Abstract
The CRS1–YhbY domain (also called the CRM domain) is represented as a stand-alone protein in Archaea and Bacteria, and in a family of single- and multidomain proteins in plants. The function of this domain is unknown, but structural data and the presence of the domain in several proteins known to interact with RNA have led to the proposal that it binds RNA. Here we describe a phylogenetic analysis of the domain, its incorporation into diverse proteins in plants, and biochemical properties of a prokaryotic and eukaryotic representative of the domain family. We show that a bacterial member of the family, Escherichia coli YhbY, is associated with pre-50S ribosomal subunits, suggesting that YhbY functions in ribosome assembly. GFP fused to a single-domain CRM protein from maize localizes to the nucleolus, suggesting that an analogous activity may have been retained in plants. We show further that an isolated maize CRM domain has RNA binding activity in vitro, and that a small motif shared with KH RNA binding domains, a conserved “GxxG” loop, contributes to its RNA binding activity. These and other results suggest that the CRM domain evolved in the context of ribosome function prior to the divergence of Archaea and Bacteria, that this function has been maintained in extant prokaryotes, and that the domain was recruited to serve as an RNA binding module during the evolution of plant genomes.
Keywords: CRS1–YhbY, group II intron, ribosome assembly, RNA binding domain, UPF0044
INTRODUCTION
Among the fundamental insights to emerge from large-scale genome sequencing projects are the large number of genes whose functions are unknown in even the most intensively studied organisms, and the great degree to which genes are conserved across the kingdoms of life. Consequently, insights into the functions of conserved genes can come from unanticipated directions. Exploration of protein-facilitated group II intron splicing in chloroplasts led to the initial functional data for a conserved domain represented in Archaea, Bacteria, and plants designated variously as UPF0044 (INTERPRO database), COG1534 (COG database), or CRS1–YhbY (Pfam database). Three maize proteins, CRS1, CAF1, and CAF2, each with multiple copies of the domain, are required for the splicing of group II introns in chloroplasts and are bound specifically to their intron targets in vivo (Till et al. 2001; Ostheimer et al. 2003). These prior findings, together with results presented here, point to participation of this domain in the assembly of two classes of catalytic ribonucleoprotein particle: group II intron particles and the large ribosomal subunit. We suggested the name chloroplast RNA splicing and ribosome maturation (CRM) domain (Ostheimer et al. 2003) to reflect these functions, and we use this name to refer to the domain below.
CRM domains in prokaryotes exist as stand-alone proteins encoded by single-copy ORFs of ∼100 amino acids; we refer to these as YhbY orthologs after the name assigned in Escherichia coli. Structural features of bacterial YhbY orthologs (Ostheimer et al. 2002; Willis et al. 2002; Liu and Wyss 2004) and biochemical and genetic data for the CRM group II intron splicing factors suggested that CRM domains might bind RNA, but activities associated with isolated CRM domains have not been documented. In this report we present a biochemical and phylogenetic description of this conserved domain family. We show that an isolated CRM domain from maize binds RNA, that a small structural motif shared between CRM and KH RNA binding domains contributes to RNA binding activity, that a bacterial CRM domain protein is associated in vivo with pre-50S ribosomal subunits, and that a single-domain plant CRM protein localizes to the nucleolus. These results establish the CRM domain as an RNA binding domain. They suggest further that bacterial CRM proteins function in the assembly of the large ribosomal subunit and that a ribosome-assembly function may have been retained among the CRM family in plants. When considered together with the phylogenetic analysis and genomic context of CRM domain coding regions, these findings suggest that prokaryotic CRM proteins existed as ribosome-associated proteins prior to the divergence of Archaea and Bacteria, and that they were co-opted in the plant lineage as RNA binding modules by incorporation into diverse protein contexts.
RESULTS
Phyletic distribution of the CRM domain
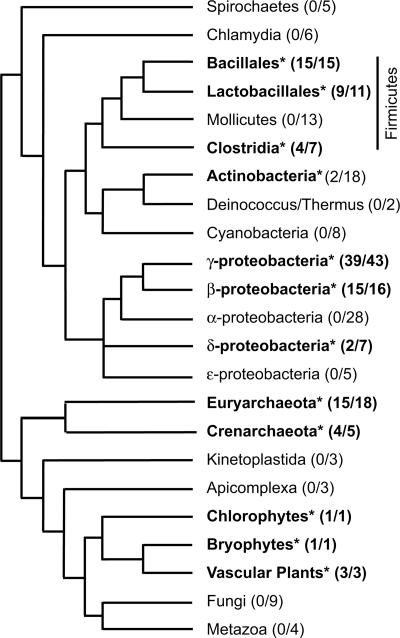
CRM domains are represented in Archaea and Bacteria as single-domain YhbY orthologs, whereas in plants they are found in a family of single- and multidomain proteins. Figure 1 shows an alignment of phylogenetically diverse prokaryotic YhbY orthologs and isolated CRM domains from several plant proteins. Bacterial YhbY orthologs consist of the core CRM domain and little else (Fig. 1; Ostheimer et al. 2002). YhbY orthologs are widely distributed throughout the Euryarchaeota and Crenarchaeota, but in the Bacteria they are found only within the Firmicutes, Actinobacteria, and Proteobacteria (Fig. 2). They are absent from sequenced metazoan, protozoan, and fungal genomes. Phylograms of prokaryotic YhbY orthologs (see Supplemental Fig. 1; http://rna.uoregon.edu/crm/BarkanSuppData.pdf) mimic the organismal phylogeny, consistent with the possibility that a YhbY ortholog was present in the common ancestor of Bacteria and Archaea, and that it has been lost independently in a subset of bacterial lineages. In accordance with this view, YhbY orthologs are among the pool of genes identified as sharing a common history and used to build a consensus supertree of the prokaryotes (Daubin et al. 2002). Nonetheless, the possibility of lateral transfer into several bacterial lineages cannot be excluded.
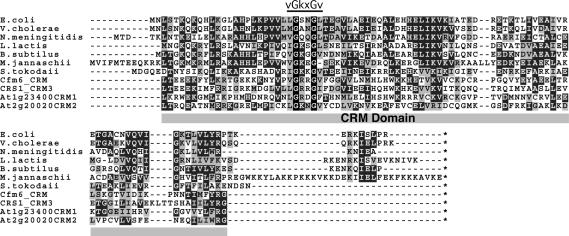
FIGURE 1.
Alignment of representative YhbY orthologs and plant CRM domains. The complete sequences of seven prokaryotic YhbY orthologs are aligned with four CRM domains excerpted from larger plant proteins. The domain boundary was chosen according to the structural core of E. coli, Haemophilus influenzae, and Staphylococcus aureus YhbY (Ostheimer et al. 2002; Willis et al. 2002; Liu and Wyss 2004). Identical residues are shaded in black, and similar residues in gray (similarity threshold of 0.4 for shading). CFM6 and CRS1 are maize proteins with one and three CRM domains, respectively. At1g23400 and At2g20020 are Arabidopsis proteins with two CRM domains each; the position of the CRM domain in the multi-CRM proteins is stated in the domain name. The conserved vGkxGv motif, which is similar to a motif found in KH RNA binding domains, is indicated.
FIGURE 2.
Phyletic distribution of YhbY orthologs and CRM domains. The organismal tree is a composite based on trees in Daubin et al. (2002) and Pennisi (2003). Taxa with CRM domains are indicated with bold text and an asterisk. The number of species within each group harboring CRM domains are indicated as the fraction of the number of fully sequenced genomes that were analyzed for the presence of the domain.
Among eukaryotes, CRM domains are restricted to the plant lineage, where they occur even in basal species such as the chlorophyte Chlamydomonas reinhardtii and the liverwort Marchantia polymorpha. CRM domains in vascular plants are found in a family of proteins, most of which contain multiple copies of the domain. The 33 CRM domains in the predicted Arabidopsis proteome form two clades (Supplemental Figs. 1,2; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). The two domains in the smaller clade most closely resemble prokaryotic YhbY orthologs; these likely represent the basal branch in the plant CRM domain lineage because they cluster with the single CRM ORF in the predicted C. reinhardtii proteome (Supplemental Fig. 5; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). YhbY orthologs are absent in sequenced cyanobacterial and α-proteobacterial genomes, so the presence of CRM domains in plant genomes is unlikely to have originated with the endosymbiotic events that led to chloroplasts and mitochondria. From these data it seems equally plausible that CRM domains were present in the last common eukaryotic ancestor and subsequently lost during the evolution of Fungi, Metazoa, Kinetoplastida, and Apicomplexa, or that they were acquired laterally into the plant lineage, prior to the divergence of Chlorophytes.
E. coli YhbY is bound in vivo to precursors of 50S ribosomal subunits
A role for YhbY orthologs in translation was suggested by their genomic context in the Archaea, where they are typically embedded in predicted operons that encode ribosomal proteins and translation factors (data not shown; ftp://ftp.ncbi.nih.gov/pub/koonin/gene_neighborhoods/; Rogozin et al. 2002). E. coli yhbY is monocistronically transcribed, but it is adjacent to and divergently transcribed with ftsJ/rrmJ, which encodes a 23S rRNA methyl-transferase (Bugl et al. 2000; Caldas et al. 2000a). These genomic contexts motivated us to explore the possibility that E. coli YhbY functions in translation. To facilitate these studies, we generated an antibody to YhbY and an E. coli mutant with a deletion of the YhbY ORF (ΔyhbY). The ΔyhbY strain is viable but grows more slowly than its yhbY + progenitor (Supplemental Fig. 3A; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). The antibody detected YhbY on immunoblots of E. coli extract as an abundant cytoplasmic protein of ∼10 kDa (predicted molecular weight is 10.8 kDa) that is absent in the ΔyhbY strain (Supplemental Fig. 3B; http://rna.uoregon.edu/crm/BarkanSuppData.pdf).
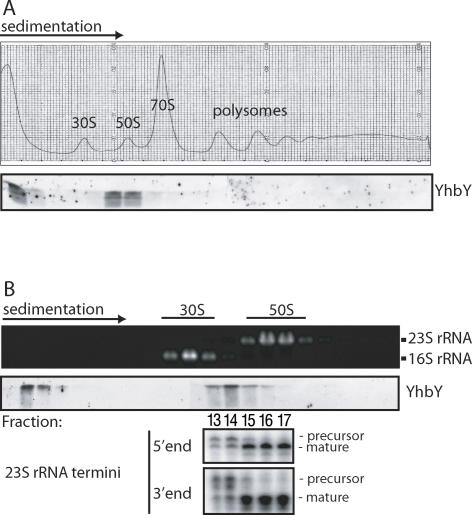
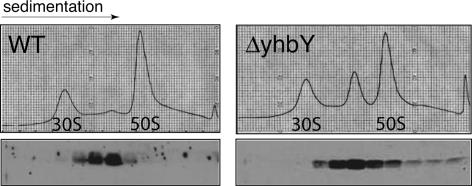
When E. coli extract was sedimented through sucrose gradients under conditions that resolve polysomes from free ribosomal subunits, YhbY was found in two peaks: one peak sedimented slightly behind 50S ribosomal subunits; the second was near the top of the gradient, likely representing a pool of free YhbY (Fig. 3A). When extract was centrifuged under conditions that promote the dissociation of ribosomes into 30S and 50S subunits and that yield increased resolution, YhbY was well resolved from the 50S peak, sedimenting at ∼40S (Fig. 3B). Incompletely processed 23S rRNA, a hallmark of intermediates in the assembly of 50S ribosomal subunits (Srivastava and Schlessinger 1988; Hage and Alix 2004), was enriched in the YhbY peak fractions (Fig. 3B). The well-defined YhbY peak at ∼40S was distinct from the major peaks of absorbance at 260 nm, indicating that this sedimentation behavior is not due to nonspecific interactions with RNA.
FIGURE 3.
YhbY cosediments with pre-50S ribosomal subunits. (A) E. coli lysates prepared under conditions that maintain polysome integrity were sedimented through sucrose gradients. The A260 profile and an immunoblot of gradient fractions probed with anti-YhbY antibody are shown in the upper and lower panels, respectively. (B) E. coli lysates were sedimented through sucrose gradients under conditions that dissociate 70S ribosomes into 30S and 50S subunits, and that increase resolution in the 30S to 50S range. RNA extracted from gradient fractions was analyzed by agarose gel electrophoresis and ethidium bromide staining (upper panel). YhbY was detected in gradient fractions by probing an immunoblot with anti-YhbY antibody (middle panel). The termini of 23S rRNA in fractions 13 through 17 were mapped with RNAse-protection assays (bottom panel).
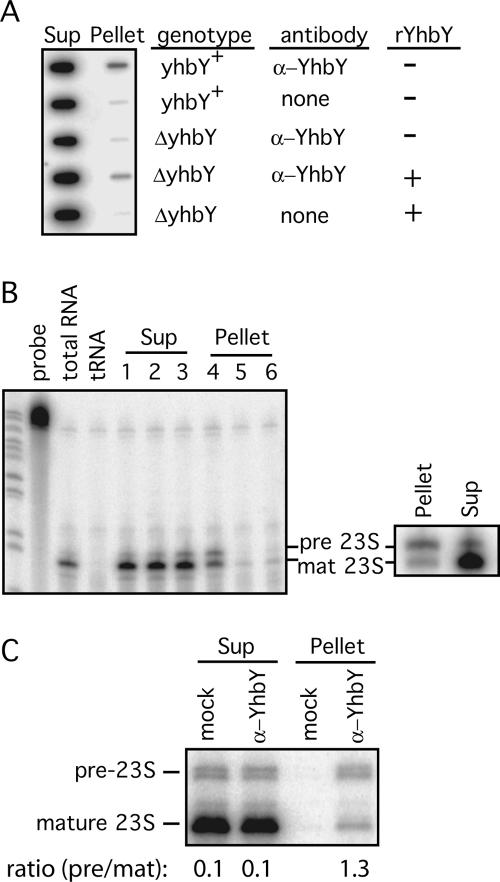
The genomic clustering of prokaryotic YhbY genes with translation-related genes together with the sedimentation of YhbY at ∼40S suggested that YhbY is bound to particles related to 50S ribosomal subunits. To test this possibility, we determined whether 23S rRNA coimmunoprecipitates with YhbY from E. coli extract (Fig. 4). Slot-blot hybridization detected more 23S rRNA in the pellets of immunoprecipitations containing α-YhbY antiserum and yhbY+ cell extract than in control reactions lacking antiserum or using ΔyhbY cell extract (Fig. 4A). Addition of recombinant YhbY to ΔyhbY extract restored the ability of the antibody to coimmunoprecipitate 23S rRNA. Only a small fraction of the 23S rRNA was recovered in the immunoprecipitation pellet, consistent with the fact that YhbY sediments more slowly than the highly abundant population of mature 50S ribosomal subunits.
FIGURE 4.
Pre-50S ribosomal subunits coimmunoprecipitate with YhbY. (A) Anti-YhbY antibody was used in immunoprecipitation reactions with cytoplasmic extract of yhbY+ or ΔyhbY E. coli cells. RNA extracted from the immunoprecipitation pellets and supernatants (Sup) was applied to a slot blot, and hybridized with a probe for the 23S rRNA. An equal proportion of the total pellet and supernatant RNA was analyzed in each slot. Addition of recombinant YhbY (rYhbY) to ΔyhbY extract reconstituted the coimmunoprecipitation of 23S rRNA, indicating that rYhbY can associate with a 50S subunit-related particle present in ΔyhbY cells. Similar results were obtained in four repetitions of this experiment involving different extract preparations (data not shown). (B) RNAse-protection analysis of the 5′ ends of 23S rRNA in the coimmunoprecipitation samples. Lanes 1 and 4: RNA from yhbY + extract immunoprecipitated with anti-YhbY serum; lanes 2 and 5: RNA from a mock immunoprecipitation with yhbY + extract; lanes 3 and 6: RNA from ΔyhbY extract immunoprecipitated with α-YhbY serum. The panel on the right shows the results of a replicate experiment with yhbY + extract and α-YhbY serum. Pre-23S and mat-23S are the products of protection by the precursor and mature 23S rRNAs, respectively. (C) Quantification of the ratio of precursor to mature 23S rRNA in YhbY immunoprecipitations. 5′ Termini were mapped by primer extension, using a radiolabeled primer complementary to the 5′ region of mature 23S rRNA; comparison to a sequencing ladder showed the bands to correspond to the established 5′ termini of mature and immature 23S rRNA (data not shown). The ratios of precursor to mature 5′ termini were quantified with a PhosphorImager and are shown below. This experiment involved a different immunoprecipitation reaction than those in B to illustrate the reproducible enrichment of pre-23S rRNA in YhbY coimmunoprecipitates. The mock assay was performed in parallel and lacked antiserum.
YhbY's sedimentation at ∼40S and coimmunoprecipitation with 23S rRNA suggested that it is bound to either precursors or degradation products of 50S ribosomal subunits. To distinguish between these possibilities we took advantage of the fact that 50S subunits that have participated in translation harbor mature 23S rRNA, whereas pre-50S subunits are enriched in incompletely processed 23S rRNA (Srivastava and Schlessinger 1988; Hage and Alix 2004). Ribonuclease-protection (Fig. 4B) and primer extension (Fig. 4C) assays were used to map the 5′ ends of the 23S rRNA that coimmunoprecipitated with YhbY. In three independent experiments, immature 23S rRNA was enriched with respect to mature 23S rRNA in YhbY immunoprecipitation pellets (Fig. 4B, left panel, cf. lanes 1 and 4; compare the pellet to the supernatant samples in the right panels of Figs. 4B, 4C). Furthermore, the ratio of precursor to mature 5′ ends in the YhbY coimmunoprecipitation pellets was similar to that in the pre-50S subunit peak with which YhbY cosediments (cf. lane 14 in Fig. 3B). Quantification of the well-resolved bands obtained by primer extension showed that pre-23S rRNA is enriched at least 10-fold in YhbY coimmunoprecipitations pellets, in comparison to mature 23S rRNA. The cosedimentation of YhbY with pre-50S ribosomal subunits and the coimmunoprecipitation of YhbY with pre-23S rRNA together provide very strong evidence that YhbY is associated with an intermediate in the assembly of the 50S ribosomal subunit.
The association between YhbY and pre-50S ribosomal particles is very stable, as it was maintained even in the presence of 1M ammonium chloride (Supplemental Fig. 4; http://rna.uoregon.edu/crm/BarkanSuppData.pdf), a condition that strips known nonribosomal proteins from ribosomes (Spirin 1990). Comparison of the immunoblot signal intensity between known amounts of cytoplasmic extract and recombinant YhbY leads to an estimate that YhbY comprises ∼0.1% of soluble E. coli protein during logarithmic growth at 37°C (Supplemental Fig. 3B; http://rna.uoregon.edu/crm/BarkanSuppData.pdf; data not shown). Based on the volume and ribosome content of rapidly growing E. coli cells (Donachie and Robinson 1987; Bremer and Dennis 1996; Neidhardt and Umbarger 1996), this corresponds to ∼1/10th the molar concentration of ribosomes. This is similar in concentration to RimM and RbfA, assembly factors for the 30S ribosomal subunit (Wikstrom and Bjork 1988; Bylund et al. 1998).
The ΔyhbY strain accumulated increased levels of pre-50S ribosomal subunits and reduced levels of mature 50S particles (Fig. 5). Furthermore, when recombinant YhbY was added to the mutant lysate, it cosedimented with the pre-50S peak (Fig. 5). These results are consistent with the possibility that YhbY binds to a specific pre-50S ribosomal particle and promotes its maturation. However, interpretation of these results is complicated by the fact that the “–35” region for the more upstream of two promoters driving expression of the adjacent, divergently transcribed ftsJ/rrmJ gene (Herman et al. 1995) maps within the YhbY ORF and was deleted in this strain. RrmJ mutants show ribosome defects that resemble those in the ΔyhbY strain (Bugl et al. 2000; Caldas et al. 2000b), suggesting that reduced rrmJ expression might contribute to the ΔyhbY phenotype. However, RrmJ protein accumulated to near normal levels in the ΔyhbY strain, and introduction of an RrmJ expression plasmid into ΔyhbY cells did not fully restore their growth and ribosome assembly defects (data not shown). Taken together, the fact that YhbY associates tightly and specifically with pre-50S ribosomal subunits and the phenotype of the ΔyhbY strain strongly suggest that YhbY functions in ribosome maturation, but proof for such a role will require construction of a new mutant strain and is beyond the scope of this study.
FIGURE 5.
Aberrant ribosome accumulation in ΔyhbY mutant. Lysates of wild-type and ΔyhbY cells were resolved in sucrose gradients under conditions that dissociate 70S ribosomes into 30S and 50S subunits. Recombinant YhbY was added to the mutant lysate prior to sedimentation (right panels). Upper panels show A260 profiles; lower panels show immunoblots of gradient fractions probed with anti-YhbY antibody. The A260 profile of the mutant lysate was unchanged by the addition of recombinant YhbY (data not shown). The immunoblot signal in the mutant lysate derives only from the added recombinant YhbY, as the antibody detected no protein in unsupplemented mutant lysate (Supplemental Fig. 3B; http://rna.uoregon.edu/crm/BarkanSuppData.pdf; data not shown).
Expansion and diversification of the CRM domain family in plants
In contrast to prokaryotes where CRM domains are represented solely as stand-alone proteins from single-copy genes, plant genomes encode multiple CRM domain proteins, most of which have several copies of the domain. Queries of the complete predicted proteomes of Arabidopsis thaliana (Arabidopsis) and Oryza sativa (rice) with YhbY and the maize group II splicing factors CRS1 and CAF1 detected 16 Arabidopsis and 14 rice proteins containing one or more CRM domain (Fig. 6). The Pfam (version 18) profile for the CRS1–YhbY domain (ID PF01985) detects this same set of proteins.
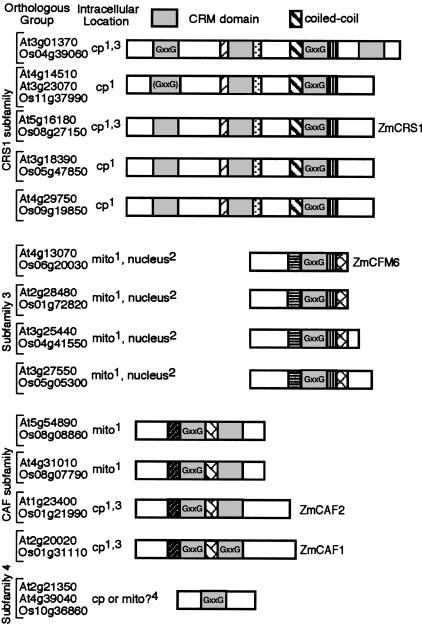
FIGURE 6.
The CRM domain family in plants. Orthologous groups were assigned based on the results of mutual best hit BLAST comparisons among the complete proteomes of rice and Arabidopsis, and are supported by the phylogram shown as Supplemental Figure 5 (http://rna.uoregon.edu/crm/BarkanSuppData.pdf). The orthologous groups corresponding to maize (Zm) CRS1, CAF1, CAF2, and CFM6 are indicated. The proteins are grouped into four subfamilies based on their domain organization and their clustering in the phylogram. CRM domains are indicated by gray boxes, with other regions of similarity represented by distinct pattern fills. CRM domains harboring the “GxxG” motif are indicated and are conserved in both species, except where shown in parentheses. The coiled-coil motif that is characteristic of the CRS1 subfamily is shown, and is the only functional motif detected in these proteins other than the CRM domain itself. Intracellular locations were based on consensus predictions and/or experimental data, according to the following key: 1Prediction with TargetP (Emanuelsson and Heijne, 2001) and/or Predotar (Small et al. 2004) in both rice and Arabidopsis; 2Prediction with two of the three nuclear predictors PredictNLS (Cokol et al. 2000), NucPred (http://www.sbc.su.se/~maccallr/nucpred/), or PSORTII (http://psort.ims.u-tokyo.ac.jp/) in both rice and Arabidopsis; 3Maize ortholog established to be in chloroplast (data not shown; Till et al. 2001; Ostheimer et al. 2003); 4Weak predictions that differ between species.
The Arabidopsis and rice CRM proteins were placed into 14 orthologous groups (Fig. 6) based on reciprocal best hits in whole-proteome BLAST comparisons (see http://plantrbp.uoregon.edu); these groups are supported by their clustering in a phylogram of the Arabidopsis and rice CRM proteins (Supplemental Fig. 5; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). The phylogram shows that the structure of the family was established prior to the divergence of monocot and dicot plants, and suggests that two members of the family were subsequently duplicated in the Arabidopsis lineage. CRM domain proteins in plants can be divided into four subfamilies based on their domain organization and on the degree of sequence similarity both within and flanking the CRM domains (Fig. 6; Supplemental Figs. 1,5; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). A search for known functional motifs detected a predicted coiled-coil domain preceding the third CRM domain in all members of the CRS1 subfamily (Fig. 6); this region might mediate homo- or heterodimerization, including, potentially, the homodimerization reported for recombinant CRS1 (Ostersetzer et al. 2005).
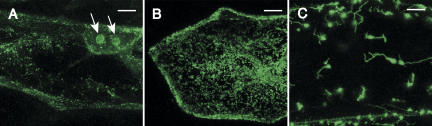
Targeting prediction algorithms (Nakai and Horton 1999; Cokol et al. 2000; Emanuelsson and Heijne 2001; Small et al. 2004) suggest that CRM domain proteins in plants are found in the nucleus, mitochondrion, and chloroplast (Fig. 6). Proteins in subfamily 3 are predicted to localize to the nucleus and resemble YhbY in that they contain a single CRM domain and little else, suggesting that they might function in nucleolar ribosome biogenesis. In support of this possibility, GFP fused to one member of this subfamily, maize CFM6 (see Fig. 6), localized to the nucleolus in transient expression assays (Fig. 7). This fusion protein also localized to mitochondria (Fig. 7), suggesting that CFM6 may function in the metabolism of the divergent ribosomes within the nucleolus and mitochondrion.
FIGURE 7.
Nucleolar localization of CFM6–GFP in onion epidermal cells. (A) Full-length maize CFM6 (40 kDa) was fused at its carboxy-terminus to GFP and transiently expressed in onion root epidermal cells. The arrows show nucleolar localization of GFP. The speckled fluorescence is similar to that shown in B for mitochondrial-targeted GFP, and is therefore likely to be mitochondrion-localized CFM6–GFP. (B) Mitochondrial targeting of GFP fused to the targeting peptide of mitochondrial FDH. (C) Chloroplast targeting of GFP fused to the targeting peptide of chloroplast RecA. Bars = 20 μm.
In vitro RNA binding activity of an isolated CRM domain
Several lines of evidence support the idea that CRM domains bind RNA: (1) all three characterized CRM domain proteins in plants (CRS1, CAF1, and CAF2) are associated with RNA in vivo and influence RNA metabolism (Till et al. 2001; Ostheimer et al. 2003); (2) recombinant CRS1 binds with high affinity to its cognate group II intron RNA in vitro (Ostersetzer et al. 2005); (3) E. coli YhbY is bound in vivo to pre-50S ribosomal subunits (see above), whose surface is composed largely of RNA (Ban et al. 2000; Yusupov et al. 2001); and (4) structural studies of several YhbY orthologs revealed structural similarity to known RNA binding domains and a putative RNA binding surface (Ostheimer et al. 2002; Willis et al. 2002; Aravind et al. 2003; Liu and Wyss 2004). In addition, CRM domains share an intriguing similarity with the KH RNA binding domain: a six amino acid motif, “GxxG” flanked by large aliphatic residues, within which one “x” is typically a basic residue (Fig. 1), and which is presented as a loop extending from the structural core of the domain (Ostheimer et al. 2002; Willis et al. 2002; Liu and Wyss 2004). The GxxG loop in KH domains contributes to their RNA binding activity (Musco et al. 1996; Lewis et al. 2000). A role for the GxxG motif in CRM domain function is supported by the fact that the motif is almost universally conserved among prokaryotic YhbY orthologs and that it is present in at least one of the CRM domains in each member of the rice/Arabidopsis CRM family. However, in proteins with multiple CRM domains, it is typical that only one of the domains has retained the motif (Fig. 6). Analogously, only a subset of the KH domains in multi-KH proteins typically retain the “invariable” GxxG motif (Musco et al. 1996); it was suggested that this degeneracy might be important to prevent excessive affinity for RNA.
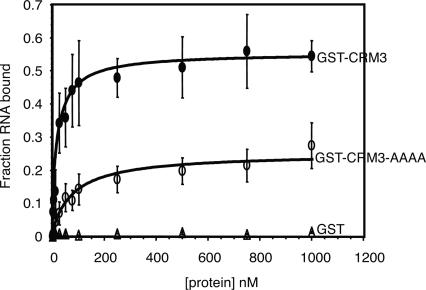
Despite this suggestive evidence, RNA binding activity has not been reported for an isolated CRM domain. We chose the third CRM domain from CRS1 to assay for RNA binding activity because CRS1 has been shown to bind RNA in vitro and its third CRM domain has maintained the GxxG motif. This domain was expressed in E. coli as a GST-fusion protein (GST–CRM3), purified, and used in filter-binding assays with CRS1’s native substrate, atpF intron RNA (Fig. 8). GST–CRM3 bound RNA with high affinity (apparent Kd ∼21 nM). However, the isolated domain lacked sequence specificity (L. Klipcan and O. Ostersetzer, in prep.), unlike intact CRS1, which binds to specific sites within atpF intron domains 1 and 4 (Ostersetzer et al. 2005). Mutation of the four residues in CRM3′s GxxG loop to alanine, which is not expected to disrupt the folding of the protein, decreased the affinity for RNA considerably (apparent Kd ∼79 nM; see GST–CRM3–AAAA in Fig. 8), supporting the notion that the GxxG loop contributes to RNA binding activity. It seems plausible that the CRM domains that have retained the GxxG motif in multi-CRM proteins bear the primary responsibility for high-affinity RNA binding, with the degenerate CRM domains performing an accessory role by contributing to specificity.
FIGURE 8.
RNA binding activity of an isolated CRM domain. Filter binding assays were performed with a trace amount of 32P-labeled atpF intron RNA and increasing concentrations of GST–CRM3, GST–CRM3–AAAA, or GST. Values represent the means, ±1 standard deviation, of nine experiments involving four different protein preparations. Single-site binding isotherms were fit to the data using the equation: Fraction RNA bound = (maximum RNA bound*[protein])/(Kd+[protein]). GST–CRM3 and GST–CRM3–AAAA bound the atpF intron RNA with apparent Kds of 21.4 ± 4.3 and 79.3 ± 14.4 nM, respectively.
DISCUSSION
Results presented here provide insight into the phylogenetic history, biological functions, and biochemical activities of the CRM/CRS1–YhbY domain, a conserved domain represented as a free-standing ORF in prokaryotes and in a family of single- and multidomain proteins in plants. The structures of bacterial YhbY orthologs (Ostheimer et al. 2002; Willis et al. 2002; Liu an Wyss 2004) and the established functions of three plant CRM proteins in group II intron splicing (Till et al. 2001; Ostheimer et al. 2003) suggested that CRM domains might bind RNA. Here we have confirmed this prediction by showing that an isolated CRM domain has RNA binding activity in vitro. Additionally, we add to the functions described for this family by showing that E. coli YhbY is bound to pre-50S ribosomal subunits in vivo, suggesting a role in ribosome assembly. Archaeal YhbY orthologs are typically embedded in operons devoted to translation, supporting the notion that a ribosome-associated function is conserved in Archaea.
Two proteins that associate with bacterial pre-50S ribosomal subunits and that promote their maturation have been described previously: the DEAD-box helicases CsdA and SrmB (Charollais et al. 2003, 2004). The pre-50S particle to which YhbY is bound is similar in size to those bound by CsdA and SrmB; furthermore, the YhbY-bound particle harbors immature 23S rRNA (Fig. 4) and mature 5S rRNA (data not shown), as do the 40S particles that accumulate in the absence of CsdA and SrmB (Charollais et al. 2003, 2004). Like YhbY, CsdA and SrmB are necessary for optimal growth but not for cellular viability (Jones et al. 1996; Charollais et al. 2003). YhbY differs from these assembly factors in that it is bound tightly to the precursor particle and it is not predicted to harbor helicase activity (Ostheimer et al. 2002; Willis et al. 2002; Liu and Wyss 2004). Thus, it seems possible that SrmB and/or CsdA promote rearrangements during late steps in 50S subunit maturation that lead to release of YhbY.
Among the plant CRM-domain family, only maize CRS1, CAF1, and CAF2 have been characterized; all three of these proteins associate with, and promote the splicing of specific chloroplast group II introns in vivo (Till et al. 2001; Ostheimer et al. 2003). There are 14 proteins harboring CRM domains in rice and 16 in Arabidopsis, with CRS1, CAF1, and CAF2 orthologs identifiable by phylogenetic analysis in both species (Supplemental Fig. 5; http://rna.uoregon.edu/crm/BarkanSuppData.pdf). Functions for uncharacterized members of the family are suggested by their predicted intracellular locations and by their strong resemblance to specific characterized CRM proteins (Fig. 6). For example, two proteins with striking similarity to maize CAF1 and CAF2 are predicted to localize to mitochondria; these are excellent candidates for mitochondrial group II intron splicing factors. All members of the CRS1 subfamily are predicted to localize to chloroplasts; these, like CRS1, may promote the splicing of specific chloroplast group II introns. Single-domain CRM proteins (subfamilies 3 and 4 in Fig. 6), which most closely resemble prokaryotic YhbY orthologs, are predicted to reside in the mitochondrion, chloroplast, and nucleus; perhaps these, like YhbY, are pre-ribosome binding proteins. In accordance with this possibility, a GFP fusion with one such protein localizes to both the nucleolus and the mitochondrion (Fig. 7).
These findings, when considered in the context of the phylogenetic data presented here, suggest that CRM domains evolved in the context of ribosome maturation early in the evolution of prokaryotic organisms, that this function was retained in extant prokaryotes and possibly in the nucleolar compartment of plant cells, and that the domain was recruited to serve as an RNA binding module during the evolution of plant genomes. The expansion of the CRM family in the plant lineage occurred after divergence of the chlorophytes, as the fully sequenced genome of the chlorophyte C. reinhardtii encodes just one CRM protein, with just a single CRM domain. The available genome sequence data are consistent with the possibility that the CRM family expanded early in the evolution of the Streptophyta, in concert with the acquisition of group II introns in their chloroplast genomes (Turmel et al. 2002).
It is noteworthy that all of the established substrates for CRM domain proteins (large ribosomal subunits and group II introns) have catalytic RNAs at their core. This trend is strengthened by our recent finding that one member of the CRS1 subfamily in maize is associated in vivo with the sole group I intron in the chloroplast (Y. Asakura and A. Barkan, in prep.). Thus, CRM proteins seem to have a propensity to interact with highly structured, catalytic RNAs. The challenges associated with the productive folding of such RNAs have been extensively discussed (Herschlag 1995; Weeks 1997; Woodson 2000; Treiber and Williamson 2001; Schroeder et al. 2004). It will be interesting to explore whether the CRM domain is particularly well suited to guide the folding of highly structured RNAs, whether these various RNA substrates share structural motifs that are recognized by CRM domains, or whether these correlations are merely fortuitous.
Materials and Methods
Phylogenetic analysis
The phyletic distribution was determined through BLAST searches of the predicted proteomes of fully sequenced genomes available at NCBI. In addition, annotations of fully sequenced prokaryotic genomes were queried for “CRS1–YhbY,” “COG1534,” and “UPF0044,” which revealed several YhbY orthologs that did not emerge from the BLAST searches. Hits were discarded if they lacked several highly conserved residues and motifs that are characteristic of CRM domains. Trees were built in PAUP version 4.0, based on an alignment using 108 characters that was generated in T-Coffee and manually edited. The alignment is available as Supplemental Figure 2 (http://rna.uoregon.edu/crm/BarkanSuppData.pdf).
Production of YhbY antiserum and deletion mutant
Full-length YhbY was expressed in E. coli using the vector pET28, and used for polyclonal antibody production in rabbits. Sera were affinity purified against the same antigen prior to use. E. coli strains EMG2 and K38 deleted for the yhbY ORF (all codons except for the start and stop codons) were generated with the replacement vector pKO3 according to the method of Link et al. (1997).
Sucrose gradient fractionation of E. coli extract
E. coli cultures were grown in LB medium at 37°C to an OD600 = 0.4, pelleted, resuspended in a minimal volume of lysis buffer (20 mM HEPES–KOH pH 7.5, 6 mM MgCl2, 30 mM ammonium chloride, 4 mM β-mercaptoethanol, 0.75 mg/mL lysozyme), and lysed via two freeze–thaw cycles in liquid N2. Insoluble material was pelleted by centrifugation at 15,000 rpm in a microfuge for 45 min at 4°C. Aliquots (∼6 A260 units) were layered onto 10%–40% sucrose gradients prepared in either polysome buffer (Fig. 3A: 20 mM Tris–HCl pH 7.8, 10 mM MgCl2, 100 mM ammonium chloride, 200 μg/mL heparin) or dissociation buffer (Fig. 3B: 20 mM Tris–HCl pH 7.8, 1 mM MgCl2, 100 mM ammonium chloride). Gradients were centrifuged in a Beckman SW41 rotor at 35,000 rpm for 2.5 h (Fig. 3A) or 7 h (Fig. 3B). RNA was purified from gradient fractions by addition of SDS to 0.5% and EDTA to 10 mM, followed by phenol-chloroform extraction and ethanol precipitation.
Coimmunoprecipitation experiments
E. coli extracts were prepared as described for the sucrose gradient analyses and incubated with affinity-purified α-YhbY antibody. The procedures for immunoprecipitation, RNA extraction, and slot-blot hybridizations were as described in Ostheimer et al. (2003). The probe for 23S rRNA was a 226 bp PCR product encompassing 120 base pairs (bp) upstream and 106 bp downstream of the 5′ end of mature 23S rRNA.
RNAse protection and primer extension assays
RNAse protection assays were performed as described previously for analysis of chloroplast RNAs (Barkan et al. 1994). The 5′ end of 23S rRNA was mapped with a probe encompassing 120 nucleotides (nt) upstream and 106 nt downstream of the mature 5′ end; the 3′ end was mapped with a probe encompassing the terminal 50 nt of mature 23S rRNA and 96 nt of downstream sequence. Probes were generated by in vitro transcription of PCR products containing a T7 promoter; transcription reactions included 30 ng template DNA, 0.5 mM ATP, GTP and CTP, 0.25 mM UTP, and 20 μCi 32[P]-UTP (800 Ci/mmol), in transcription buffer supplied by the manufacturer; 150,000 cpm of radiolabeled RNA was used per reaction. RNAse digestions were performed with 20 μg/mL RNAse A and 60 U RNase T1 at 30°C for 1 h. Primer extension reactions were performed as described in Watkins et al. (1994), with a 5′-end-labeled oligonucleotide primer complementary to 23S rRNA ∼100 nt downstream of the mature 5′ end: 5′-GGTTATAACGGTTCATATCACC-3′.
Expression and purification of GST–CRM domain fusion proteins
A PCR fragment encoding the third CRM domain of CRS1 was generated with primers CRM3–5′ (5′-AAAGTCGACAACACTTGACAGAAGAGGAA-3′) and CRM3–3′ (5′-TTTGCGGCCGCATTGCTGGGCGGCGATA-3′) using a crs1cDNA clone as a template. Mutation of the GxxG motif was achieved by overlap extension PCR as follows: (1) a 5′ fragment was generated with the CRM3–5′ primer and a 3′ primer encoding the mutated GxxG residues (5′-CGCCGCCGCCGCTAGGAGAACAAGCCCATCCAT-3′); an overlapping 3′ fragment was generated with the primer (5′- GCGGCGGCGGCGATCTTTGATGGTGTAATTGAAGAG-3′) together with the CRM3–3′ primer. The intact mutant CRM3-encoding DNA was generated with a third PCR reaction using both PCR products as templates, together with the CRM3–5′ and CRM3–3′ primers. The wild-type and mutant CRM3-encoding PCR products were digested with SalI and NotI and cloned into pGEX-4T1 (Pharmacia-Amersham), such that GST was fused in-frame to the CRM domain; the proteins encoded by the resulting plasmids were named GST–CRM3 and GST–CRM3–AAAA.
Plasmids were introduced into E. coli strain XL1-Blue (Stratagene). Cultures were grown to an OD600 of 0.8, and protein expression was induced by the addition of 1 mM IPTG for 16 h at 22°C. Cells were pelleted and resuspended in 40 mL ice-cold PBS, lysed with a French Press, and cleared by centrifugation for 15 min at 10,000×g at 4°C. The lysates were applied to glutathione-Sepharose in high salt buffer (50 mM Tris–HCl, 750 mM NaCl, 0.1% Triton X-100, pH 8.0). The beads were washed once in PBS and proteins were eluted by incubation for 5 min in 0.5 mL 100 mM Tris–HCl pH 8.0, 100 mM NaCl, and 20 mM reduced glutathione. The beads were pelleted by centrifugation for 2 min at 10,000×g at 4°C, and the supernatant was dialyzed against 50% glycerol, 50 mM HEPES–KOH pH 7.0, 500 mM KCl, 0.1% Triton X-100, 5 mM βME, and stored at –20°C. The wild-type and mutant proteins were prepared and assayed in parallel.
RNA binding assays
Filter binding assays were performed as described previously (Ostersetzer et al. 2005). The atpF intron RNA substrate included the complete intron plus 22 nt of exon 1 and 24 nt of exon 2, and was body labeled during transcription in vitro with T7 RNA polymerase (2.5 mM each of ATP, GTP, CTP, 0.25 mM UTP, 20 μCi [α32P]-UTP 3000 Ci/mmol). The RNA was gel purified, subjected to phenol-chloroform extraction and ethanol precipitation, and stored in ddH2O at –20°C. Immediately before each assay, RNA was denatured by heating to 95°C for 2 min in 10 mM Tris–HCl pH 7.0, 1 mM EDTA, and folded by slow cooling to 55°C in the presence of 0.15 M KOAc and 10 mM MgOAc. Binding reactions (20 μL) contained 25 pM-labeled RNA, 10 mM Tris–HCl pH 7.0, 150 mM KOAc, 10 mM MgOAc, 5 mM DTT, 10 μg/mL BSA, 1 U/μL RNAse inhibitor (Fermentas), and between 0 and 1 μM protein. After a 15-min incubation at 25°C, reactions were chilled on ice and passed through sandwiched nitrocellulose and positively charged nylon membranes by vacuum filtration with a slot-blot manifold. Slots were washed once with 100 μL of 50 mM Tris–HCl pH 7.0, 150 mM KOAc, 10 mM MgCl2. Radioactivity bound to each slot was quantified with a PhosphorImager and ImageQuant software (Molecular Dynamics). The fraction of RNA bound was calculated as the ratio between RNA captured by the nitrocellulose and the total RNA captured by both membranes. Apparent dissociation constants were determined by using ORIGIN 7.5 (Microcal Software Inc.) to fit single-site binding isotherms to the data, using the equation: Fraction RNA Bound = (maximum RNA bound*[protein])/(Kd+[protein]). When data were fit to the Hill equation, the Hill coefficients were close to 1, indicating lack of cooperative binding under these binding conditions.
Localization of ZmCfm6–GFP fusion protein in a transient expression assay
The orthologous group containing rice Os06g20030 and Arabidopsis At4g13070 was named CRM family member 6 (cfm6). Maize sequences with high nucleotide identity to rice Cfm6 (Os06g20030) were identified by querying public databases. This sequence was used to design the following primers for amplification of a cDNA encoding the maize Cfm6 open reading frame from a seedling leaf cDNA library (inbred line B73): Cfm6 F(NheI): 5′-CCTGCTAGCATGGCAGCTCTCGCGCCGTGG-3′ and Cfm6 R(XhoI): 5′-CCTCTCGAGCTTTAGAATCTGAGGTAGTTGC-3′. The product was cloned into pGEM-T (Promega) to yield pGEM–Cfm6. The coding region was excised from pGEM–Cfm6 by digestion with NheI and XhoI and cloned into the NheI and SalI sites of pOL–LT (Peeters et al. 2000), creating pCfm6–GFP. The ZmCfm6 cDNA and deduced protein sequences are deposited in GenBank under accession DQ402046. Rice Cfm6 (Os06g20030) is the top hit when ZmCfm6 nucleotide or protein sequence is used to query the rice genome/proteome. pOL–LT, pRecA–GFP, and pFDH–GFP were kindly supplied by Dr. I. Small (INRA).
pCfm6–GFP, pRecA–GFP (encoding a chloroplast-targeted protein), and pFDH–GFP (encoding a mitochondrial-targeted protein) were coated onto 1.675 μm M25 tungsten particles as follows. Tungsten particles were sterilized in ethanol and washed three times with distilled water. Five micrograms of DNA (10 μL) were precipitated onto 50 μL of a particle suspension (60 mg/mL) by addition of 50 μL of 2.5 M CaCl2 and 20 μL of 1 M spermidine for 10 min on ice. After removing 80 μL of the supernatant, 10 μL of the remaining particle suspension was placed on the grid of a 13-mm Swinney filter holder (Gelman Sciences). An inner layer of an onion bulb (Allium cepa) was placed on moist paper towels (inner side up) in a Petri dish at a distance of 6 cm from a helium microprojectile particle device. Bombardment was initiated by drawing a vacuum down to 27 in. Hg, and applying a helium pulse to a 900-psi rupture disk; a mesh screen ahead of the rupture disk was used to distribute the microprojectiles. Samples were sealed with parafilm and incubated in the dark for 2 d at room temperature. The epidermal layer was peeled off and observed using a confocal laser-scanning microscope (Bio-Rad Radiance2100 MP; Bio-Rad). GFP fluorescence was measured as emission at 515 nm.
ACKNOWLEDGMENTS
We are especially grateful to Peggy Saks, Joe Thornton, Julie Toplin, and Christian Schmitz-Linneweber for help with phylogenetic analyses. We also thank George Church for providing plasmid pKO3 and E. coli strain EMG2, Ian Small for providing plasmids for GFP fusion protein analysis, Ursula Jakob for providing an antibody and expression construct for RrmJ, Susan Belcher and Maureen Hanson for advice regarding onion epidermal cell bombardments, and Roz Williams-Carrier for help with figure preparation. This work was supported by grants to A.B. from the National Science Foundation (MCB-0314597 and DBI-0421799) and from the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (2003-35318-13578). T.K. was supported in part by a predoctoral fellowship from the American Heart Association.
NOTE ADDED IN PROOF
YhbY was recently shown to be associated with a particle of ∼40S by Jiang et al. (2006).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.139607.
REFERENCES
- Aravind, L., Iyer, L.M., Anantharaman, V. The two faces of Alba: The evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P., Steitz, T. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., Johnson, D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms. EMBO J. 1994;13:3170–3181. doi: 10.1002/j.1460-2075.1994.tb06616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, H., Dennis, P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F.C., editor. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 2. ASM Press; Washington, DC: 1996. pp. 1553–1569. [Google Scholar]
- Bugl, H., Fauman, E., Staker, B., Zheng, F., Kushner, S., Saper, M., Bardwell, J., Jakob, U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Bylund, G., Wipemo, L., Lundberg, L., Wikstrom, P. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli . J. Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., Costa, A., Desgres, J., Richarme, G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 2000a;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., Richarme, G. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 2000b;271:714–718. doi: 10.1006/bbrc.2000.2702. [DOI] [PubMed] [Google Scholar]
- Charollais, J., Pflieger, D., Vinh, J., Dreyfus, M., Iost, I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli . Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- Charollais, J., Dreyfus, M., Iost, I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., Rost, B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–415. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin, V., Gouy, M., Perriere, G. A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res. 2002;12:1080–1090. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie, W.D., Robinson, A.C. Cell division: Parameter values and the process. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella typhimurium: Cellular and molecular biology. ASM Press; Washington, DC: 1987. pp. 1578–1593. [Google Scholar]
- Emanuelsson, O., Heijne, G.V. Prediction of organellar targeting signals. Biochim. Biophys. Acta. 2001;1541:114–119. doi: 10.1016/s0167-4889(01)00145-8. [DOI] [PubMed] [Google Scholar]
- Hage, A.E., Alix, J.H. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 2004;51:189–201. doi: 10.1046/j.1365-2958.2003.03813.x. [DOI] [PubMed] [Google Scholar]
- Herman, C., Thevenet, D., D'Ari, R., Bouloc, P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc. Natl. Acad. Sci. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag, D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Jiang, M., Datta, K., Walker, A., Strahler, J., Bagamasbad, P., Andrews, P.C., Maddock, J.R. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P., Mitta, M., Kim, Y., Jiang, W., Inouye, M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli . Proc. Natl. Acad. Sci. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, H., Musunuru, K., Jensen, K., Edo, C., Chen, H., Darnell, R., Burley, S. Sequence-specific RNA binding by a nova KH domain: Implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Link, A.J., Phillips, D., Church, G.M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: Application to open reading frame characterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D., Wyss, D.F. Solution structure of the hypothetical protein SAV1595 from Staphylococcus aureus, a putative RNA binding protein. J. Biomol. NMR. 2004;29:391–394. doi: 10.1023/B:JNMR.0000032524.62229.93. [DOI] [PubMed] [Google Scholar]
- Musco, G., Stier, G., Joseph, C., Morelli, M., Nilges, M., Gibson, T., Pastore, A. Three-dimensional structure and stability of the KH domain: Molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- Nakai, K., Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Neidhardt, F., Umbarger, H. Chemical composition of Escherichia coli. In: Neidhardt F.C., et al., editors. Escherichia coli and Salmonella: Cellular and molecular biology. 2d ed. Vol. 1. ASM Press; Washington, DC: 1996. pp. 13–16. [Google Scholar]
- Ostersetzer, O., Watkins, K., Cooke, A., Barkan, A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer, G., Barkan, A., Matthews, B. Crystal structure of E. coli YhbY: A representative of a novel class of RNA binding proteins. Structure. 2002;10:1593–1601. doi: 10.1016/s0969-2126(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Ostheimer, G., Williams-Carrier, R., Belcher, S., Osborne, E., Gierke, J., Barkan, A. Group II intron splicing factors derived by diversification of an ancient RNA binding module. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., Small, I. Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 2000;50:413–423. doi: 10.1007/s002390010044. [DOI] [PubMed] [Google Scholar]
- Pennisi, E. Drafting a tree. Science. 2003;300:1694. doi: 10.1126/science.300.5626.1694. [DOI] [PubMed] [Google Scholar]
- Rogozin, I.B., Makarova, K.S., Murvai, J., Czabarka, E., Wolf, Y.I., Tatusov, R.L., Szekely, L.A., Koonin, E.V. Connected gene neighborhoods in prokaryotic genomes. Nucleic Acids Res. 2002;30:2212–2223. doi: 10.1093/nar/30.10.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, R., Barta, A., Semrad, K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Small, I., Peeters, N., Legeai, F., Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics. 2004;4:1581–1590. doi: 10.1002/pmic.200300776. [DOI] [PubMed] [Google Scholar]
- Spirin, A. Ribosome preparation and cell-free protein synthesis. In: Hill W., et al., editors. The ribosome: Structure, function, and evolution. ASM Press; Washington, DC: 1990. pp. 56–70. [Google Scholar]
- Srivastava, A., Schlessinger, D. Coregulation of processing and translation: Mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. 1988;85:7144–7148. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till, B., Schmitz-Linneweber, C., Williams-Carrier, R., Barkan, A. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7:1227–1238. doi: 10.1017/s1355838201010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber, D.K., Williamson, J.R. Beyond kinetic traps in RNA folding. Curr. Opin. Struct. Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- Turmel, M., Otis, C., Lemieux, C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: Insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl. Acad. Sci. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, K.P., Dungan, J.M., Agabian, N. Identification of a small RNA that interacts with the 5′ splice site of the Trypanosoma brucei spliced leader RNA in vivo . Cell. 1994;76:171–182. doi: 10.1016/0092-8674(94)90181-3. [DOI] [PubMed] [Google Scholar]
- Weeks, K.M. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- Wikstrom, P., Bjork, G. Noncoordinate translation-level regulation of ribosomal and non-ribosomal protein genes in the Escherichia coli trmD operon. J. Bacteriol. 1988;170:3025–3031. doi: 10.1128/jb.170.7.3025-3031.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis, M., Krajewski, W., Chalamasetty, V., Reddy, P., Howard, A., Herzberg, O. Structure of HI1333 (YhbY), a putative RNA-binding protein from Haemophilus influenzae . Proteins. 2002;49:423–426. doi: 10.1002/prot.10225. [DOI] [PubMed] [Google Scholar]
- Woodson, S.A. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 2000;57:796–808. doi: 10.1007/s000180050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M., Yusupova, G., Baucom, A., Lieberman, K., Earnest, T., Cate, J., Noller, H. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]