Abstract
Tropomyosin is a well-characterized regulator of muscle contraction. It also stabilizes actin filaments in a variety of muscle and non-muscle cells. Although these two functions of tropomyosin could have different impacts on actin cytoskeletal organization, their functional relationship has not been studied in the same experimental system. Here, we investigated how tropomyosin stabilizes actin filaments and how this function is influenced by muscle contraction in Caenorhabditis elegans body wall muscle. We confirmed the antagonistic role of tropomyosin against UNC-60B, a muscle-specific ADF/cofilin isoform, in actin filament organization using multiple UNC-60B mutant alleles. Tropomyosin was also antagonistic to UNC-78 (AIP1) in vivo and protected actin filaments from disassembly by UNC-60B and UNC-78 in vitro, suggesting that tropomyosin protects actin filaments from the ADF/cofilin-AIP1 actin disassembly system in muscle cells. A mutation in the myosin heavy chain caused greater reduction in contractility than tropomyosin depletion. However, the myosin mutation showed much weaker suppression of the phenotypes of ADF/cofilin or AIP1 mutants than tropomyosin depletion. These results suggest that muscle contraction has only minor influence on the tropomyosin’s protective role against ADF/cofilin and AIP1, and that the two functions of tropomyosin in actin stability and muscle contraction are independent of each other.
Keywords: tropomyosin, ADF/cofilin, AIP1, myofibrils, actin dynamics
INTRODUCTION
Proper regulation of disassembly and stabilization of actin filaments are generally important for assembly, maintenance, and reorganization of actin cytoskeletal structures. Myofibrils, stable contractile apparatuses in muscle, are also subjected to constant exchange of actin subunits within the thin filaments [Imanaka-Yoshida et al., 1993; Komiyama et al., 1993; Littlefield et al., 2001], which is probably required for maintaining their organized structures. Actin depolymerizing factor (ADF)/cofilin enhances actin dynamics by severing actin filaments and accelerating monomer dissociation from the pointed ends [Bamburg, 1999; Bamburg et al., 1999], and its functional significance in muscle cells have been demonstrated in several organisms [Obinata et al., 1997; Ono 2003a]. On the other hand, tropomyosin is one of the major F-actin binding proteins in muscle and provides stability to actin filaments [Cooper, 2002; Gunning et al., 2005]. Tropomyosin slows down dissociation of actin monomers from actin filaments [Lal and Korn, 1986; Hitchcock-DeGregori et al., 1988; Broschat et al., 1989; Broschat, 1990] and protects actin filaments from severing by gelsolin [Fattoum et al., 1983; Ishikawa et al., 1989a,b; Nyakern-Meazza et al., 2002] and ADF/cofilin (see below).
Several biochemical studies have demonstrated that ADF/cofilin and tropomyosin compete for binding to F-actin and antagonistically regulate actin filament dynamics in vitro [Bernstein and Bamburg, 1982; Mabuchi, 1982; Nishida et al., 1984, 1985]. A genetic study in the nematode Caenorhabditis elegans showed that tropomyosin has an inhibitory role for ADF/cofilin-mediated actin filament dynamics in the body wall muscle in vivo [Ono and Ono, 2002]. In C. elegans,the unc-60 gene encodes two ADF/cofilin isoforms, UNC-60A and UNC-60B, which are generated by alternative pre-mRNA splicing [McKim et al., 1994; Anyanful et al., 2004]. UNC-60B is specifically expressed in body wall muscle, and mutations in unc-60B cause severe disorganization of actin filaments [Waterston et al., 1980; Zengel and Epstein, 1980; Ono et al., 1999, 2003]. C. elegans tropomyosin (CeTM) is encoded by the lev-11/tmy-1 gene which produces multiple splice variants [Williams and Waterston, 1994; Kagawa et al., 1995]. RNA interference of CeTM induces disorganization of actin filaments in wild-type back-ground, but has minimal effects on an unc-60B mutant [Ono and Ono, 2002], suggesting that CeTM protects actin from disassembly by UNC-60B. Actin disassembly is also regulated by the unc-78 gene encoding actin interacting protein 1 (AIP1) in body wall muscle [Ono, 2001; Mohri et al., 2006]. AIP1 is a conserved regulator of actin dynamics and enhances disassembly of actin filaments only in the presence of ADF/cofilin [Ono, 2003b]. The UNC-78 protein has very strong activity to enhance disassembly of UNC-60B-bound actin filaments in vitro [Mohri and Ono, 2003; Ono et al., 2004]. However, a regulatory mechanism of the activity of UNC-78 is unknown.
Tropomyosin is also well-characterized as a regulator of muscle contraction [Perry, 2003; Brown and Cohen, 2005]. In striated muscle, tropomyosin anchors troponin to the thin filaments [Ebashi and Kodama, 1966] and transmits a calcium signal to the thin filaments to activate actomyosin interaction [Ebashi, 1984; Squire and Morris, 1998; Gordon et al., 2000]. The actomyosin contractile activity is necessary for proper assembly of striated myofibrils [Soeno et al., 1999; De Deyne, 2000; Ramachandran et al., 2003; Kagawa et al., 2006]. However, excessive contraction can be a mechanical stress that disrupts the myofibril organization [Lieber et al., 1991; Friden and Lieber, 1992]. In C. elegans, disorganization of muscle actin filaments due to mutations in unc-87, a calponin-repeat protein [Goetinck and Waterston, 1994] or RNA interference of kettin, an immunoglobulin-like repeat protein [Ono et al., 2006] can be suppressed by a myosin mutation that reduces muscle contraction, suggesting that these F-actin-binding proteins also stabilize actin filaments. Thus, tropomyosin physically stabilizes actin filaments, but it also activates cycling of contraction and relaxation, which could lead to a mechanical stress to the myofibrils. However, these two functions of tropomyosin have not been examined in parallel in the same experimental system.
In this study, we investigated how tropomyosin functionally interacts with the ADF/cofilin-AIP1 actin disassembly system and how muscle contraction affects this interaction. We found that tropomyosin is antagonistic to ADF/cofilin and AIP1 in vitro and in vivo, and that muscle contraction has only a weak effect on this interaction in vivo. The results suggest that tropomyosin has two independent roles as a stabilizer of actin filaments and as a regulator of muscle contraction.
MATERIALS AND METHODS
C. elegans Strains and Culture
N2 (wild-type) and unc-54(s95) [Moerman et al., 1982] were obtained from Caenorhabditis Genetics Center (Minneapolis, MN). unc-60B(r398) and unc-60B(s1309) [McKim et al., 1988] were provided by Dr. David L. Baillie (Simon Fraser University, Burnaby, Canada). The original unc-60B(r398) strain was outcrossed three times with N2 by Dr. Myeongwoo Lee (Baylor University, Waco, TX) and provided to us. unc-78(gk27) [Ono, 2001] was provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia (Vancouver, Canada). Double mutant strains, unc-54(s95);unc-60B(r398), unc-54(s95);unc-60B(s1309), and unc-54(s95);unc-78(gk27) were generated by standard crosses, and homozygosity of the mutant alleles was confirmed by complementation tests. All mutants were examined as homozygotes. Nematodes were grown under standard conditions at 20°C as described previously [Brenner, 1974].
RNA Interference Experiments
Nematodes were treated with RNA interference (RNAi) for CeTM by feeding Escherichia coli expressing double-stranded RNA under conditions described previously [Ono and Ono, 2002]. Control experiments were performed with the E. coli strain HT115 (DE3) that was transformed with an empty RNAi vector L4440 (provided by Dr. Andrew Fire, Stanford University, Stanford, CA) [Timmons and Fire, 1998; Timmons et al., 2001]. Two vectors for RNAi of CeTM, TM1 for CeTMI, II(RNAi) (targeting two CeTM isoforms) and TM2 for CeT-MI,II,III,IV(RNAi) (targeting four CeTM isoforms), were described previously [Ono and Ono, 2002, 2004]. Phenotypes were analyzed in their F1 generation.
Worm Motility Assay
Worm motility was quantified as described [Epstein and Thomson, 1974]. Briefly, adult worms were placed in M9 buffer. Then, one beat was counted when a worm swung its head to either right or left. The total number of beats in 30 s was recorded.
Phalloidin Staining
Staining of worms with tetramethylrhodamine-phalloidin was performed as described previously [Ono, 2001]. Samples were viewed by epifluorescence using a Nikon Eclipse TE2000 inverted microscope with a CFI Plan Fluor ELWD 40× objective (dry, N.A.= 0.60). Images were captured by a SPOT RT Monochrome CCD camera (Diagnostic Instruments, Sterling Heights, MI) and processed by the IPLab imaging software (Scanalytics, Rockville, MD) and Adobe Photoshop 6.0.
Actin Pelleting Assay
Rabbit muscle actin [Pardee and Spudich, 1982], CeTM [Ono and Ono, 2002], recombinant UNC-60B [Ono and Benian, 1998], and recombinant glutathione S-transferase (GST)-tagged UNC-78 [Mohri et al., 2004] were purified as described previously. Actin pelleting assays were performed as described previously [Mohri et al., 2004] with slight modifications. F-actin (10 μM) was pre-incubated with or without 5 μM CeTM for 30 min in a buffer containing 0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-NaOH, 1 mM dithiothreitol, pH 7.5. Then, final 10 μM UNC-60B and 0-2 μM GST-UNC-78 were added to the reactions, and they were incubated for 15 min. They were ultracentrifuged at 80,000 rpm (285,000× g) for 20 min in a Beckman TLA-100 rotor. The supernatants and pellets were adjusted to the same volumes and analyzed by SDS-PAGE. Gels were stained with Coomassie brilliant blue R-250 (National Diagnostics, Atlanta, GA) and scanned by a UMAX PowerLook III scanner at 300 dpi, and the band intensity was quantified by Scion Image Beta 4.02 (Scion, Frederick, MD).
RESULTS AND DISCUSSION
Re-Evaluation of the Effect of CeTM-RNA Interference on unc-60B ADF/Cofilin Mutants
We previously demonstrated that RNA interference (RNAi) of C. elegans tropomyosin (CeTM) caused disorganization of actin filaments in body wall muscle and sterility due to ovulation defects in wild-type back-ground. This treatment did not worsen the actin organization in body wall muscle [Ono and Ono, 2002] or alter the ovulation process [Ono and Ono, 2004] in unc-60B(r398), a weak loss-of-function mutant of unc-60B (a muscle-specific ADF/cofilin isoform). However, preliminary experiments by Dr. M. Lee’s group (Baylor University, Waco, TX, personal communication) suggested that the unc-60B(r398) strain might have an RNAi-defective phenotype. Therefore, we compared the original unc-60B(r398) strain and the unc-60B(r398) strain that had been outcrossed three times with wild-type for their phenotypes after the CeTM-RNAi treatment.
Worm motility was measured to quantify the contractile activity of body wall muscle [Epstein and Thomson, 1974]. Motility of the original unc-60B(r398) strain was not greatly affected by the CeTM-RNAi treatments [Ono and Ono, 2002], while that of the outcrossed unc-60B(r398) strain was reduced 25 and 50% by CeTMI,II (RNAi) (targeting two CeTM isoforms) and CeT-MI,II,III,IV (RNAi) (targeting four CeTM isoforms), respectively (Fig. 1). The original unc-60B(r398) strain showed nearly normal ovulation and produced progeny even after the CeTM-RNAi treatments [Ono and Ono, 2004], whereas the outcrossed unc-60B(r398) strain became sterile by the CeTM-RNAi treatments as shown by the absence of embryos on the culture plates (Figs. 2E and 2F). Organization of actin filaments in the body wall muscle was not significantly modified by the CeTM-RNAi treatments in the original unc-60B(r398) strain [Ono and Ono, 2002]. However, in the outcrossed strain, the actin organization was improved by the CeTM-RNAi treatments with less actin aggregates and more striated myofibrillar actin filaments (Figs. 3D-3F). CeTMI,II,III,IV (RNAi) had stronger suppression effects on the actin organization than CeTMI,II (RNAi) (Fig. 3, compare E and F), which is in reverse correlation to their effects on worm motility (Fig. 1).
Fig. 1.
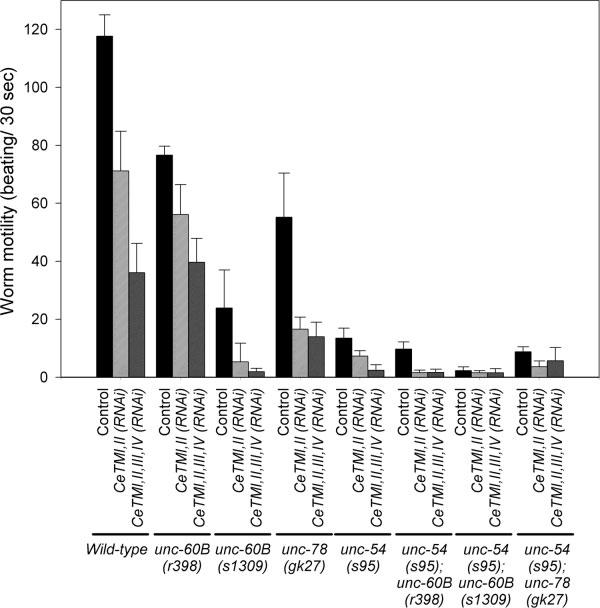
Effects of CeTM-RNAi and mutations in unc-60B, unc-78,and unc-54 on worm motility. Motility of wild-type and other mutant worms with treatments with control RNAi, CeTMI,II (RNAi),or CeTMI,II,III,IV (RNAi) was quantified as beating frequency. Data are means ± standard deviations (n = 10).
Fig. 2.
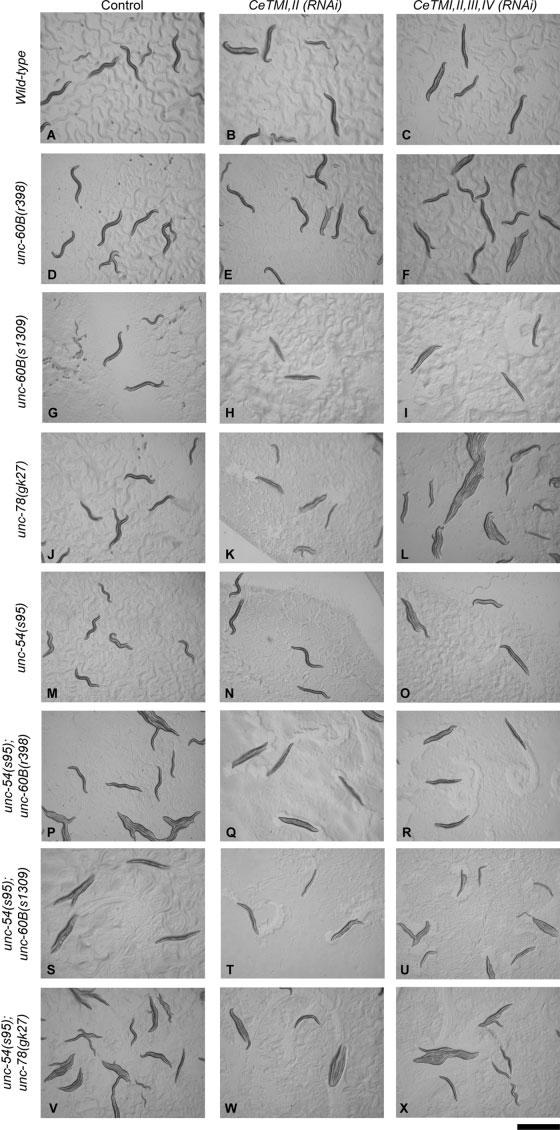
Effects of CeTM-RNAi and mutations in unc-60B, unc-78, and unc-54 on worm movement. Micrographs of live adult worms on culture plates are shown. Wild-type worms showed typical sinusoidal movement (A). However, reduction in muscle contractility and paralysis were represented by increase in the appearance of more straight worms. Bar, 1.0 mm.
Fig. 3.
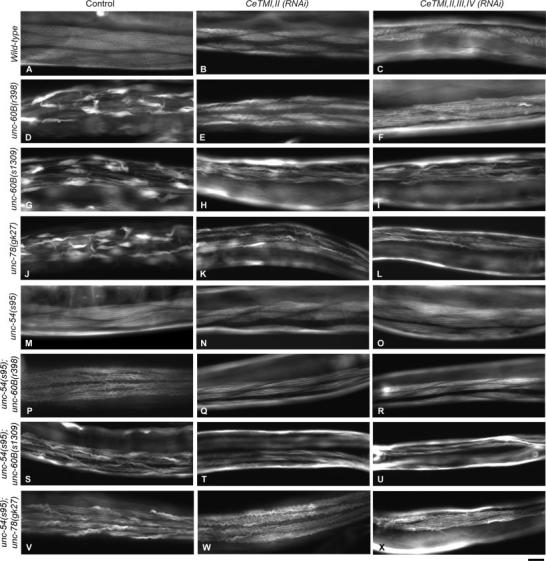
Effects of CeTM-RNAi and mutations in unc-60B, unc-78, and unc-54 on actin organization in the body wall muscle. Adult worms were stained with tetramethylrhodamine-phalloidin to visualize F-actin. Wild-type muscle showed organized striated pattern of actin filaments (A). CeTMI,II (RNAi) or CeTMI,II,III,IV (RNAi) increased formation of wavy actin bundles in wild-type (B and C), while mutations in unc-60B (D and G) or unc-78 (J) caused aggregation of actin filaments. The actin aggregates in unc-60B or unc-78 mutants were suppressed by CeTMI,II (RNAi) (E, H, and K) or CeT-MI,II,III,IV (RNAi) (F, I, and L). The unc-54 mutation partially suppressed the phenotype in the unc-60B (P and S) or unc-78 (V) mutants. CeTMI,II (RNAi) or CeTMI,II,III,IV suppressed formation of actin aggregates in the unc-54;unc-60B (Q, R, T, and U) or unc-54;unc-78 (W and X) double mutants. Bar, 20 μm.
We conclude that the original unc-60B(r398) strain had a background mutation that weakens the effects of RNAi treatments, and that outcrosses successfully removed this mutation. Although the nature of the background mutation is unknown, the effect appears to be tissue- or gene-specific, because the protein levels of CeTM in total worm extracts were reduced in the original unc-60B(r398) strain by the CeTM-RNAi treatments [Ono and Ono, 2002], and RNAi of pat-10 troponin C caused paralysis in the original unc-60B(r398) strain in a similar manner to wild-type [Ono and Ono, 2004]. Therefore, we need to re-evaluate our two previous conclusions based on the results obtained with the outcrossed unc-60B(r398) strain. First, we previously concluded that CeTM and UNC-60B antagonistically regulate the actin organization in the body wall muscle [Ono and Ono, 2002]. Our new results actually confirmed this conclusion with much more marked in vivo effects. in vitro, UNC-60B enhances actin filament dynamics [Ono and Benian, 1998; Ono et al., 1999; Yamashiro et al., 2005], while CeTM stabilizes actin filaments [Ono and Ono, 2002]. Thus, the result that RNAi of CeTM suppressed the unc-60B loss-of-function phenotype is consistent with their antagonistic roles on actin filament dynamics. Second, we previously concluded that CeTM and UNC-60B also antagonistically function during ovulation [Ono and Ono, 2004]. However, the new result that CeTM-RNAi caused sterility in the outcrossed unc-60B(r398) strain strongly suggests that the absence of ovulation defects in the original unc-60B(r398) strain was likely due to the background mutation but not the unc-60B mutation.
To determine if the suppression of the Unc-60B phenotype by the CeTM-RNAi treatments is an allele- or strain-specific event, we examined the effect of CeTM-RNAi on a different unc-60B allele. unc-60B(s1309) is a stronger loss-of-function allele than unc-60B(r398) [McKim et al., 1988; Ono et al., 1999]. unc-60B(r398) produces a mutant UNC-60B protein with normal G-actin-binding but defective F-actin-binding and severing activities in vitro [Ono et al., 1999, 2001], while the mutant UNC-60B protein in unc-60B(s1309) shows severe defects in both G- and F-actin-binding and severing activities in vitro [Ono et al., 1999]. The unc-60B(s1309) homozygotes moved slower than unc-60B(r398) (Fig. 1, compare controls for unc-60B(r398) and unc-60B (s1309)). CeTMI,II (RNAi) significantly reduced motility of unc-60B(s1309) (Fig. 1 and Fig. 2H), and CeT-MI,II,III,IV (RNAi) caused nearly complete paralysis (Fig. 1, Fig. 2I). In contrast, actin organization in the body wall muscle of unc-60B(s1309) was improved by the CeTM-RNAi treatments (Figs. 3G-3I). Although the myofibrils are still disorganized, both CeTMI,II (RNAi) and CeTMI,II,III,IV (RNAi) similarly reduced formation of actin aggregates and enhanced organization of actin into a striated pattern (Figs. 3H and 3I). Thus, the antagonistic effects of CeTM and UNC-60B were also confirmed by a strong loss-of-function unc-60B allele, suggesting that their functional interaction is not allele- or strain-specific.
Interestingly, RNAi of CeTM and unc-60B mutations, in particular with a strong loss-of-function allele unc-60B(s1309), synergistically reduced worm motility, (Fig. 1 and Figs. 2G-2I), while actin organization was improved by a combination of CeTM-RNAi and an unc-60B mutation (Figs. 3D-3I). These apparently paradoxical phenotypes suggest uncoupling of the regulation of actin organization from the regulation of muscle contraction. Although the combination of CeTM-RNAi and unc-60B mutations improved actin organization in the myofibrils (Figs. 3D-3I), other myofibrillar components that are required for contraction, such as troponin, might not be properly recruited to the thin filaments when CeTM is knocked down. Also, even if the actin filaments are apparently organized, dynamics of the myofibrillar proteins may not be properly regulated when CeTM and UNC-60B are impaired. In addition to dynamic exchange of actin subunits within myofibrils [Imanaka-Yoshida et al., 1993; Komiyama et al., 1993; Littlefield et al., 2001], even tropomyosin and troponin undergo constant turnover within mature myofibrils [Michele et al., 1999]. Such protein dynamics might be dependent on the activities of CeTM and UNC-60B to maintain normal contractile activity.
Suppression of the unc-78 AIP1 Mutant Phenotype by CeTM-RNAi
UNC-78/AIP1 enhances disassembly of UNC-60B-bound actin filaments and is required for organized assembly of actin filaments in body wall muscle [Ono, 2001; Mohri and Ono, 2003; Mohri et al., 2006]. Therefore, we reasoned that UNC-78 might also be antagonistic to CeTM for actin organization in the body wall muscle. We tested functional interaction between UNC-78 and CeTM by examining CeTM-RNAi phenotypes in an unc-78 mutant background. unc-78(gk27) is a null allele, and homozygous animals do not express the UNC-78 protein [Ono, 2001; Mohri and Ono, 2003]. CeTMI,II (RNAi) and CeTMI,II,III,IV (RNAi) reduced worm motility of unc-78(gk27) (Fig. 1), and the treated worms were nearly paralyzed on the culture plates (Figs. 2K and 2L). On the other hand, actin aggregates in the body wall muscle were diminished and striated organization was improved in the CeTM-RNAi-treated unc-78(gk27) worms (Figs. 3J-3L). CeTMI,II,III,IV (RNAi) caused a stronger suppression effect than CeTMI,II (RNAi) (Fig. 3, compare K and L). These results are similar to the antagonism between CeTM and UNC-60B and strongly suggest that CeTM is also antagonistic to UNC-78 for organized assembly of actin filaments in the body wall muscle. Our results are consistent with the recently reported antagonism between AIP1 and tropomyosin for stability of actin cables in budding yeast [Okada et al., 2006]. Therefore, the antagonistic roles of tropomyosin and the ADF/cofilin-AIP1 actin disassembly system might be a conserved mechanism to regulate assembly and maintenance of actin cytoskeletal structures.
The antagonistic role of CeTM against the UNC-60B-UNC-78 system was further characterized in vitro by an actin pelleting assay (Fig. 4). Native CeTM was purified from a crude thin filament fraction [Ono and Ono, 2002]. Therefore, the CeTM preparation is likely to be a mixture of multiple CeTM isoforms. However, the purified CeTM was resolved as a single band on SDS-PAGE, which corresponded to the size of high-molecular-weight isoforms, CeTMI and/or CeTMII [Kagawa et al., 1995]. F-actin (10 μM) was pre-incubated for 30 min with buffer alone or with 5 μM CeTM that should saturate F-actin binding [Ono and Ono, 2002], and then, UNC-60B and UNC-78 were added to the mixture. After incubating 15 min, extent of actin disassembly was determined by a pelleting assay as described in Materials and Methods. In the absence of CeTM in the pre-incubation, increasing concentrations of GST-UNC-78 (0-2 μM) enhanced disassembly of actin filaments and decreased the amounts of actin in the pellets as shown by SDS-PAGE (Fig. 4A, -CeTM; compare intensity of the actin bands in p). Densitometric quantification of actin in the pellets showed that less than 50% of actin sedimented in the presence of 2 μM GST-UNC-78, while more than 80% of actin was present in the pellet in the absence of GST-UNC-78 (Fig. 4B, black circles). However, when actin was pre-incubated with CeTM, actin disassembly was significantly inhibited (Fig. 4A, +CeTM; see that majority of actin was fractionated in p in the presence of 1.0 or 2.0 μM GST-UNC-78). Densitometric quantification indicated that more than 70% of actin sedimented in the presence of 2 μM GST-UNC-78 (Fig. 4B, white circles). The differences in the amounts of pelletable actin with and without CeTM were statistically significant (P < 0.01). Thus, these biochemical results show that CeTM binds to F-actin and protects it from actin disassembly by UNC-60B and UNC-78.
Fig. 4.
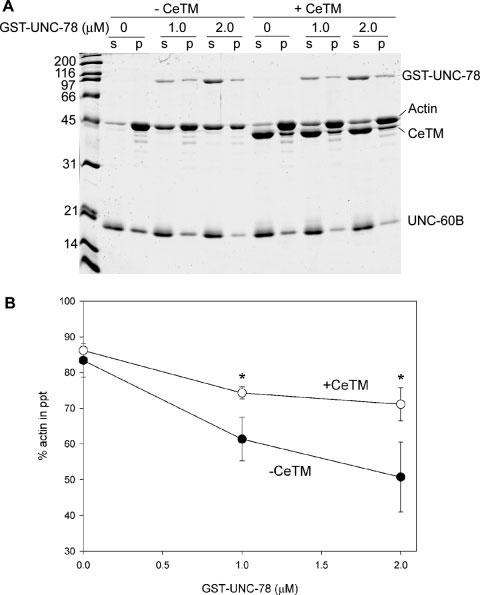
An inhibitory effect of CeTM on actin disassembly by UNC-60B and UNC-78. (A) 10 μM F-actin was pre-incubated with or without 5 μM CeTM for 30 min, and then, final 10 μM UNC-60B and 0-2 μM GST-UNC-78 were added. After incubating 15 min, the reactions were ultracentrifuged and fractionated into supernatants (s) and pellets (p). (B) Quantitative data of the actin pelleting assays. Percentages of actin in the pellets are plotted as a function of the concentrations of GST-UNC-78. Black circles are data without CeTM, and white circles are data with 5 μM CeTM. Data are means ± standard deviations of three experiments. Asterisks indicate P < 0.01 by the t-test.
The biochemical results on the protective role of CeTM against actin disassembly by UNC-60B (ADF/cofilin) and UNC-78 (AIP1) are consistent with the in vivo data on the antagonism between CeTM and UNC-60B or UNC-78. AIP1 enhances disassembly of actin filaments in the presence of ADF/cofilin, while it has negligible effects on actin in the absence of ADF/cofilin [Aizawa et al., 1999; Okada et al., 1999; Rodal et al., 1999; Balcer et al., 2003; Mohri and Ono, 2003; Ono et al., 2004]. AIP1 has both actin- and ADF/cofilin-binding sites that are predicted to be necessary for its specific interaction with ADF/cofilin-bound actin filaments [Rodal et al., 1999; Mohri et al., 2004; Clark et al., 2006; Mohri et al., 2006; Okada et al., 2006]. Therefore, CeTM (tropomyosin) prevented decoration of actin filaments with UNC-60B (ADF/cofilin) as described previously [Ono and Ono, 2002], and, as a result, UNC-78 (AIP1) was unable to bind and disassemble CeTM-decorated actin filaments. Thus, one of the physiological functions of tropomyosin is to stabilize actin filaments by preventing them from disassembly by ADF/cofilin and AIP1.
Weak Suppression of the unc-60B and unc-78 Phenotypes by a Myosin Mutation
Tropomyosin not only stabilizes actin filaments but also regulates muscle contraction. Reduction of motility in the CeTM-RNAi-treated worms is likely due to defective regulation of cycling of muscle contraction and relaxation. However, muscle contraction can cause a mechanical stress and destabilize the contractile apparatuses. Therefore, the suppression of the Unc-60B and Unc-78 phenotypes by CeTM-RNAi could also be due to reduced muscle contraction in addition to alteration in the actin dynamics. To examine the effect of muscle contraction on actin stability, muscle contraction was reduced by a mutation of the UNC-54 myosin heavy chain independently of CeTM, and its effect on the unc-60B or Unc-78 phenotypes was examined.
UNC-54 is the major myosin heavy chain in the body wall muscle [MacLeod et al., 1977; Miller et al., 1983]. The unc-54(s95) mutation impairs muscle contraction without affecting the structure of myofibrils [Moerman et al., 1982]. A missense mutation in unc-54(s95) converts Gly-118 to Arg that is located near the ATP-binding site of the myosin head [Dibb et al., 1985], suggesting that this mutation alters myosin ATPase activity but not myosin’s ability to bind to actin and to assemble into thick filaments. This myosin mutation has been shown to suppress disorganization of myofibrils due to loss of thin filament proteins, UNC-87 [Goetinck and Waterston, 1994] and kettin [Ono et al., 2006]. CeT-MI,II (RNAi) and CeTMI,II,III,IV (RNAi) reduced worm motility of unc-54(s95) (Fig. 1) with a stronger effect by CeTMI,II,III,IV (RNAi) than CeTMI,II (RNAi) (Fig. 1 and compare Figs. 2N and 2O). CeTMI,II (RNAi) and CeTMI,II,III,IV (RNAi) induced only minor disorganization of actin filaments in unc-54(s95) (Figs. 3N and 3O) as compared to wild-type in which CeTMI,II (RNAi) and CeTMI,II,III,IV (RNAi) caused formation of wavy actin bundles (Figs. 3B and 3C). These results suggest that disorganization of actin filaments by the CeTM-RNAi treatments is partly due to actomyosin contractility, and that this phenotype is suppressed by reduced myosin activity. This is also supported by the observation that CeTMI,II (RNAi) caused stronger actin disorganization in unc-54(s95) than CeTMI,II,III,IV (RNAi) (Fig. 3, compare N and O), which is in reverse correlation to their effects on worm motility (Fig. 1).
The unc-54(s95) mutation reduced worm motility of unc-60B or unc-78 mutants (Fig. 1) and weakly suppressed disorganization of the actin filaments (Figs. 3M, 3P, 3S, and 3V). The actin organization was improved with diminished actin aggregates in unc-54(s95);unc-60B(r398) (Fig. 3, compare D and P), while it was only moderately improved and smaller actin bundles were still formed in unc-54(s95);unc-60B(s1309) (Fig. 3, compare G and S) or unc-54(s95);unc-78(gk27) (Fig. 3, compare J and V). The phenotype of unc-60B(r398) was weaker than that of unc-60B(s1309), which is probably why the extent of suppression by the myosin mutation was more noticeable in unc-60B(r398) than in unc-60B(s1309). Importantly, worm motility was reduced by the myosin mutation to greater extents than by the CeTM-RNAi treatments (Fig. 1), yet the myosin mutation had much weaker suppression effects on actin organization than the CeTM-RNAi treatments (Figs. 3E, 3F, 3H, 3I, 3K, and 3L). This strongly suggests that reduced muscle contraction can only weakly suppress disorganized myofibrils when the actin disassembly system is defective, and that RNAi of CeTM suppresses this phenotype by altering the stability of actin filaments rather than by reducing contractile activity.
To further test the role of CeTM as a stabilizer of actin filaments, the unc-54;unc-60B or unc-54;unc-78 double mutants were treated with CeTMI,II (RNAi) or CeTMI,II, III,IV (RNAi). The double mutants were already paralyzed under the control conditions (Fig. 1 and Figs. 2P, 2S, and 2V), and either of the CeTM-RNAi treatments further reduced motility of unc-54(s95);unc-60B(r398) (Fig. 1 and Figs. 2P-2R) but did not have additional effects on motility of unc-54(s95);unc-60B(s1309) and unc-54(s95);unc-78(gk27) (Fig. 1 and Figs. 2S-2X). In all three double mutant strains, CeTM-RNAi reduced formation of actin aggregates and enhanced striated organization of actin filaments (Figs. 3P-3X). In the unc-54(s95);unc-60B(r398) mutant, wavy actin bundles in control worms (Fig. 3P) were diminished and actin filaments were more clearly assembled into a striated pattern by CeTMI,II (RNAi) (Fig. 3Q) or CeTMI,II,III,IV (RNAi) (Fig. 3R). In the unc-54(s95); unc-60B(s1309) double mutant, CeT-MI,II,III,IV (RNAi) (Fig. 3U) showed a slightly stronger effect than CeTMI,II (RNAi) (Fig. 3T) in reducing long wavy actin aggregates that are found in the control worms (Fig. 3S). In the unc-54(s95); unc-78(gk27) double mutant, the CeTMI,II (RNAi) worms had thinner wavy actin bundles than the control worms (Fig. 3, compare W and V), while CeTMI,II (RNAi) apparently induced more extensive waviness to the actin bundles (Fig. 3W). However, CeTMI,II,III,IV (RNAi) significantly improved the actin organization with fewer abnormal actin bundles (Fig. 3, compare X and V).
These results demonstrate that RNAi of CeTM in already paralyzed double mutant worms can still cause significant suppression of the phenotypes in actin organization in the unc-60B and unc-78 mutants. We observed several different patterns of actin disorganization. Typically, abnormal actin filaments became large aggregates (Figs. 3D, 3G, and 3J) or thin and wavy actin bundles (Figs. 3B, 3P, 3S, and 3W). Although we do not know how these abnormal structures are formed, there is tendency that severe defects in actin organization result in large actin aggregates, while modest defects cause thin and wavy bundles. For example, formation of large actin aggregates in unc-60B(r398) (Fig. 3D) could be transformed into wavy actin bundles by CeTMI,II (RNAi) (Fig. 3E) or the unc-54(s95) mutation (Fig. 3P), suggesting that these phenotypes indicate severity in the defects of the actin-regulatory system. However, we cannot rule out the possibility that these phenotypes are caused by different mechanisms. Taken together, these observations support the function of CeTM as a stabilizer of actin filaments that is antagonistic to the ADF/cofilin-AIP1 actin disassembly system independently of its function as a regulator of muscle contraction.
CONCLUSION
Based on the current data and previous observations, we present a model of the relationship between actin dynamics and muscle contraction in organization of myofibrils (Fig. 5). Previous biochemical studies have shown that UNC-60B (ADF/cofilin) and UNC-78 (AIP1) cooperatively enhance actin dynamics by disassembling actin filaments [Ono et al., 1999; Mohri et al., 2006], while CeTM (tropomyosin) antagonistically stabilizes actin filaments [Ono and Ono, 2002]. These factors are expected to maintain the balance between dynamic and stable actin filaments (Fig. 5A), and stable actin filaments might be assembled into organized myofibrils by unknown mechanisms (Fig. 5A). As a separate function, CeTM activates actomyosin interaction and enhances muscle contraction (Fig. 5A). Moderate actomyosin contractility facilitates assembly of actin into myofibrils as shown in other experimental systems [Soeno et al., 1999; De Deyne, 2000; Ramachandran et al., 2003; Kagawa et al., 2006], although it has not been demonstrated in C. elegans. However, excessive contraction could disrupt the myofibril structure [Lieber et al., 1991; Friden and Lieber, 1992], and the actin filaments might be subjected to disassembly by the ADF/cofilin-AIP1 system (Fig. 5A).
Fig. 5.
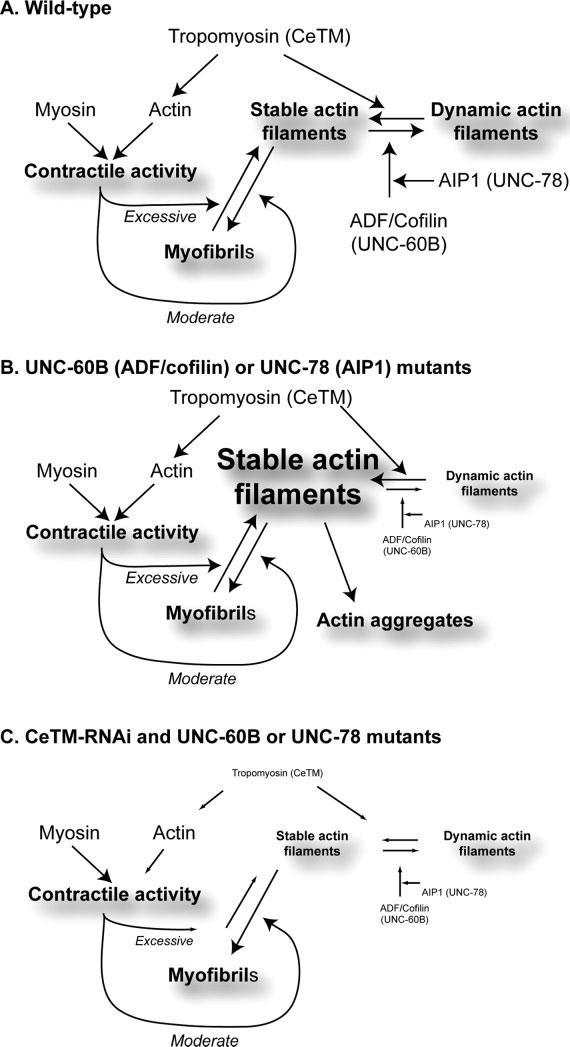
Models of the dual functions of tropomyosin in actin stability and actomyosin contractility in assembly and maintenance of myofibrils. Models are proposed for wild-type (A), UNC-60B (ADF/cofilin) or UNC-78 (AIP1) mutants, or CeTM-RNAi and UNC-60B or UNC-78 mutants (C).
When UNC-60B (ADF/cofilin) or UNC-78 (AIP1) is defective (Fig. 5B), the activity to disassemble actin is decreased, and stable actin filaments might be increased by CeTM. Although actin could still be assembled into myofibrils, excessive actin filaments may not be depolymerized and may form actin aggregates (Fig. 5B). Actomyosin contractility could partially disrupt myofibrils, and dissociated actin filaments may be incorporated into the actin aggregates. This hypothesis is supported by our results that the unc-54(s95) myosin mutation partially suppresses formation of actin aggregates in the unc-60B and unc-78 mutants (Figs. 3P, 3S, and 3V). The mechanism of formation of actin aggregates is not known. However, actin aggregates in unc-60B mutants contain CeTM [Ono and Ono, 2002], but not α-actinin or vinculin [Ono et al., 2003], suggesting that CeTM might stabilize actin filaments in the aggregates. When CeTM is depleted in unc-60B or unc-78 mutants (Fig. 5C), stable vs. dynamic actin filaments are better balanced, and proper amounts of actin might be integrated into myofibrils. Depletion of CeTM also reduces muscle contraction, which could reduce mechanical disruption of myofibrils. These models still need to be tested rigorously in C. elegans and other organisms. In particular, dynamics of actin filaments and other myofibrillar proteins are measurable in live cells using greenfluorescent protein-tagged proteins [Dabiri et al., 1999] and advanced microscopy techniques, such as fluorescence recovery after photobleaching [Lippincott-Schwartz et al., 2003]. These live cell studies should be very useful for testing these models.
Tropomyosin is associated with thin filaments in all muscle types and modulates contraction [Perry, 2003; Brown and Cohen, 2005]. In particular, in striated muscle, tropomyosin, together with troponin, is a core component of the actin-linked regulatory system for contraction [Ebashi, 1984; Squire and Morris, 1998; Gordon et al., 2000]. ADF/cofilin is also commonly expressed in muscle cells and believed to enhance reorganization of the actin filaments [Ono, 2003a]. Importantly, musclespecific ADF/cofilin isoforms are adapted for functions in muscle cells in mammals [Ono et al., 1994; Mohri et al., 2000; Thirion et al., 2001; Vartiainen et al., 2002; Nakashima et al., 2005] and C. elegans [Ono and Benian, 1998; Ono et al., 1999; Yamashiro et al., 2005]. Therefore, tropomyosin is expected to be a common inhibitor of the ADF/cofilin-dependent actin dynamics in other muscle systems. In chick cardiac myocytes, tropomyosin and tropomodulin protect thin filaments from depolymerization [Mudry et al., 2003], most likely by inhibiting the action of ADF/cofilin. Mutations in tropomyosin are associated with several muscle diseases, including nemaline myopathy, hypertrophic cardiomyopathy, and dilated cardiomyopathy [Michele and Metzger, 2000; Tubridy et al., 2001; Clarkson et al., 2004; Chang and Potter, 2005]. Biochemical studies on some of the pathogenic mutations in tropomyosin showed that its activity to control the actomyosin interaction is impaired by the mutations [Golitsina et al., 1997; Moraczewska et al., 2000]. However, some of the pathogenic mutations may alter tropomyosin’s activity to stabilize actin filaments. As a result, actin filaments might be exposed to the ADF/cofilin-AIP1 actin disassembly system, and the myofibrils might be disorganized. Thus, the dual functions of tropomyosin in contraction and actin stability could be crucial for maintaining normal muscle activity.
Tropomyosin is also associated with contractile actomyosin structures in non-muscle cells, such as stress fibers [Pittenger et al., 1994; Lin et al., 1997; Gunning et al., 2005]. In motile cells, tropomyosin is generally absent from the leading edge where ADF/cofilin is concentrated [DesMarais et al., 2002], with exception of several tropomyosin isoforms [Bryce et al., 2003; Hillberg et al., 2006]. However, tropomyosin localizes to lamella, which is the proximal region of membrane protrusions, and is proposed to recruit myosin and promote cell migration [Gupton et al., 2005]. Thus, during cell migration, the dual functions of tropomyosin might be important to convert dynamic actin filaments into a contractile structure and support actomyosin contraction. Further studies in other experimental systems might reveal general significance of the functions of tropomyosin in actin stability and actomyosin contractility.
ACKNOWLEDGMENTS
The authors thank Kanako Ono and Sawako Yamashiro for technical assistance. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institute of Health National Center for Research Resources.
Footnotes
Contract grant sponsor: National Science Foundation; Contract grant number: MCB-0110464; Contract grant sponsor: National Institute of Health; Contract grant number: R01 AR48615.
REFERENCES
- Aizawa H, Katadae M, Maruya M, Sameshima M, Murakami-Murofushi K, Yahara I. Hyperosmotic stress-induced reorganization of actin bundles in Dictyostelium cells over-expressing cofilin. Genes Cells. 1999;4:311–324. doi: 10.1046/j.1365-2443.1999.00262.x. [DOI] [PubMed] [Google Scholar]
- Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in Caenorhabditis elegans. J Cell Biol. 2004;167:639–647. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr Biol. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Bamburg JR. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, McGough A, Ono S. Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 1999;9:364–370. doi: 10.1016/s0962-8924(99)01619-0. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broschat KO. Tropomyosin prevents depolymerization of actin filaments from the pointed end. J Biol Chem. 1990;265:21323–21329. [PubMed] [Google Scholar]
- Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28:8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: How structure illuminates function. Adv Protein Chem. 2005;71:121–159. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AN, Potter JD. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail Rev. 2005;10:225–235. doi: 10.1007/s10741-005-5252-6. [DOI] [PubMed] [Google Scholar]
- Clark MG, Teply J, Haarer BK, Viggiano SC, Sept D, Amberg DC. A genetic dissection of Aip1p’s interactions leads to a model for Aip1p-cofilin cooperative activities. Mol Biol Cell. 2006;17:1971–1984. doi: 10.1091/mbc.E05-10-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson E, Costa CF, Machesky LM. Congenital myopathies: Diseases of the actin cytoskeleton. J Pathol. 2004;204:407–417. doi: 10.1002/path.1648. [DOI] [PubMed] [Google Scholar]
- Cooper J. Actin dynamics: Tropomyosin provides stability. Curr Biol. 2002;12:R523–R525. doi: 10.1016/s0960-9822(02)01028-x. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Ayoob JC, Turnacioglu KK, Sanger JM, Sanger JW. Use of green fluorescent proteins linked to cytoskeletal proteins to analyze myofibrillogenesis in living cells. Methods Enzymol. 1999;302:171–186. doi: 10.1016/s0076-6879(99)02017-0. [DOI] [PubMed] [Google Scholar]
- De Deyne PG. Formation of sarcomeres in developing myotubes: Role of mechanical stretch and contractile activation. Am J Physiol Cell Physiol. 2000;279:C1801–C1811. doi: 10.1152/ajpcell.2000.279.6.C1801. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Ichetovkin I, Condeelis J, Hitchcock-DeGregori SE. Spatial regulation of actin dynamics: A tropomyosin-free, actin-rich compartment at the leading edge. J Cell Sci. 2002;115:4649–4660. doi: 10.1242/jcs.00147. [DOI] [PubMed] [Google Scholar]
- Dibb NJ, Brown DM, Karn J, Moerman DG, Bolten SL, Waterston RH. Sequence analysis of mutations that affect the synthesis, assembly and enzymatic activity of the unc-54 myosin heavy chain of Caenorhabditis elegans. J Mol Biol. 1985;183:543–551. doi: 10.1016/0022-2836(85)90170-6. [DOI] [PubMed] [Google Scholar]
- Ebashi S. Ca2+ and the contractile proteins. J Mol Cell Cardiol. 1984;16:129–136. doi: 10.1016/s0022-2828(84)80701-4. [DOI] [PubMed] [Google Scholar]
- Ebashi S, Kodama A. Interaction of troponin with F-actin in the presence of tropomyosin. J Biochem (Tokyo) 1966;59:425–426. doi: 10.1093/oxfordjournals.jbchem.a128320. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature. 1974;250:579–580. doi: 10.1038/250579a0. [DOI] [PubMed] [Google Scholar]
- Fattoum A, Hartwig JH, Stossel TP. Isolation and some structural and functional properties of macrophage tropomyosin. Biochemistry. 1983;22:1187–1193. doi: 10.1021/bi00274a031. [DOI] [PubMed] [Google Scholar]
- Friden J, Lieber RL. Structural and mechanical basis of exercise-induced muscle injury. Med Sci Sports Exerc. 1992;24:521–530. [PubMed] [Google Scholar]
- Goetinck S, Waterston RH. The Caenorhabditis elegans UNC-87 protein is essential for maintenance, but not assembly, of bodywall muscle. J Cell Biol. 1994;127:71–78. doi: 10.1083/jcb.127.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golitsina N, An Y, Greenfield NJ, Thierfelder L, Iizuka K, Seidman JG, Seidman CE, Lehrer SS, Hitchcock-DeGregori SE. Effects of two familial hypertrophic cardiomyopathy-causing mutations on alpha-tropomyosin structure and function. Biochemistry. 1997;36:4637–4642. doi: 10.1021/bi962970y. [DOI] [PubMed] [Google Scholar]
- Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: Divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Gupton SL, Anderson KL, Kole TP, Fischer RS, Ponti A, Hitchcock-DeGregori SE, Danuser G, Fowler VM, Wirtz D, Hanein D, Waterman-Storer CM. Cell migration without a lamellipodium: Translation of actin dynamics into cell movement mediated by tropomyosin. J Cell Biol. 2005;168:619–631. doi: 10.1083/jcb.200406063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillberg L, Zhao Rathje L-S, Nyakern-Meazza M, Helfand B, Goldman RD, Schutt CE, Lindberg U. Tropomyosins are present in lamellipodia of motile cells. Eur J Cell Biol. 2006;85:399–409. doi: 10.1016/j.ejcb.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Sampath P, Pollard TD. Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry. 1988;27:9182–9185. doi: 10.1021/bi00426a016. [DOI] [PubMed] [Google Scholar]
- Imanaka-Yoshida K, Sanger JM, Sanger JW. Contractile protein dynamics of myofibrils in paired adult rat cardiomyocytes. Cell Motil Cytoskeleton. 1993;26:301–312. doi: 10.1002/cm.970260405. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Annealing of gelsolin-severed actin fragments by tropomyosin in the presence of Ca2+. Potentiation of the annealing process by caldesmon. J Biol Chem. 1989a;264:16764–16770. [PubMed] [Google Scholar]
- Ishikawa R, Yamashiro S, Matsumura F. Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J Biol Chem. 1989b;264:7490–7497. [PubMed] [Google Scholar]
- Kagawa H, Sugimoto K, Matsumoto H, Inoue T, Imadzu H, Takuwa K, Sakube Y. Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J Mol Biol. 1995;251:603–613. doi: 10.1006/jmbi.1995.0459. [DOI] [PubMed] [Google Scholar]
- Kagawa M, Sato N, Obinata T. Effects of BTS (N-benzyl-p-toluene sulphonamide), an inhibitor for myosin-actin interaction, on myofibrillogenesis in skeletal muscle cells in culture. Zool Sci. doi: 10.2108/zsj.23.969. (in press) [DOI] [PubMed] [Google Scholar]
- Komiyama M, Kouchi K, Maruyama K, Shimada Y. Dynamics of actin and assembly of connectin (titin) during myofibrillogenesis in embryonic chick cardiac muscle cells in vitro. Dev Dyn. 1993;196:291–299. doi: 10.1002/aja.1001960412. [DOI] [PubMed] [Google Scholar]
- Lal AA, Korn ED. Effect of muscle tropomyosin on the kinetics of polymerization of muscle actin. Biochemistry. 1986;25:1154–1158. doi: 10.1021/bi00353a031. [DOI] [PubMed] [Google Scholar]
- Lieber RL, Woodburn TM, Friden J. Muscle damage induced by eccentric contractions of 25% strain. J Appl Physiol. 1991;70:2498–2507. doi: 10.1152/jappl.1991.70.6.2498. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol. 1997;170:1–38. doi: 10.1016/s0074-7696(08)61619-8. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH. Photobleaching and photoactivation: Following protein dynamics in living cells. Nat Cell Biol. 2003;5(Suppl):S7–S14. [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- Mabuchi I. Effects of muscle proteins on the interaction between actin and an actin-depolymerizing protein from starfish oocytes. J Biochem (Tokyo) 1982;92:1439–1447. doi: 10.1093/oxfordjournals.jbchem.a134068. [DOI] [PubMed] [Google Scholar]
- MacLeod AR, Waterston RH, Fishpool RM, Brenner S. Identification of the structural gene for a myosin heavy-chain in Caenorhabditis elegans. J Mol Biol. 1977;114:133–140. doi: 10.1016/0022-2836(77)90287-x. [DOI] [PubMed] [Google Scholar]
- McKim KS, Heschl MF, Rosenbluth RE, Baillie DL. Genetic organization of the unc-60 region in Caenorhabditis elegans. Genetics. 1988;118:49–59. doi: 10.1093/genetics/118.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- Michele DE, Metzger JM. Physiological consequences of tropomyosin mutations associated with cardiac and skeletal myopathies. J Mol Med. 2000;78:543–553. doi: 10.1007/s001090000161. [DOI] [PubMed] [Google Scholar]
- Michele DE, Albayya FP, Metzger JM. Thin filament protein dynamics in fully differentiated adult cardiac myocytes: Toward a model of sarcomere maintenance. J Cell Biol. 1999;145:1483–1495. doi: 10.1083/jcb.145.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Moerman DG, Plurad S, Waterston RH, Baillie DL. Mutations in the unc-54 myosin heavy chain gene of Caenorhabditis elegans that alter contractility but not muscle structure. Cell. 1982;29:773–781. doi: 10.1016/0092-8674(82)90439-1. [DOI] [PubMed] [Google Scholar]
- Mohri K, Ono S. Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J Cell Sci. 2003;116:4107–4118. doi: 10.1242/jcs.00717. [DOI] [PubMed] [Google Scholar]
- Mohri K, Takano-Ohmuro H, Nakashima H, Hayakawa K, Endo T, Hanaoka K, Obinata T. Expression of cofilin isoforms during development of mouse striated muscles. J Muscle Res Cell Motil. 2000;21:49–57. doi: 10.1023/a:1005682322132. [DOI] [PubMed] [Google Scholar]
- Mohri K, Vorobiev S, Fedorov AA, Almo SC, Ono S. Identification of functional residues on Caenorhabditis elegans actininteracting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:31697–31707. doi: 10.1074/jbc.M403351200. [DOI] [PubMed] [Google Scholar]
- Mohri K, Ono K, Yu R, Yamashiro S, Ono S. Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol Biol Cell. 2006;17:2190–2199. doi: 10.1091/mbc.E05-11-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraczewska J, Greenfield NJ, Liu Y, Hitchcock-DeGregori SE. Alteration of tropomyosin function and folding by a nemaline myopathy-causing mutation. Biophys J. 2000;79:3217–3225. doi: 10.1016/S0006-3495(00)76554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudry RE, Perry CN, Richards M, Fowler VM, Gregorio CC. The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J Cell Biol. 2003;162:1057–1068. doi: 10.1083/jcb.200305031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Sato N, Nakagaki T, Abe H, Ono S, Obinata T. Two mouse cofilin isoforms, muscle-type (MCF) and non-muscle type (NMCF), interact with F-actin with different efficiencies. J Biochem (Tokyo) 2005;138:519–526. doi: 10.1093/jb/mvi152. [DOI] [PubMed] [Google Scholar]
- Nishida E, Maekawa S, Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- Nishida E, Muneyuki E, Maekawa S, Ohta Y, Sakai H. An actin-depolymerizing protein (destrin) from porcine kidney. Its action on F-actin containing or lacking tropomyosin. Biochemistry. 1985;24:6624–6630. doi: 10.1021/bi00344a049. [DOI] [PubMed] [Google Scholar]
- Nyakern-Meazza M, Narayan K, Schutt CE, Lindberg U. Tropomyosin and gelsolin cooperate in controlling the microfilament system. J Biol Chem. 2002;277:28774–28779. doi: 10.1074/jbc.M203360200. [DOI] [PubMed] [Google Scholar]
- Obinata T, Nagaoka-Yasuda R, Ono S, Kusano K, Mohri K, Ohtaka Y, Yamashiro S, Okada K, Abe H. Low molecularweight G-actin binding proteins involved in the regulation of actin assembly during myofibrillogenesis. Cell Struct Funct. 1997;22:181–189. doi: 10.1247/csf.22.181. [DOI] [PubMed] [Google Scholar]
- Okada K, Obinata T, Abe H. XAIP1: A Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J Cell Sci. 1999;112:1553–1565. doi: 10.1242/jcs.112.10.1553. [DOI] [PubMed] [Google Scholar]
- Okada K, Ravi H, Smith EM, Goode BL. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: A coordinated mechanism for severing and capping filaments. Mol Biol Cell. 2006;17:2855–2868. doi: 10.1091/mbc.E06-02-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell. 2004;15:2782–2793. doi: 10.1091/mbc.E04-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J Cell Sci. 2003;116:2073–2085. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- Ono K, Yu R, Mohri K, Ono S. Caenorhabditis elegans kettin, a large immunoglobulin-like repeat protein, binds to filamentous actin and provides mechanical stability to the contractile apparatuses in body wall muscle. Mol Biol Cell. 2006;17:2722–2724. doi: 10.1091/mbc.E06-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. The Caenorhabditis elegansunc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J Cell Biol. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. Function of ADF/cofilin in muscle cells: An important regulator of actin cytoskeletal dynamics in myofibril assembly and muscle diseases. In: Fagan J, Davidson JN, Shimizu N, editors. Recent Developments in Cell Research. Research Signpost; Kerala, India: 2003a. pp. 31–44. [Google Scholar]
- Ono S. Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: New blades for twisted filaments. Biochemistry. 2003b;42:13363–13370. doi: 10.1021/bi034600x. [DOI] [PubMed] [Google Scholar]
- Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–3783. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Minami N, Abe H, Obinata T. Characterization of a novel cofilin isoform that is predominantly expressed in mammalian skeletal muscle. J Biol Chem. 1994;269:15280–15286. [PubMed] [Google Scholar]
- Ono S, Baillie DL, Benian GM. UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol. 1999;145:491–502. doi: 10.1083/jcb.145.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, McGough A, Pope BJ, Tolbert VT, Bui A, Pohl J, Benian GM, Gernert KM, Weeds AG. The C-terminal tail of UNC-60B (ADF/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- Ono S, Mohri K, Ono K. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J Biol Chem. 2004;279:14207–14212. doi: 10.1074/jbc.M313418200. [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85:164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Perry SV. What is the role of tropomyosin in the regulation of muscle contraction? J Muscle Res Cell Motil. 2003;24:593–596. doi: 10.1023/b:jure.0000009811.95652.68. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Kazzaz JA, Helfman DM. Functional properties of non-muscle tropomyosin isoforms. Curr Opin Cell Biol. 1994;6:96–104. doi: 10.1016/0955-0674(94)90122-8. [DOI] [PubMed] [Google Scholar]
- Ramachandran I, Terry M, Ferrari MB. Skeletal muscle myosin cross-bridge cycling is necessary for myofibrillogenesis. Cell Motil Cytoskeleton. 2003;55:61–72. doi: 10.1002/cm.10113. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Tetreault JW, Lappalainen P, Drubin DG, Amberg DC. Aip1p interacts with cofilin to disassemble actin filaments. J Cell Biol. 1999;145:1251–1264. doi: 10.1083/jcb.145.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno Y, Shimada Y, Obinata T. BDM (2,3-butanedione monoxime), an inhibitor of myosin-actin interaction, suppresses myofibrillogenesis in skeletal muscle cells in culture. Cell Tissue Res. 1999;295:307–316. doi: 10.1007/s004410051237. [DOI] [PubMed] [Google Scholar]
- Squire JM, Morris EP. A new look at thin filament regulation in vertebrate skeletal muscle. FASEB J. 1998;12:761–771. doi: 10.1096/fasebj.12.10.761. [DOI] [PubMed] [Google Scholar]
- Thirion C, Stucka R, Mendel B, Gruhler A, Jaksch M, Nowak KJ, Binz N, Laing NG, Lochmuller H. Characterization of human muscle type cofilin (CFL2) in normal and regenerating muscle. Eur J Biochem. 2001;268:3473–3482. doi: 10.1046/j.1432-1327.2001.02247.x. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tubridy N, Fontaine B, Eymard B. Congenital myopathies and congenital muscular dystrophies. Curr Opin Neurol. 2001;14:575–582. doi: 10.1097/00019052-200110000-00005. [DOI] [PubMed] [Google Scholar]
- Vartiainen MK, Mustonen T, Mattila PK, Ojala PJ, Thesleff I, Partanen J, Lappalainen P. The three mouse actin-depolymerizing factor/cofilins evolved to fulfill cell-type-specific requirements for actin dynamics. Mol Biol Cell. 2002;13:183–194. doi: 10.1091/mbc.01-07-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Thomson JN, Brenner S. Mutants with altered muscle structure of Caenorhabditis elegans. Dev Biol. 1980;77:271–274. doi: 10.1016/0012-1606(80)90475-3. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Mohri K, Ono S. The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry. 2005;44:14238–14247. doi: 10.1021/bi050933d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]


