4), have an effect on translational accuracy similar to that of the aminoglycoside antibiotics, causing increased ribosomal ambiguity (i.e., decreased accuracy) (Gorini 1971). The effect of rpsD mutations on several misreading errors has been demonstrated in E. coli. For example, a rpsD mutation increased the third position G•G misreading of the UGG tryptophan codon by tRNACys GCA during translation of ribosomal protein S6 (Bouadloun et al. 1983) and increased the misincorporation of cysteine into the T7 0.3 protein as well (Rice et al. 1984). A first position U•G misreading of GGC (Gly) by tRNASer GCU was also significantly increased by the rpsD mutation (Toth et al. 1988). It has, however, also been reported that the rpsD mutation had no effect on the first position A•C misreading of CGU (Arg) by tRNACys GCA (Bouadloun et al. 1983) or on the misincorporation of histidine into the Qβ protein (Khazaie et al. 1984).
We decided to test the effect of an rpsD mutation on the full spectrum of mutations at the K529 codon. We measured the F-luc activity of each near-cognate codon in rpsD12 mutants to determine the effect on each type of misreading event (Fig. 6). This rpsD12 mutation is a deletion of nucleotides C528-A532, which leads to a truncated 180 amino acid S4 protein (Dahlgren and Ryden-Aulin 2000). The rpsD12 mutation significantly increased misreading of a subset of the codon 529 mutations that responded to the aminoglycoside antibiotics. There were statistically significant increases in misreading only from the codons UAG, AGA, AGG, AAU, and AAC (P < 0.05), but little or no effect on the remaining codons. The codon AAU had the largest increase in misreading, to 1.4 × 10−2 per codon, or 8.4-fold more than in a wild-type background (Table 1). This effect was higher than the effect of the aminoglycoside antibiotics, but the effect on the other four codons was substantially lower. The average increase for all other codons tested was only 1.8-fold compared with a 3.4-fold effect by streptomycin and a 5.0-fold effect of paromomycin on the same codons.
FIGURE 6.
A mutation in rpsD increases misreading only of error-prone codons. The F-luc activities relative to wild type of the indicated K529 codon replacements are shown from a wild-type strain (black bars) and a strain carrying the rpsD12 mutation (gray bars).
An rpsL mutation reduces all misreading of error-prone codons to near-background levels
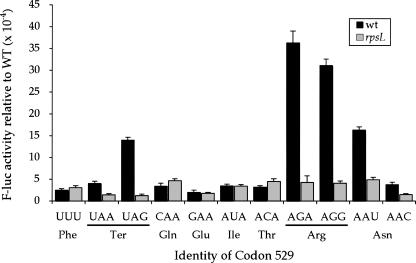
Several studies have demonstrated codon-specific hyperaccuracy caused by mutant forms of rpsL, which encodes rpS12. For example, Parker and colleagues reported decreased misreading of the AAN asparagine codons by tRNALys UUU and U•U and C•U misreading of phenylalanine codons by tRNALeu UAA (Parker and Friesen 1980; Parker et al. 1980, 1983; Johnston et al. 1984; Parker and Holtz 1984; Precup et al. 1989). Other studies reported decreased misreading in rpsL mutants of arginine, tryptophan, and phenylalanine codons by tRNACys (GCA) (Bouadloun et al. 1983; Laughrea et al. 1987) and of unspecified codons by tRNACys (Rice et al. 1984). Yet, an rpsL mutation had little or no effect on misreading of some arginine, tryptophan, and phenylalanine codons by tRNACys (Laughrea et al. 1987) or on misreading of the GGC glycine codon by tRNASer (GCU) (Toth et al. 1988).
We measured the F-luc activity of the K529F(UUU) mutant and of 10 mutants with near-cognate substitutions at codon 529 in bacteria carrying the mutation rpsL141, which introduces a K42N mutation in rpS12 (Andersson et al. 1982; Ruusala et al. 1984). This mutation confers a hyperaccurate phenotype. K42 of rpS12 hydrogen bonds to C912 in Helix 18 of the 16S rRNA, a component of the ribosomal A site, and is close to the codon•anticodon complex. This mutation reduced misreading of the six error-prone near-cognate codons, UAA, UAG, AGA, AGG, AAU, and AAC, but had little or no effect on the others (Fig. 7). In the rpsL141 background, none of the tested codons had a frequency of misreading higher than 5 × 10−4 and three of the codons whose misreading is affected (UAA, UAG, and AAC) showed extremely low error frequencies of 1.3–1.5 × 10−4 (Table 1). Misreading at the affected codons dropped an average of sixfold with the largest affect, on UAG, being 10-fold. We conclude that rpsL141 reduces the frequency of misreading of error-prone near-cognate codons to or below the background level typical of other codons, but that it does not reduce this background level of error.
FIGURE 7.
A mutation in rpsL reduces misreading only of error-prone codons. The F-luc activities relative to wild type of the indicated K529 codon replacements are shown from a wild-type strain (black bars) and a strain carrying the rpsL141 mutation (gray bars).
Discussion
Here we report the whole range of possible misreading errors by a single tRNA species at a single site. The results of this work provide an unbiased comparison of relative frequencies of all the possible errors. Using mutant forms of the firefly luciferase gene with point mutations in an essential lysine residue (K529), we have found that misreading events by tRNALys UUU occur at frequencies that vary from as high as 3.6 × 10−3 (at an AGA codon) to a low of no more than 2.0 × 10−4 (at a GAA codon), a range of 18-fold. The median rate of misreading on the 24 codons tested was no more than 3.4 × 10−4 per codon.
We had expected frequent misreading by mispairing at the wobble position with codon-specific effects on misreading frequencies (Dix and Thompson 1989). Previous work has shown that tRNALys UUU frequently misreads AAU and AAC, AAU four- to ninefold more frequently than AAC (Parker et al. 1983; Parker and Holtz 1984; Precup and Parker 1987). In agreement with these data, the frequency of Lys for Asn misreading of AAU in our system is 4.2-fold greater than AAC (see Table 1). Surprisingly, the most frequently misread codons were the two Arg codons, AGA and AGG. In vitro studies have shown that U•G mispairs of the kind necessary for tRNALys UUU to decode AGA or AGG are much less destabilizing to RNA helices than had previously been expected, implying a higher misreading frequency for this type of error (Grosjean et al. 1978; Sugimoto et al. 1986). However, a study in the yeast Saccharomyces cerevisiae using an F-luc mutant approach similar to ours found misreading frequencies at AGA were not elevated (Salas-Marco and Bedwell 2005). So, frequent error is not simply a feature of codon•anticodon pairing. Rather, competition between cognate and near-cognate tRNAs present at different concentrations must also be a major factor in the frequency of misreading errors in E. coli, where the abundance of the tRNAs decoding AGA, tRNAArg UCU, and AGG, tRNAArg CCU is very low (Ikemura 1981). Misreading occurs in direct competition with correct decoding, so this effect indirectly results in higher error rates.
Poor availability of cognate competitor tRNAs appears to play a major role in misreading in general. The codons showing significant misreading in our analysis fall into two groups. The codons AAU and AAC are recognized by a major tRNA isoacceptor (Ikemura 1981). These are the only codons that tRNALys UUU can decode by wobble misreading, which is known to be frequent. All other codons that were error prone in one or more of our experiments are decoded by minor isoacceptors (AGA, AGG, and AUA) or have no cognate tRNAs (UAA, UAG). Though other considerations may affect misreading frequency, lack of an abundant cognate might be necessary for frequent errors at codon position 1 or 2. Determining whether this is true will require analysis of misreading errors by other tRNAs.
The solution of high-resolution structures of several bacterial ribosomes has provided insights into the mechanism of error correction. The structures involving ribosomes in error-prone or hyperaccurate states are particularly relevant to this study, including 30S ribosomal subunits bound to paromomycin or streptomycin (Carter et al. 2000; Ogle et al. 2001, 2002). Paromomycin binds to the small ribosomal subunit and displaces A1492 and A1493 into a conformation similar to that induced by a cognate tRNA (Ogle et al. 2001). In this conformation, the two nucleotides can interact with and recognize cognate codon•anticodon complexes. Cognate complexes provide the energy required to force this conformational shift, and paromomycin may compensate for the lack of this energy for near or noncognate complexes (for review, see Ogle et al. 2003; Ogle and Ramakrishnan 2005). In so doing, paromomycin may promote near or noncognate acceptance in the A site. In vitro studies show that paromomycin increases the rates of GTPase activation and accommodation, stabilizing the tRNA in the A site regardless of whether it is cognate or near cognate (Pape et al. 1999; Rodnina and Wintermeyer 2001), which is consistent with this model.
Streptomycin alters the ribosome's structure in a similar manner by binding near the ribosomal A site and inducing a movement of the shoulder domain. This movement resembles one induced by a cognate codon•anticodon helix, though the movement is not as great (Ogle et al. 2003). Streptomycin also appears to alter the kinetics of GTP hydrolysis during tRNA selection by reducing the rates of GTPase activation and accommodation for a cognate tRNA and increasing those rates for a near-cognate tRNA (Rodnina et al. 2002; Gromadski and Rodnina 2004b). Because the difference in the rate of GTPase activiation is a governing kinetic step in tRNA acceptance (Rodnina et al. 2005), this change would tend to increase the probability of near-cognate acceptance.
Our data suggest that the effect of these antibiotics is far less general than the structural and in vitro studies imply. Rather than having a blanket effect of stimulating all types of near-cognate misreading, paromomycin and streptomycin both stimulate frequent errors at a subset of near-cognate codons. Three of these codons, UAA, UAG, and AUA, make the least energetically unfavorable near-cognate interaction, involving a relatively stable U•U mismatch. Misreading of two others, AGA and AGG, requires a nearly as favorable U•G pair, but depends on the inability of their low-abundance cognate tRNA to compete for binding. The last two codons, AAU and AAC, mismatch only at the wobble position, which the ribosome does not monitor as tightly as the other two, even in the absence of the antibiotic (Ogle et al. 2001). Importantly, the drugs have no effect on misreading of the other near-cognate codons. Misreading of these codons would require first or second position C•U mismatches (CAA, CAG, ACA, ACG) or first position U•G mismatches (GAA, GAG). Either the antibiotics cannot overcome the energetic barrier to forming these pairs or the abundant cognate tRNA for these codons out-competes tRNALys UUU for decoding these codons.
Many rpsL mutations occur in residues of ribosomal protein S12 that interact with Helices 27 and 44 of the 16S rRNA after 30S subunit closure (Ogle et al. 2002). It has been suggested then that the closed form of the 30S subunit is weakened by such mutations, leading to hyperaccuracy during decoding (Ogle et al. 2002, 2003). Our data are consistent with this hypothesis. The rpsL141 mutation reduced misreading of all near-cognate codons to a low background level. However, rpsL141 had no measurable effect on the less error-prone near-cognate codons. This may signal a specific effect of rpS12 on the error-prone codons or, more likely, the noise from other types of errors (e.g., transcriptional or tRNA charging) may make it impossible to measure more subtle effects using our system.
Mutations in rpsD occur at residues that reside in an interface between ribosomal proteins S4 and S5 that is broken during closure of the 30S subunit (Carter et al. 2000). Consequently, such mutations may lower the number of bonds that must break during closure. It has therefore been suggested that rpsD mutations increase the likelihood that near-cognate tRNAs can induce closure of the 30S subunit (Ogle et al. 2002, 2003). It is possible to imagine then that if all near-cognate tRNAs have an increased probability of inducing 30S closure in rpsD mutants, misreading of all near-cognate substrates should increase. This, however, is not what our data show. Unexpectedly, only those near-cognate codons frequently misread during normal translation showed increased misreading in rpsD mutants. Again, the failure to observe increased errors at other codons may reflect a signal-to-noise problem, but we note that the rpsD effect (Fig. 6) appears far more specific than even the effect of the aminoglycoside antibiotics (Fig. 7). The only misreading error greatly affected by the rpsD mutant was wobble misreading of AAU, which increased almost ninefold, while the effect on the other codons was no more than twofold. These data appear inconsistent with the 30S closure model.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Bacterial strains used in this study were: DH5α (endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ[lacZYA-argF] [φ80lacZΔM15]) (Invitrogen), Xac (ara Δ[lacproAB] gyrA rpoB argE[amber]), US157 (Xac rpsL141 zcg-174::Tn10), and UD131 (Xac rpsD12) (Andersson et al. 1982). The identity of the mutational changes in rpsL and rpsD were determined by direct sequencing of PCR-amplified DNA from each mutant strain. Cultures were grown at 37°C in 5 mL of Luria-Bertani (LB) medium (10 g NaCl, 10 g tryptone, and 5 g yeast extract per L) supplemented with 50 μg/mL ampicillin or 12 μg/mL tetracycline, where required.
Plasmids
We constructed the dual luciferase reporter plasmid pEK4 by PCR amplifying the dual luciferase reporter gene (R-luc–F-luc) from plasmid p2luci (Grentzmann et al. 1998) and subcloning it into the HindIII/NcoI sites of plasmid pTrc99A, a high-copy bacterial plasmid with a regulated Ptac promoter (Amann et al. 1988). QuikChange Site-Directed Mutagenesis (Stratagene) was used to make all mutations in the AAA lysine codon at position 529 in the firefly luciferase gene (Fig. 1). For overexpression of tRNAArg UCU, we used the plasmid pAlter-Ex2/argU (Ruzicka et al. 2000), which encodes the AGA and AGG-decoding tRNAArg UCU, coexpressing it with the AGA and AGG mutant constructs.
Preparation of cell lysates and dual luciferase assays
Plasmids were introduced into bacterial cells either by transformation using the Z-Competent Transformation system (Zymo Research) or electroporation (Ausubel et al. 1997). Cultures of individual clones grown to midlog phase at 37° were pelleted and resuspended in 200 μL, 1 mg/mL lysozyme/10 mM TrisCl at pH 8.0/1 mM EDTA. Cells were incubated on ice for 10 min, frozen on dry ice, and thawed on ice (de Wet et al. 1985). Five microliter samples of this extract were assayed for firefly (F-luc) and Renilla (R-luc) luciferase activities using the Dual-Luciferase Reporter Assay System (Promega). Luminescence was measured using an MLX Microtiter Plate Luminometer (Dynex Technologies). Three colonies of each construct were assayed in triplicate. The R-luc activity was used as an internal standard and standardized F-luc activity was calculated as the ratio of the F-luc to R-luc activity expressed in relative light units (RLU).
ACKNOWLEDGMENTS
We thank Monica Rydén-Aulin for the generous gift of bacterial strains. This work was supported by a grant from the National Institute of General Medical Sciences (GM 29480).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.294907.
REFERENCES
- Amann, E., Ochs, B., Abel, K.J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli . Gene. 1988;69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- Andersson, D.I., Bohman, K., Isaksson, L.A., Kurland, C.G. Translation rates and misreading characteristics of rpsD mutants in Escherichia coli . Mol. Gen. Genet. 1982;187:467–472. doi: 10.1007/BF00332630. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struhl, K. Current protocols in molecular biology. John Wiley & Sons; New York: 1997. [Google Scholar]
- Bouadloun, F., Donner, D., Kurland, C.G. Codon-specific missense errors in vivo. EMBO J. 1983;2:1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchini, B.R., Murtiashaw, M.H., Magyar, R.A., Anderson, S.M. The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. doi: 10.1021/bi9928804. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- Cornut, B., Willson, R.C. Measurement of translational accuracy in vivo: Missense reporting using inactive enzyme mutants. Biochimie. 1991;73:1567–1572. doi: 10.1016/0300-9084(91)90192-4. [DOI] [PubMed] [Google Scholar]
- Dahlgren, A., Ryden-Aulin, M. A novel mutation in ribosomal protein S4 that affects the function of a mutated RF1. Biochimie. 2000;82:683–691. doi: 10.1016/s0300-9084(00)01160-3. [DOI] [PubMed] [Google Scholar]
- de Wet, J.R., Wood, K.V., Helinski, D.R., DeLuca, M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli . Proc. Natl. Acad. Sci. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix, D.B., Thompson, R.C. Codon choice and gene expression: Synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. 1989;86:6888–6892. doi: 10.1073/pnas.86.18.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann, P., Gallant, J. Mistranslation in E. coli . Cell. 1977;10:131–137. doi: 10.1016/0092-8674(77)90147-7. [DOI] [PubMed] [Google Scholar]
- Ellis, N., Gallant, J. An estimate of the global error frequency in translation. Mol. Gen. Genet. 1982;188:169–172. doi: 10.1007/BF00332670. [DOI] [PubMed] [Google Scholar]
- Galas, D., Kirkwood, T., Rosenberger, R. An introduction to the problem of accuracy. In: Kirkwood T., et al., editors. Accuracy in molecular processes: Its control and relevance to living systems. Chapman and Hall; New York: 1986. pp. 1–16. [Google Scholar]
- Gorini, L. Ribosomal discrimination of tRNAs. Nat. New Biol. 1971;234:261–264. doi: 10.1038/newbio234261a0. [DOI] [PubMed] [Google Scholar]
- Grentzmann, G., Ingram, J.A., Kelly, P.J., Gesteland, R.F., Atkins, J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004a;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004b;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Daviter, T., Rodnina, M.V. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- Grosjean, H., DeHenau, S., Crothers, D.M. On the physical basis for the ambiguity in genetic coding interactions. Proc. Natl. Acad. Sci. 1978;75:610–614. doi: 10.1073/pnas.75.2.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfield, J. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Johnston, T.C., Borgia, P.T., Parker, J. Codon specificity of starvation induced misreading. Mol. Gen. Genet. 1984;195:459–465. doi: 10.1007/BF00341447. [DOI] [PubMed] [Google Scholar]
- Khazaie, K., Buchanan, J.H., Rosenberger, R.F. The accuracy of Q β RNA translation. 1. Errors during the synthesis of Q β proteins by intact Escherichia coli cells. Eur. J. Biochem. 1984;144:485–489. doi: 10.1111/j.1432-1033.1984.tb08491.x. [DOI] [PubMed] [Google Scholar]
- Kurland, C., Hughes, D., Ehrenberg, M. Limitations of translational accuracy. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C.: 1996. pp. 979–1004. [Google Scholar]
- Laughrea, M., Latulippe, J., Filion, A.M., Boulet, L. Mistranslation in twelve Escherichia coli ribosomal proteins. Cysteine misincorporation at neutral amino acid residues other than tryptophan. Eur. J. Biochem. 1987;169:59–64. doi: 10.1111/j.1432-1033.1987.tb13580.x. [DOI] [PubMed] [Google Scholar]
- Loftfield, R.B., Vanderjagt, D. The frequency of errors in protein biosynthesis. Biochem. J. 1972;128:1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio, J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Ogle, J., Brodersen, D., Clemons W.M., Jr., Tarry, M., Carter, A., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Ramakrishnan, V. Structural insights into translational fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Carter, A.P., Ramakrishnan, V. Insights into the decoding mechanism from recent ribosome structures. Trends Biochem. Sci. 2003;28:259–266. doi: 10.1016/S0968-0004(03)00066-5. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M.V. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J., Friesen, J.D. “Two out of three” codon reading leading to mistranslation in vivo. Mol. Gen. Genet. 1980;177:439–445. doi: 10.1007/BF00271482. [DOI] [PubMed] [Google Scholar]
- Parker, J., Holtz, G. Control of basal-level codon misreading in Escherichia coli . Biochem. Biophys. Res. Commun. 1984;121:487–492. doi: 10.1016/0006-291x(84)90208-0. [DOI] [PubMed] [Google Scholar]
- Parker, J., Johnston, T.C., Borgia, P.T. Mistranslation in cells infected with the bacteriophage MS2: Direct evidence of Lys for Asn substitution. Mol. Gen. Genet. 1980;180:275–281. doi: 10.1007/BF00425839. [DOI] [PubMed] [Google Scholar]
- Parker, J., Johnston, T.C., Borgia, P.T., Holtz, G., Remaut, E., Fiers, W. Codon usage and mistranslation. In vivo basal level misreading of the MS2 coat protein message. J. Biol. Chem. 1983;258:10007–10012. [PubMed] [Google Scholar]
- Pauling, L. The probability of errors in the process of synthesis of protein molecules. In: Birkhauser A., editor. Festschrift Arthur Stoll. Birkhauser; Basel, Switzerland: 1957. pp. 597–602. [Google Scholar]
- Precup, J., Parker, J. Missense misreading of asparagine codons as a function of codon identity and context. J. Biol. Chem. 1987;262:11351–11355. [PubMed] [Google Scholar]
- Precup, J., Ulrich, A., Roopnarine, O., Parker, J. Context specific misreading of phenylalanine codons. Mol. Gen. Genet. 1989;218:397–401. doi: 10.1007/BF00332401. [DOI] [PubMed] [Google Scholar]
- Rice, J.B., Libby, R.T., Reeve, J.N. Mistranslation of the mRNA encoding bacteriophage T7 0.3 protein. J. Biol. Chem. 1984;259:6505–6510. [PubMed] [Google Scholar]
- Rodnina, M.V., Wintermeyer, W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem. Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Daviter, T., Gromadski, K., Wintermeyer, W. Structural dynamics of ribosomal RNA during decoding on the ribosome. Biochimie. 2002;84:745–754. doi: 10.1016/s0300-9084(02)01409-8. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Gromadski, K.B., Kothe, U., Wieden, H.J. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Ruusala, T., Andersson, D., Ehrenberg, M., Kurland, C.G. Hyper-accurate ribosomes inhibit growth. EMBO J. 1984;3:2575–2580. doi: 10.1002/j.1460-2075.1984.tb02176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka, F.J., Lieder, K.W., Frey, P.A. Lysine 2,3-aminomutase from Clostridium subterminale SB4: Mass spectral characterization of cyanogen bromide-treated peptides and cloning, sequencing, and expression of the gene kamA in Escherichia coli . J. Bacteriol. 2000;182:469–476. doi: 10.1128/jb.182.2.469-476.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco, J., Bedwell, D.M. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 2005;348:801–815. doi: 10.1016/j.jmb.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Sankaranarayanan, R., Moras, D. The fidelity of the translation of the genetic code. Acta Biochim. Pol. 2001;48:323–335. [PubMed] [Google Scholar]
- Schulman, L.H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog. Nucleic Acid Res. Mol. Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Söll, D. The accuracy of aminoacylation—Ensuring the fidelity of the genetic code. Experientia. 1990;46:1089–1096. doi: 10.1007/BF01936918. [DOI] [PubMed] [Google Scholar]
- Sugimoto, N., Kierzek, R., Freier, S.M., Turner, D.H. Energetics of internal GU mismatches in ribooligonucleotide helixes. Biochemistry. 1986;25:5755–5759. doi: 10.1021/bi00367a061. [DOI] [PubMed] [Google Scholar]
- Thompson, R. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem. Sci. 1988;13:91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Toth, M.J., Murgola, E.J., Schimmel, P. Evidence for a unique first position codon-anticodon mismatch in vivo. J. Mol. Biol. 1988;201:451–454. doi: 10.1016/0022-2836(88)90152-0. [DOI] [PubMed] [Google Scholar]
- Ulrich, A.K., Li, L.Y., Parker, J. Codon usage, transfer RNA availability and mistranslation in amino acid starved bacteria. Biochim. Biophys. Acta. 1991;1089:362–366. doi: 10.1016/0167-4781(91)90177-n. [DOI] [PubMed] [Google Scholar]
- Weickert, M.J., Apostol, I. High-fidelity translation of recombinant human hemoglobin in Escherichia coli . Appl. Environ. Microbiol. 1998;64:1589–1593. doi: 10.1128/aem.64.5.1589-1593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]