Abstract
Previous studies of episodic memory report a greater extent of blood-oxygenation-level-dependent (BOLD) response in non-demented older adults with the apolipoprotein E epsilon-4 (APOE ε4) allele than in those without the allele. We conducted a functional MRI study to investigate whether APOE genotype is related to brain response to verbal paired-associate encoding and consolidation, particularly in the right hemisphere, among non-demented older adults. Structurally segmented volumes and BOLD response were measured in 13 non-ε4 and 12 ε4 subjects. The ε4 group displayed greater activation than the non-ε4 group in multiple right hemisphere regions for previously encoded word pairs relative to fixation. Activation within manually outlined hippocampal regions of interest also displayed genotype-specific dissociations consistent with whole brain analyses. Furthermore, this differential BOLD response occurred in the presence of equivalent behavioral and neuropsychological performances as well as comparable hippocampal and overall structural segmentation volumes between groups. Results implicate a widely distributed and interconnected network of right hemisphere brain regions that may be involved in compensating for APOE ε4-related deficiencies associated with verbal episodic memory encoding and consolidation.
Keywords: Verbal paired-associate encoding, Hippocampus, Apolipoprotein E, BOLD, Episodic memory, Non-demented older adults
1. Introduction
Subtle changes in episodic memory can be an early indicator of the development of Alzheimer's disease (AD) in non-demented older adults [1,2,5,10,21,23,24,27,43]. These early “preclinical” memory changes are more likely to occur in those with a genetic susceptibility marker for AD, the apolipoprotein E (APOE) ε4 allele [11,37], than in those without this risk factor. In parallel with these findings, structural [32,44] and functional [4,6,15,20,34,42] neuroimaging alterations are more evident in non-demented elderly with the ε4 allele than in those without this allele type.
These results suggest that the combined use of functional neuroimaging and genetic assessments may offer a promising avenue for early and accurate recognition of preclinical AD. The need for such sophisticated and sensitive multimodal approaches for early detection has increased with the advent of treatments that may be most effective when applied in the earliest stages of AD, before significant neuronal loss has accrued [31]. A few studies have investigated the relationship between APOE genotype and functional neuroimaging of memory processes in non-demented elderly, but the interpretation of these results has been confounded by a number of factors. These include: (1) poorer memory in the ε4 group than in the non-ε4 group [6], (2) questions of interpretability of signal change via the subtraction method, particularly when contrasting two higher-level conditions such as novel versus familiar items, (3) no assessment for differential atrophy [6,20], or (4) the use of general standard space region of interest (ROI) analyses rather than specific individualized native space renderings [4,20] (see also Vandenbroucke et al. [46] for discussion).
The present study served to address these concerns by examining APOE genotype-related differences in functional brain response to verbal paired-associate encoding and consolidation by controlling for equivalent behavioral performances and examining structural volumetry, including manually outlined hippocampal ROIs defined in native space, in ε4 and non-ε4 non-demented older adults. Given that elderly with the ε4 allele are more likely than those without the allele to be in a preclinical stage of AD, we hypothesized that the ε4 group would require more brain activation than the non-ε4 group to maintain the same level of memory performance. In addition, we expected an over-recruitment of right medial temporal lobe (MTL) activation during verbal paired-associate encoding and consolidation consistent with [26] finding that greater right MTL engagement is associated with better verbal list learning, and with the findings from a growing number of fMRI studies of at-risk older adults [4,15,16,20,25,26]. Thus, in the context of equivalent learning performance, we predicted that more right hemisphere response would be required by ε4 individuals.
2. Methods
2.1. Subjects
Twenty-five healthy and independently living older adults were recruited with institutional review board-approved procedures from a larger pool of volunteers participating in the UCSD Alzheimer's Disease Research Center as well as from a longitudinal study of aging. Participants were selected without regard to ethnicity, race, or socioeconomic status. The recruitment and study procedures were approved by the UCSD institutional review board and written informed consent was obtained from all subjects. All subjects were verified as non-demented based on extensive medical, neurological, laboratory, and neuropsychological evaluations. Participants with a reported history of alcohol and drug abuse, learning disability, neurological, or psychiatric illness (including depression) were routinely excluded from the pool of subjects.
All participants were genotyped for APOE allele type using a polymerase chain reaction based method [37]. The 25 participants were selected for scanning based on their APOE genotype and to ensure matching on demographic variables. There were 12 ε4 (one ε4/ε4, 11 ε3/ε4) and 13 non-ε4 (11 ε3/ε3, two ε2/ε3) participants. All but one of the ε4 and one of the non-ε4 participants were right-hand dominant for writing. As shown in Table 1, the two groups did not differ in mean age, education, gender distribution, or on global cognitive functioning (all p-values >0.16). Furthermore, the two groups did not differ in memory performance as measured by key indices from the Wechsler Memory Scale—Revised [47], the California Verbal Learning Test (CVLT) [14], or the Memory subscale of the Dementia Rating Scale [29].
Table 1.
Demographic, global cognitive, and learning and memory characteristics of the APOE ε4 and non-ε4 groups
| Variables | Apolipoprotein E genotype |
t | p | |||
|---|---|---|---|---|---|---|
| ε4(n = 12) |
non-ε4(n = 13) |
|||||
| Mean | S.D. | Mean | S.D. | |||
| Demographics/general condition | ||||||
| Age | 77.1 | 6.1 | 77.5 | 6.4 | 0.18 | 0.86 |
| Education | 15.1 | 1.9 | 16.2 | 2.9 | 1.10 | 0.28 |
| Gender (women/men) | 4/8 | 9/4 | 1.85 | 0.16 | ||
| Dementia rating scale | 140.3 | 3.4 | 139.5 | 2.6 | 0.66 | 0.52 |
| Learning and memory | ||||||
| DRS memory subscale | 24.3 | 0.5 | 23.9 | 1.1 | −1.44 | 0.16 |
| WMS-R immediate recall | 21.8 | 6.8 | 25.3 | 7.6 | 1.15 | 0.26 |
| WMS-R delayed recall | 19.1 | 8.9 | 21.5 | 9.5 | 0.61 | 0.55 |
| CVLT list 1–5 total recall | 42.2 | 15.7 | 49.2 | 10.8 | 1.30 | 0.21 |
| CVLT long delay free recall | 9.7 | 3.7 | 9.8 | 3.7 | 0.12 | 0.90 |
| CVLT recognition memory (%) | 89.9 | 10.6 | 92.2 | 8.4 | 0.61 | 0.55 |
| Post-MRI cued recall memory accuracy for word pairs (%) | 62.2 | 18.1 | 67.8 | 19.7 | 0.73 | 0.47 |
2.2. Materials and procedure
Task parameters were identical to those described by Fleisher et al. [20]. During functional MRI scanning, participants were instructed to learn 32 pairs of associated nouns presented for 5 s each, 16 of which they had previously learned to a criterion of 10 out of 16 correct on cured recall (OLD) and 16 of which were unfamiliar (NEW). No participant required more than three repetitions of the list to reach criterion prior to scanning (ε4: mean = 2.42, S.D. = 0.58; non-ε4: mean = 2.00, S.D. = 0.79; t = −1.51, p = 0.14). In addition to the learning instructions, subjects were also told to press a button in response to each word pair to indicate which word was capitalized in order to track attention to stimuli. Learning trials were presented in blocks of four trials each and interspersed with blocks of fixation (cross-hair visual depiction of “+”) that varied in length between 8 and 16 s. The blocks were presented in a fixed, pseudo-random order. Stimuli were presented to the subject via an LCD projector, back-projected onto a screen at the participant's feet. After scanning, self-paced cued recall of OLD and NEW words was assessed.
2.3. Imaging analysis
2.3.1. Anatomic and whole brain imaging
All scans were whole brain acquisitions conducted with a 1.5 T GE model Signa imager. High-resolution T1 weighted anatomical images were collected with an SPGR sequence (124 slices acquired in the sagittal plane; 1.2 mm slice thickness; 256 × 256 matrix; FOV = 250 mm; resulting in a 0.98 mm by 0.98 mm in-plane resolution). Functional data from 69 whole brain images of blood oxygen level dependent (BOLD) signal intensity were acquired axially using a gradient-recalled echo planar imaging (EPI) sequence. Four millimeter-thick slices sampled the entire brain (29 slices acquired in the axial plane; TR = 4000 ms; TE = 40 ms; flip angle = 90°; 64 × 64 matrix; FOV = 250 mm; 3.906 mm2 in-plane resolution).
2.3.2. Individual participant data analysis path
All analyses were conducted with Analysis of Functional NeuroImages (AFNI) Software [12], and all participant datasets were analyzed in the same scripted manner. A three-dimensional (3D) brick was created from the structural scan slices, and then they were warped into Talairach and Tournoux [45] standardized coordinate space. A 3D brick of image data was also created for each TR through the time course of the EPI scanning sequence. Small movement effects were minimized with the AFNI 3D volume registration program. Very occasionally when excessive motion occurred, the mean value of adjacent repetitions in voxels with residual motion was inserted in place of the motion-corrupted scans. A signal-intensity threshold-value was used to exclude low intensity values usually located outside the brain.
Each of the learning conditions (NEW and OLD items) was compared to a lower level baseline fixation condition (FIX) in order to circumvent some of the difficulties seen in prior studies in interpretability of BOLD signal changes from novel to familiar items. Thus, the comparisons of interest for the whole brain analyses and hippocampus-specific analyses were the difference in activation levels while viewing new word pairs versus fixation (i.e. NEW–FIX) and old word pairs versus fixation (i.e. OLD–FIX). A trapezoidal reference function was cross-correlated with the motion-corrected MR time course data in each voxel within the 3D functional brick. The fit coefficients for each contrast at each voxel were then resampled into isotropic voxels 4 mm per side and written into Talairach and Tournoux space [45]. Finally, functional datasets were blurred with an 8 mm FWHM kernel to reduce noise and account for correlations between adjacent voxels and variations in anatomy.
For native space hippocampal manual tracing analyses, gradient-recalled echo (GRE) scans from all subjects were reprocessed according to the criteria outlined above with the exception of blurring. Since blurring would significantly distort the functional data observed in the manually traced hippocampi, the 8 mm FWHM kernel was not used in order to preserve the most accurate individual variation in functional activation. GRE scans were carefully visually inspected for possible signal loss, and no noticeable signal loss was observed within our regions of interest. Datasets were aligned parallel to the anterior commissure–posterior commissure line (consistent with manual tracing, see below), but not warped into Talairach atlas space. Datasets were then resampled into isotropic voxels 4 mm per side and the mean fit coefficient across voxels within the left and right hemispheres of the hippocampal ROI was calculated separately.
2.3.3. Group data analysis path
T-statistic maps were created for each APOE genotype group to determine whether the mean intensity value difference between the NEW versus FIX and OLD versus FIX conditions in each voxel was significantly different than zero. Between-group t-maps were also generated to determine whether the pattern of average intensity value differences between the word pair conditions differed between the two APOE genotype groups. A commonly employed fMRI cluster thresholding technique (AlphaSim) was used to determine which areas of activation on the t-maps were significant by thresholding at a p-value of 0.025. The cluster size was predetermined as a region of at least 15 contiguous significant voxels (i.e. 960 mm3 volume), which controlled for a whole brain two-sided p-value of 0.05. This correction stringently protects the hypothesis that when no activation is present anywhere in the brain, a chance volume of activation will occur <2.5% of the time.
2.3.4. Structural data analysis path
2.3.4.1. Tissue segmentation
Whole brain segmentation of same session structural MRI scans was undertaken. Following N3 bias correction of field inhomogeneities [40], each scan was processed with one or both of the following automated skull-stripping methods: FreeSurfer's Hybrid Watershed Algorithm [38] and Brain Surface Extractor (Version 3.3) [36,39]. Both methods have been shown to be particularly effective when working with older adult images [19]. Scans were also manually edited when necessary to remove residual non-brain material. Tissue segmentation was performed using FSL's FAST (FMRIB's Automated Segmentation Tool), which segments a 3D image of the brain into gray matter, white matter, and CSF compartments using a hidden Markov random field model and an associated Expectation-Maximization algorithm [48]. Whole brain volume was also derived and proportions of each compartment were calculated.
2.3.4.2. Manually outlined hippocampal masks created in native space
Bilateral hippocampi were delineated via visual inspection and manual tracing performed in the coronal plane, perpendicular to the anterior–posterior commission line, using an approach adapted from previously published methods [30]. Tracings were performed by an experienced operator (AJJ), who was blind to participant identity and group, with established high levels of intra- and inter-rater reliability (intraclass correlation coefficients (ICC) > 0.90). Using a stereotactic approach, the anterior boundary of the hippocampus was chosen as the coronal slice through the fullest portion of the mammillary bodies. At this level, the hippocampus was delineated from the amygdala by a horizontal line from the anterior temporal horn to the alveus. Dorsally and laterally, the hippocampus continued to be bound by the temporal horn and alveus. The ventral boundary was demarcated by the parahippocampal white matter. Medially, the hippocampal boundary was defined by the ambient cistern. The posterior hippocampal boundary was traced on the last coronal slice on which the superior colliculi could be visualized. Bilateral hippocampal regions of interest were created from these tracings for each individual by resampling the identified voxels within these boundaries down to the 4 mm × 4mm × 4 mm resolution of the functional MRI data.
3. Results
3.1. Behavioral performances
There were no significant differences in performance between those with and without an APOE ε4 allele on post-MRI cued recall accuracy for the word pairs (Table 1). Both groups averaged better than 60% accuracy for cued recall following their scanning sessions (APOE ε4 group = 62.2%, S.D. = 18.1; APOE non-ε4 group = 67.8%, S.D. = 19.7; p = 0.47). Additionally, of those pairs correctly recalled, there were no significant differences in performance between groups with respect to correctly recalled new word pairs (APOE ε4 group = 31.8%, S.D. = 10.0; APOE non-ε4 group = 38.4%, S.D. = 7.37; p = 0.08) or correctly recalled old word pairs (APOE ε4 group = 68.2%, S.D. = 10.0; APOE non-ε4 group = 61.6%, S.D. = 7.37; p = 0.47).
3.2. Functional imaging
3.2.1. Whole brain analyses
3.2.1.1. Within-group contrasts
In response to new words relative to fixation, the ε4 group displayed significant clusters of activation in the left postcentral gyrus (123,392 mm3), right precentral gyrus (35,328 mm3), right inferior occipital gyrus (18,688 mm3), right culmen (11,328 mm3), right middle frontal gyrus (1984 mm3), right middle geniculum body (1792 mm3), left pyramis (1472 mm3), and right thalamus (1344 mm3). The non-ε4 group displayed activation in left precentral gyrus (115,712 mm3), right inferior occipital gyrus (36,096 mm3), right caudate tail (2176 mm3), and left inferior frontal gyrus (1152 mm3).
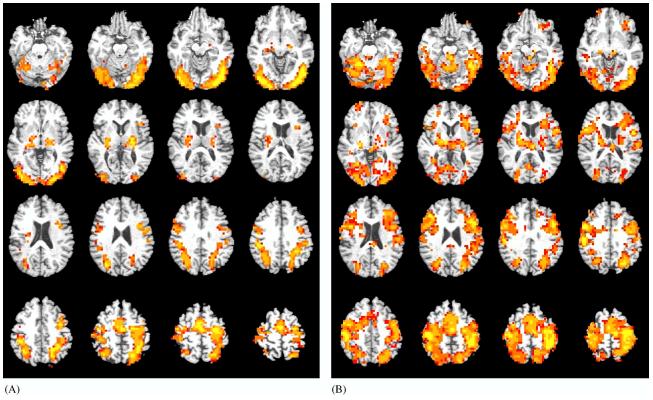
In response to old words relative to fixation, within-group analyses revealed diffuse bilateral activation extending into all lobes for the ε4 group with peak activation in the left inferior parietal lobule (379,584 mm3). This widespread pattern of activation observed for ε4 subjects met criteria for a single cluster. For the non-ε4 group, smaller significant clusters of activation were observed in left superior parietal lobule (54,016 mm3), left middle occipital gyrus (34,240 mm3), right cuneus (31,040 mm3), right inferior occipital gyrus (28,224 mm3), right (6464 mm3) and left (5184 mm3) putamen and lateral globus pallidus (Table 2 and Fig. 1).
Table 2.
Clusters of significantly greater brain activation during encoding of old word pairs versus fixation in non-demented older adults
| Brain region of maximum intensity voxel (MIV) | Volume (mm3) | Coordinates of MIV | η2 for OLD–FIX |
|---|---|---|---|
| APOE ε4 group | |||
| Widespread (left inferior parietal lobule maximum) | 379584 | −42, −37, 48 | 0.74 |
| APOE Non-ε4 group | |||
| Left superior parietal lobule | 54016 | −22, −61, 44 | 0.74 |
| Left middle occipital gyrus | 34240 | −42, −65, −4 | 0.73 |
| Right cuneus | 31040 | −30, 73, 28 | 0.72 |
| Right inferior occipital gyrus | 28224 | 38, −85, −4 | 0.72 |
| Right putamen and lateral globus pallidus | 6464 | 26, −13, 4 | 0.71 |
| Left putamen and lateral globus pallidus | 5184 | −22, −9, 4 | 0.74 |
| ε4 > Non-ε4 | |||
| Right anterior cingulate | 5504 | 22, 43, 8 | 0.50 |
| Right lingual gyrus | 2368 | 26, −57, 0 | 0.49 |
| Right middle temporal/parahippocampal gyri | 1856 | 50, −69, 24 | 0.52 |
| Right middle frontal gyrus | 1472 | 30, 31, 40 | 0.51 |
| Right posterior cingulate/precuneus | 1280 | 6, −61, 20 | 0.48 |
| Right cerebellar tonsil | 960 | 6, −45, −36 | 0.52 |
For the within-group analysis, ε4 subjects displayed widely diffuse and bilateral whole brain activation which met criteria for a single cluster (see Fig. 1), while non-ε4 subjects showed smaller significant clusters (note volumes for each cluster per group). For the between-group analysis, no significant clusters were observed such that non-ε4 > ε4.
Fig. 1.
Magnitude and direction of voxel-level activation to the task superimposed onto axial slices of a representative image in Taliarach space (slices span from 19 inferior to 56 superior in 4 mm increments). Activation displayed includes voxels significant at p < 0.025 that are contained within a cluster of 15 or more voxels. Color scale represents effect sizes for the within-subject difference between OLD items and FIXATION as measured by η2 (red voxels: 40 < η2 < 60; orange voxels: 60 < η2 < 80; yellow voxels: 80 < η2 < 100 [η2 indexes the effect size for the magnitude of the difference between the observed response and 0]). See also Table 2 for areas of significant activation. Images are presented in radiological view. (A) Non-ε4 group. (B) ε4 group.
3.2.1.2. Between-group contrasts
Between-group analyses for the NEW–FIX contrast revealed only a small cluster of greater activation in the right superior frontal gyrus (1216 mm3)inthe ε4 group than in the non-ε4 group. Analyses for the OLD–FIX contrast also only revealed greater areas of activation for the ε4 group than for the non-ε4 group, and all were within the right hemisphere (Table 2 and Fig. 2). These areas included the anterior cingulate (5504 mm3), lingual gyrus (2368 mm3), middle temporal and parahippocampal gyri (1856 mm3), middle frontal gyrus (1472 mm3), posterior cingulate and precuneus (1280 mm3), and cerebellar tonsil (960 mm3)(Fig. 3).
Fig. 2.
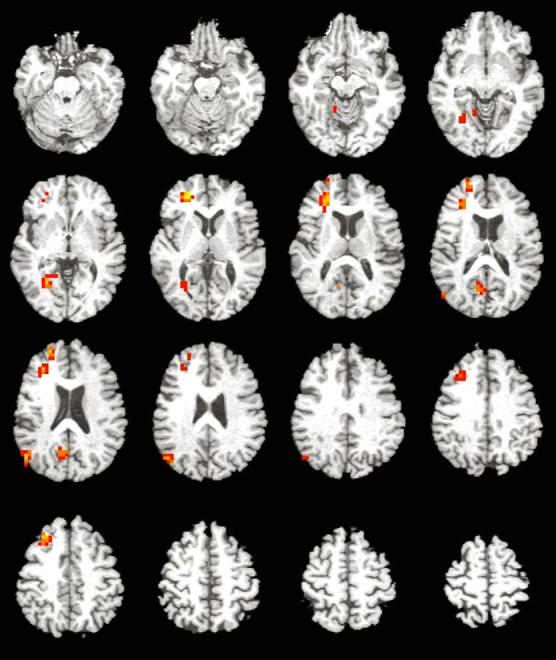
Magnitude and direction of voxel-level activation to the task superimposed onto axial slices of a representative image in Taliarach space (slices span from 19 inferior to 56 superior in 4 mm increments). Activation displayed includes voxels significant at p < 0.025 that are contained within a cluster of 15 or more voxels. Color scale represents effect sizes for the between-subject difference between OLD items and FIXATION as measured by η2 (red voxels: 40 < η2 < 60; orange voxels: 60 < η2 < 80; yellow voxels: 80 < η2 < 100 [η2 indexes the effect size for the magnitude of the difference between the observed response and 0]). See also Table 2 for areas of significant activation. Images are presented in radiological view.
Fig. 3.
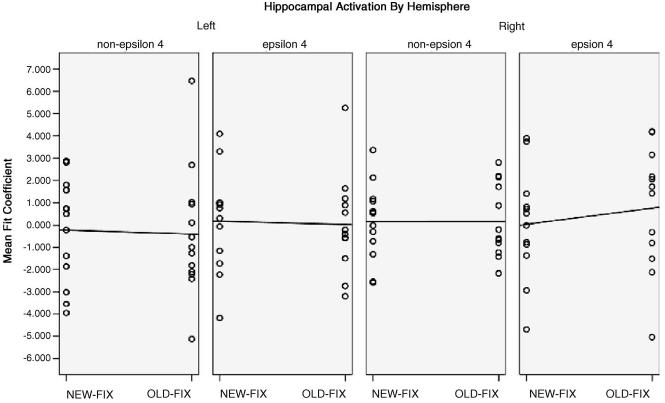
Scatterplots and linear regression estimations of mean fit coefficients for both left and right hippocampus according to NEW–FIX and OLD–FIX contrasts and APOE genotype.
3.2.2. Hippocampal activity
Although whole brain analyses did not reveal group differences in the hippocampus, these analyses can lack power to detect subtle effects due to the necessity for spatial blurring and corrections for multiple comparisons. In addition, it is difficult to test explicitly for laterality differences on a whole brain level. We therefore sought to test our a priori hypothesis about right hemisphere overactivation within the anatomically based, native space ROIs. Based on the previous observation of the hippocampus displaying an adaptation effect in response to previously presented material [25], it was expected that the magnitude of the NEW–FIX contrast would be greater than that of the OLD–FIX contrast for both groups in both hemispheres, with the exception that ε4 carriers would actually show an inverse pattern in the right hemisphere. In the left hemisphere, a small adaptation effect was observed as the NEW–FIX response was slightly greater than OLD–FIX response in both groups, although in the non-ε4 group this reflected a smaller negative response to NEW words (OLD: mean = −0.40, S.D. = 2.83; NEW: mean = −0.22, S.D. = 2.33) and in the ε4 group it reflected a greater positive response (OLD: mean = 0.03, S.D. = 2.21; NEW: mean = 0.17, S.D. = 2.29). For the right hemisphere, the non-ε4 group failed to show any evidence of an adaptation effect (NEW: mean = 0.15, S.D. = 1.69; OLD: mean = 0.16, S.D. = 1.60), whereas the ε4 group showed an overactivation effect, with greater response to OLD word pairs than to NEW (NEW: mean = 0.04, S.D. = 2.46; OLD: mean = 0.76, S.D. = 2.78). A Fisher's z-transformation of the mean individual difference in activation between NEW–FIX and OLD–FIX conditions for the left and right hippocampi was significant between groups (Zdiff = −1.93, p = 0.05).
3.2.3. Segmentation
Gray matter, white matter, and CSF volumes were analyzed in a MANCOVA model with total whole brain volume entered as a covariate. Results revealed no main multivariate effects for APOE genotype (F = 0.301, p = 0.83) and no main univariate effects for any of the measures (gray matter: F = 0.54, p = 0.47; white matter: F = 0.23, p = 0.63; CSF: F = 0.002, p = 0.96). Furthermore, multivariate analysis of the volumes within the left and right hippocampal ROIs revealed no significant differences between the two APOE genotype groups (left: F = 0.85, p = 0.37; right: F = 0.62, p = 0.44). Overall, the ε4 and non-ε4 groups did not differ on any of the segmentation measures.
4. Conclusion
APOE ε4 non-demented older adults showed greater BOLD response in multiple right hemisphere brain regions (anterior cingulate, lingual gyrus, middle temporal gyrus, middle frontal gyrus, posterior cingulate, precuneus, and cerebellar tonsil) during a verbal paired-associate learning task than their demographically similar non-ε4 counterparts. These differences were most salient in the OLD–FIX condition likely due to more successful encoding of familiar word pairs versus novel word pairs, as confirmed by the relatively poor post-scanning cued recall rates for new word pairs among all subjects. Results with the OLD–FIX contrast also suggest greater effort with consolidation processes among the APOE ε4 group. Furthermore, these differences in BOLD activation were seen in the absence of differences on multiple neuropsychological measures of learning and memory (DRS, WMS-R, CVLT, post-scanning cued recall memory) and despite comparable values for structural segmentation volumes of gray matter, white matter, CSF, and hippocampus. ROI analyses of activation in native space renderings of the hippocampi to the NEW–FIX and OLD–FIX conditions revealed similar patterns of relatively less activation in response to previously learned word pairs for the left hippocampus for both groups. However, they revealed discrepant response patterns for the right hippocampus by APOE genotype. Specifically, there was an enhanced response to OLD words among the APOE ε4 participants.
The current findings are largely congruent with previous functional neuroimaging studies investigating the role of APOE genotype on episodic memory encoding in older non-demented populations. Bookheimer et al. [6] were the first to identify increased BOLD signal in APOE ε4 participants during learning and recall, although their APOE ε4 group also showed lower performances in delayed verbal recall relative to their non-ε4 counterparts, increased variability with respect to age range, and no formal assessment for differential atrophy. The present study carefully controlled for group demographic characteristics and neuropsychological status, and special consideration was given to the possibility of structural differences in the form of gray matter, white matter, CSF, and hippocampal components. Importantly, these latter results support that our observed BOLD response differences within each of the hippocampal regions were not due to differential atrophy or partial volume effects between APOE genotype groups. We also undertook an additional learning-to-criterion procedure prior to scanning to maximize the contrast of OLD to NEW items and to focus our efforts on possible neurobiological distinctions between encoding and consolidation. This is in contrast to Bookheimer et al. [6] who examined signal change from combined learning and recall periods. Previous studies from our laboratory [4,20] also have consistently reported increased MTL signal associated with the APOE ε4 allele, and ROI analyses involving the hippocampus in these and other former studies have typically been based on a standardized anatomic atlas [12], thus not allowing for consideration of individual variation with respect to hippocampal size and consequent individual hippocampal BOLD activity. In fact, most fMRI studies average groups of different subjects into standard coordinate space, and anatomic differences in brain structure create additional variance and loss of registration accuracy [46]. We utilized native space manual outlining of individual hippocampi as structural masks for functional overlays, thereby allowing for better anatomic accuracy of hippocampal activity (see Vandenbroucke et al. [46] for discussion).
In addition, in the present study both new and old word pair conditions were contrasted against the same low level baseline condition (i.e. visual fixation) rather than with one another, thus obviating some of the difficulties seen in prior studies in interpretability of direction of BOLD signal changes from novel to familiar items. For example, prior studies – including our own – have often examined the difference in BOLD signal change between two ‘higher level’ contrasts (novel items versus familiar items), rather than contrasting either higher level condition with a lower level baseline condition such as visual fixation. It is conceivable that both higher level conditions have a good deal of neural activity associated with them and that encoding is not solely occurring during presentation of novel items. Buckner et al. [8], for example, have demonstrated encoding processes to be active during retrieval tasks as well during learning trials. Post-hoc between-group contrasts of the NEW versus OLD conditions in the present study revealed no significant differences, supporting this notion. Thus, when the method involves the higher level subtraction of novel from familiar items, either isolation of encoding processes to the novel condition or directionality of BOLD activity by APOE genotype cannot easily be inferred. The latter effect could be reflective of greater hippocampal signal change to novel items among ε4 subjects or greater signal change to familiar items among ε3 subjects. Our finding that hippocampal right hemisphere overactivation is confined to OLD word pairs among ε4 subjects is illustrative of this point.
As in previous investigations from our laboratory, results appear to be consistent with a compensatory hypothesis wherein APOE ε4 participants may require additional neurocognitive effort, as manifested by an increased BOLD response, to maintain an equivalent level of performance with non-ε4 counterparts. At-risk older persons may employ other brain regions to effectively compensate for episodic memory performance, and in so doing, actively prevent memory decline for a period of time. Our data suggest that additional frontal and temporal cortical resources are invoked for the ε4 group during episodic memory encoding, and thus implicate additional executive functions and semantic memory processes, respectively, in order to maintain comparable behavioral performances (see also Lange et al. [27]). These data are generally consistent with the HAROLD model [9], which posits a reduction in hemispheric asymmetry due to the aging process. Although left hemisphere differences were negligible, those participants at increased genetic risk for developing AD displayed greater areas of activation in the right hemisphere for previously learned stimuli. The correspondence of relatively greater activation in multiple right hemisphere regions with equivalent behavioral memory performances supports the notion of compensation through the employment of more bilateral network regions (see network view in Cabeza [9]). In all, support for the compensatory hypothesis has been demonstrated across a whole host of neuropsychological [2,10,27,41] and neuroimaging [3,4,6,22] investigations, as well as with neurochemical [13,17,35], neurotrophic [18], and mitochondrial DNA alterations [33].
Although the current study may be considered tentative support for the compensation hypothesis given the presence of APOE ε4 affiliated increases in activity in brain regions subsuming learning and memory, a necessary relationship must be established between hippocampal BOLD response and reported areas of increased activity in whole brain analyses. Hippocampal activity in the present study did show an expected adaptation effect with greater activation for NEW–FIX versus OLD–FIX in the left hippocampus, consistent with a growing body of evidence describing decreased activation in response to repeated stimuli and repetition priming paradigms in multiple cortical areas [7] as well as in the hippocampus [25]. However, the right hippocampus showed an APOE-dependent dissociation such that non-ε4 hippocampal activation remained stable and undifferentiated between the conditions whereas ε4 hippocampal activation showed a visible overactivation effect, with greater response in the OLD–FIX than the NEW–FIX contrast. Although newly developing functional connectivity analyses may further clarify the relationship between hippocampal activation and activity in other functionally related cortical regions, the consistency with regard to hemisphericity of genotypic cortical differences and hippocampal activation differences (all occurring in the right hemisphere) lends further support for the notion of a network of widely distributed, yet interconnected, cortical regions implicated in a circuit that may be over-activated to compensate for possible preclinical changes in episodic memory encoding.
Our present findings, combined with those of our prior studies [4] as well as those of Dickerson et al. [15,16] and Johnson et al. [25,26] provide converging support for alterations of the right MTL, irrespective of the modality of the encoding stimuli (i.e. verbal versus pictorial versus face-name combinations). Johnson et al. [26] have implicated right hemisphere regions as possibly facilitating more successful memory processing of verbal material in an fMRI study of CVLT performance. They extended these findings to patients with mild cognitive impairment (MCI) and showed a lack of adaptation among MCI patients to repeated stimuli in face learning [25]. In a pair of studies, Dickerson et al. [15,16] have also demonstrated specific activation patterns implicating the right medial temporal lobe in older adults. In the first of these studies, Dickerson et al. [16] showed that MCI patients recruited a larger extent of right hippocampal gyrus during picture encoding, and the greater extent of activation in this region appeared to herald subsequent decline over 2 years later. In the second of their studies, Dickerson et al. [15] found that memory performance on a face-name associative learning task was best predicted by right entorhinal cortex and hippocampal activation, and right hippocampal volume, in normally aging, MCI, and AD groups. Greater extent of entorhinal cortex activity was also shown for APOE ε4 carriers. Our prior work [4] with picture learning also demonstrated dissociations between left and right MTL activity between APOE genotype groups. Given our findings of greater activation in multiple right hemisphere regions among ε4 participants, we suggest that ε4 subjects may be more heavily recruiting such right medial temporal and association areas as a compensation strategy to maintain an equivalent level of performance.
One limitation of the present study is that cerebral blood flow was not measured, leaving open the possibility that other hemodynamic considerations might explain the current BOLD contrasts. Another limitation is the need for longitudinal follow-up of the cognitive and neurophysiologic status of the subjects to confirm whether or not present findings are truly indicative of a compensatory mechanism and predictive of subsequent cognitive decline or conversion to AD. Plans to monitor cognitive decline and possible conversion to AD in the present group are currently underway. Although group differences were significant in the right and not the left hemisphere, a third limitation is that these results may be construed as indirect evidence for a laterality effect, as this would be more stringently supported by direct comparisons of contra-lateral activation arising from ROIs of active right hemisphere regions from the current whole brain between-group analyses and their left hemisphere manually outlined native space correlates. A fourth consideration is that our results of greater activation in the ε4 subjects may also be construed as a progressive disinhibition of brain response, and therefore a precursor to a possible clinical disconnection syndrome [28]. Although a number of neuropsychological and functional neuroimaging studies of aging both within our laboratory and elsewhere provide support for viewing this greater activation as a compensatory mechanism, correlates of the present data set with longitudinal neurocognitive functioning will help elucidate which process, compensation or disinhibition, is most characteristic of the present findings. A fifth consideration is that gender may be partially influencing our results. Although there was not a significant gender difference between our two APOE groups, post-hoc analyses were conducted to investigate the effect of gender on the functional neuroimaging results. These analyses revealed no difference in hippocampal ROI activation according to gender. Small between-group gender differences were observed for whole brain analyses such that men appeared to show greater activation in right superior temporal regions and women appeared to show greater activation in left frontal regions; however, none of these regions overlapped with the reported differences in activation according to APOE genotype. Finally, the present study did not include post-hoc analyses to determine if the ε4 and non-ε4 groups differ in BOLD responses to the baseline condition. It should be noted, however, that no group baseline condition differences were observed in two previous studies from our laboratory [4,20].
To summarize, fMRI study of episodic encoding combined with genetic assessment presents a promising approach for early identification and possible treatment of preclinical AD. The present findings extend the growing literature on APOE ε4 genotypic differences in episodic memory among non-demented adults and support a compensatory hypothesis possibly targeting an interconnected network of broadly distributed right hemisphere regions. Further research is needed to clarify the issue of blood perfusion as related to APOE genotype as well as to concretely correlate the present findings with longitudinal cognitive and neurophysiologic changes.
Acknowledgements
This work was supported by National Institute on Aging Grants R01 AG12674 (MWB) and P50 AG05131 (JCB; MWB; ASF; DPS), and by the MIRECC grant from the Medical Research Service of the Department of Veterans Affairs (LTE; GGB). The authors gratefully acknowledge the assistance of staff, patients and volunteers of the UCSD Alzheimer's Disease Research Center, and the UCSD Laboratory of Cognitive Imaging.
Footnotes
There were no actual or potential conflicts of interest for the authors that could have inappropriately influenced the present work. Subjects were recruited in accordance with Internal Review Board (IRB) approved policies and procedures. Standard professional and ethical guidelines were upheld during the research study and manuscript preparation
References
- 1.Albert MS, Moss MB, Tanzi R, Jones K. Preclinical prediction of AD using neuropsychological tests. J Int Neuropsychol Soc. 2001;7:631–9. doi: 10.1017/s1355617701755105. [DOI] [PubMed] [Google Scholar]
- 2.Bäackman L, Small BJ, Fratiglioni L. Stability of the preclinical episodic memory deficit in Alzheimer's disease. Brain. 2001;124:96–102. doi: 10.1093/brain/124.1.96. [DOI] [PubMed] [Google Scholar]
- 3.Becker JT, Mintum MA, Aleva K, Wiseman MB, Nichols T, DeKosky ST. Compensatory reallocation of brain resources supporting verbal episodic memory in Alzheimer's disease. Neurology. 1996;46:692–700. doi: 10.1212/wnl.46.3.692. [DOI] [PubMed] [Google Scholar]
- 4.Bondi MW, Houston WS, Eyler LT, Brown GG. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer's disease. Neurology. 2005;64:501–8. doi: 10.1212/01.WNL.0000150885.00929.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondi MW, Monsch AU, Galasko D, Butters N, Salmon DP, Delis DC. Preclinical cognitive markers of dementia of the Alzheimer type. Neuropsychology. 1994;8:374–84. [Google Scholar]
- 6.Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, et al. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–6. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckner RL, Koutstaal W, Schacter DL, Rosen BR. Functional MRI evidence for a role of frontal and inferior temporal cortex in amodal components of priming. Brain. 2000;123:620–40. doi: 10.1093/brain/123.3.620. [DOI] [PubMed] [Google Scholar]
- 8.Buckner RL, Wheeler ME, Sheridan MA. Encoding processes during retrieval tasks. J Cogn Neurosci. 2001;13:406–15. doi: 10.1162/08989290151137430. [DOI] [PubMed] [Google Scholar]
- 9.Cabeza R. Hemisphereic asymmetry reduction in older adults: the HAROLD model. Psychol Aging. 2002;17:85–100. doi: 10.1037//0882-7974.17.1.85. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Ratcliff G, Belle SH, Cauley JA, DeKosky ST, Ganguli M. Patterns of cognitive decline in presymptomatic Alzheimer disease: a prospective community study. Arch Gen Psychiatr. 2001;58:853–8. doi: 10.1001/archpsyc.58.9.853. [DOI] [PubMed] [Google Scholar]
- 11.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 12.Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996;29:162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- 13.DeKosky ST, Ikonomovic MD, Styren SD, Beckett L, Wisniewski S, Bennett DA, et al. Upregulation of choline acetyltransferase activity in hippocampus and frontal cortex of elderly subjects with mild cognitive impairment. Ann Neurol. 2002;51:145–55. doi: 10.1002/ana.10069. [DOI] [PubMed] [Google Scholar]
- 14.Delis DC, Kramer JH, Kaplan E, Ober BA. The California verbal learning test. Psychological Corporation; New York: 1987. [Google Scholar]
- 15.Dickerson BC, Salat DH, Bates JF, Atiya M, Killiany RJ, Greve DN, et al. Medial temporal lobe function and structure in mild cognitive impairment. Ann Neurol. 2004;56:27–35. doi: 10.1002/ana.20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson BC, Salat DH, Greve DN, Chua EF, Rand-Giovannetti E, Rentz DM, et al. Increased hippocampal activation in mild cognitive impairment compared to normal aging and AD. Neurology. 2005;65:404–11. doi: 10.1212/01.wnl.0000171450.97464.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durany N, Michel T, Kurt J, Cruz-Sanchez FF, Cervas-Navarro J, Riederer P. Brain-derived neurotrophic factor and neurotrophin-3 levels in Alzheimer's disease. Int J Dev Neurosci. 2000;18:807–13. [PubMed] [Google Scholar]
- 18.Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–69. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 19.Fennema-Notestine C, Ozyurt IB, Clark CP, Morris S, Bischoff-Grethe A, Bondi MW, et al. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: effects of diagnosis, bias correction, and slice location. Human Brain Mapping. doi: 10.1002/hbm.20161. in press, doi:10.1002/hbm.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleisher AS, Houston WS, Eyler LT, Frye S, Jenkins C, Thal LJ, Bondi MW. Identification of Alzheimer disease risk by functional magnetic resonance imaging. Arch Neurol. 2005;62(12):1881–8. doi: 10.1001/archneur.62.12.1881. [DOI] [PubMed] [Google Scholar]
- 21.Fuld PA, Masur DM, Blau AD, Crystal H, Aronson MK. Object-memory evaluation for prospective detection of dementia in normal functioning elderly: Predictive and normative data. J Clin Exp Neuropsychol. 1990;12:520–8. doi: 10.1080/01688639008400998. [DOI] [PubMed] [Google Scholar]
- 22.Grady CL, McIntosh AR, Beig S, Keightley ML, Burian H, Black SE. Evidence from functional neuroimaging of a compensatory prefrontal network in Alzheimer's disease. J Neurosci. 2003;23:986–93. doi: 10.1523/JNEUROSCI.23-03-00986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grober E, Kawas C. Learning and retention in preclinical and early Alzheimer's disease. Psychol Aging. 1997;12:183–8. doi: 10.1037//0882-7974.12.1.183. [DOI] [PubMed] [Google Scholar]
- 24.Howieson DB, Dame A, Camicioli R, Sexton G, Payami H, Kaye JA. Cognitive markers preceding Alzheimer's dementia in the healthy oldest old. J Am Ger Soc. 1997;45:584–9. doi: 10.1111/j.1532-5415.1997.tb03091.x. [DOI] [PubMed] [Google Scholar]
- 25.Johnson SC, Baxter LC, Susskind-Wilder L, Connor DJ, Sabbagh MN, Caselli RJ. Hippocampal adaptation to face repetition in healthy elderly and mild cognitive impairment. Neuropsychologia. 2004;42:980–9. doi: 10.1016/j.neuropsychologia.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Johnson SC, Saykin AJ, Flashman LA, McAllister TW, Sparling MB. Brain activation on fMRI and verbal memory ability: functional neuroanatomic correlates of CVLT performance. J Int Neuropsychol Soc. 2001;7:55–62. doi: 10.1017/s135561770171106x. [DOI] [PubMed] [Google Scholar]
- 27.Lange KL, Bondi MW, Galasko DG, Delis DC, Salmon DP, Thal LJ. Decline in verbal memory during preclinical Alzheimer's disease: examination of the effect of Apolipoprotein E genotype. J Int Neuropsychol Soc. 2002;8:943–55. doi: 10.1017/s1355617702870096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B, Cabeza R, McIntosh AR, Black SE, Grady CL, Stuss DT. Functional reorganization of memory after traumatic brain injury: a study with H-sub-21-sup-50 positron emission tomography. J Neurol Neurosurg Psychiatr. 2002;73:173–81. doi: 10.1136/jnnp.73.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattis S. Dementia Rating Scale: Professional Manual. Psychological Assessment Resources; Odessa, FL: 1988. [Google Scholar]
- 30.Nagel BJ, Palmer SL, Reddick WE, Glass JO, Helton KJ, Wu S, et al. Abnormal hippocampal development in children with medulloblastoma treated with risk-adapted irradiation. Am J Neurorad. 2004;25:1575–82. [PMC free article] [PubMed] [Google Scholar]
- 31.Peterson RC, Thomas RG, Grundman M, Bennett D, Doody R, Ferris S, et al. Vitamin E and donepezil for the treatment of mild cognitive impairment. N Engl J Med. 2005;352:2379–88. doi: 10.1056/NEJMoa050151. [DOI] [PubMed] [Google Scholar]
- 32.Plassman BL, Welsh-Bohmer KA, Bigler ED, Johnson SC, Anderson CV, Helms MJ, et al. Apolipoprotein E ε4 allele and hippocampal volume in twin with normal cognition. Neurology. 1997;48:985–9. doi: 10.1212/wnl.48.4.985. [DOI] [PubMed] [Google Scholar]
- 33.Reddy PH, McWeeney S, Manczak M, Park BS, Gutala RV, Jung Y, et al. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: upregulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer's disease. Hum Mol Genet. 2004;13:1225–40. doi: 10.1093/hmg/ddh140. [DOI] [PubMed] [Google Scholar]
- 34.Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, et al. Preclinical evidence of Alzheimer's disease in persons homozygous for the ε4 allele for APOE. N Engl J Med. 1996;334:752–8. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 35.Rombouts SA, Barkhof F, van Meel CS, Scheltens P. Alterations in brain activation during cholinergic enhancement with rivastigmine in Alzheimer's disease. J Neurol Neurosurg Psychiatr. 2002;73:665–71. doi: 10.1136/jnnp.73.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandor S, Leahy R. Surface-based labeling of cortical anatomy using a deformable database. IEEE Trans Med Imag. 1997;16:41–54. doi: 10.1109/42.552054. [DOI] [PubMed] [Google Scholar]
- 37.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele ε4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–72. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 38.Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, et al. A hybrid approach to the skull stripping problem in MRI. NeuroImage. 2004;22(3):1060–75. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 39.Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13(5):856–76. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- 40.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- 41.Small SA, Chawla MK, Buonocore M, Rapp PR, Barnes CA. Imaging correlates of brain function in monkeys and rats isolates a hippocampal subregion differentially vulnerable to aging. Proc Natl Acad Sci. 2004;101:7181–6. doi: 10.1073/pnas.0400285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Small GW, Mazziotta JC, Collins MT, Baxter LR, Phelps ME, Mandelkern MA, et al. Apolipoprotein E type 4 allele and cerebral glucose metabolism in relatives at risk for familial Alzheimer disease. J Am Med Assoc. 1995;273:942–7. [PubMed] [Google Scholar]
- 43.Smith GE, Bohac DL, Waring SC, Kokmen E, Tangalos EG, Ivnik RJ, et al. Apolipoprotein E genotype influences cognitive ‘phenotype’ in patients with Alzheimer's disease but not in healthy control subjects. Neurology. 1998;50:355–62. doi: 10.1212/wnl.50.2.355. [DOI] [PubMed] [Google Scholar]
- 44.Soininen H, Partanen K, Pitkanen A, Hallikainen M, Hanninen T, Helisalmi S, et al. Decreased hippocampal volume asymmetry on MRIs in non-demented elderly subjects carrying the apolipoprotein E ε4 allele. Neurology. 1995;45:391–2. doi: 10.1212/wnl.45.2.391. [DOI] [PubMed] [Google Scholar]
- 45.Talairach J, Tournoux P. A coplanar stereotaxic atlas of the human brain. Thieme Medical Publishers; New York, NY: 1988. [Google Scholar]
- 46.Vandenbroucke MWG, Goekoop R, Duschek EJJ, Netelenbos JC, Kuijer JP, Barkhof F, et al. Interindividual differences of medial temporal lobe activation during encoding in an elderly population studied by fMRI. NeuroImage. 2004;21:173–80. doi: 10.1016/j.neuroimage.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 47.Wechsler D. Wechsler memory scale—revised. Psychological Corporation; New York: 1987. [Google Scholar]
- 48.Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation maximization algorithm. IEEE Trans Med Imag. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]