Life would not be possible without communication between the cells and the external environment. This critical capability allows cells to monitor the status of the organs and tissues and to respond to environmental threats and opportunities. The process is mediated by a group of membrane-bound proteins known as G protein-coupled receptors (GPCRs). Each GPCR monitors a specific compound or group of compounds and becomes activated when the target ligand (agonist) occupies the active site. The activated GPCR signals the presence of the ligand through interaction with a G protein that migrates within the intracellular medium and directly couples to the activated GPCR. The process provides reliable signaling with variable amplification. There are at least 700 GPCRs encoded in the human genome. Despite significant biological importance, the structure and function of most GPCRs are poorly understood, and a high-resolution crystal structure of only one GPCR, the visual pigment rhodopsin, has been reported (1–3). Visual pigments are seven transmembrane α-helical proteins that bind 11-cis retinal and initiate the light transduction signaling pathway in retinal photoreceptors (4). Although other GPCRs interact with their ligands noncovalently, the visual pigments consist of 11-cis retinal covalently attached to the protein through a conserved lysine residue in the seventh transmembrane helix (HVII, see Fig. 1). After absorption of light, the retinal chromophore isomerizes to the all-trans conformation and triggers a series of conformational changes that lead to the formation of the active state, R* or Meta II (4–9). All-trans retinal is eventually released from the vertebrate protein, and visual pigments can be regenerated from 11-cis retinal spontaneously. Although some consider a photon of light to represent the agonist, a more logical choice is all-trans retinal that is photochemically generated from the 11-cis retinal cofactor. Thus, the visual pigments have a covalently attached agonist, but one that starts out in an inactive form. The fact that the agonist is covalently attached allows studies of the activation process at a level not possible with the vast majority of the other GPCRs, where the mode and location of interaction of the agonist remain obscure.
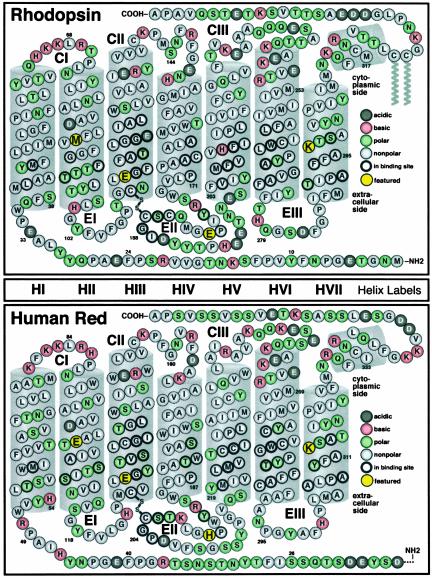
Fig. 1.
The primary sequences of bovine rhodopsin (Upper) and the human red cone (Lower) arranged to delineate the transmembrane helices (gray cylinders, labeled HI through HVII) and the loop regions. The three cytoplasmic loops (CI, CII, and CIII) and the three extracellular loops (EI, EII, and EIII) are labeled. Key binding site residues relevant to our discussion are shown in yellow filled circles; from left to right in rhodopsin these are M86, E113, E181, and K296, and in the human red cone these are E102, E129, H197, and K312. The red cone has 386 residues, and 16 of the residues on the end of the N-terminal tail are not shown.
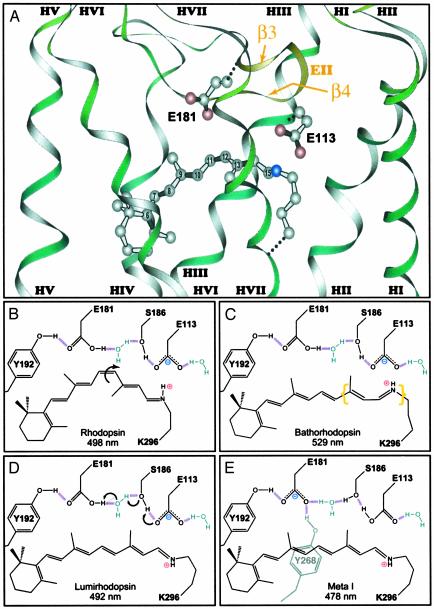
One of the more surprising findings of the rhodopsin crystal structure studies involves the loop region that connects transmembrane helices 4 and 5 (HIV and HV, Fig. 1). Because this loop is the second extracellular loop, it is called EII, and it consists of two β-strands (β3 and β4) that fold into the binding site as shown in Fig. 2A (6). A glutamic acid residue, E181, attached to the β3 sheet extends toward the center portion of the protonated chromophore, and its proximity and location suggest the possibility that it could represent a primary counterion to the chromophore. Indeed, earlier chromophore analog (10), two-photon excitation (11), and NMR (12) studies all indicated that a counterion might be located in this region. However, the two-photon studies are compatible only with a neutral chromophore-binding site (11), and site-directed mutagenesis studies have established that E113 is the primary counterion (13, 14). As a further complication, E181 occupies a node in the λmax band charge-shift electrostatic map, so that the protonation state of this residue is difficult to assign by using UV-visible (one-photon) spectroscopy (15). The role of E181 in the activation of rhodopsin is the subject of the article by Yan et al. in this issue of PNAS (16). This study comes to a dramatic and important conclusion, namely that E181 is protonated (neutral) in rhodopsin, but transfers a proton via a hydrogen-bonded network to the primary counterion, E113, during the formation of Meta I. Yan et al. further propose that this counterion switch plays a major role in the activation of the protein and may have implications for the mechanism of activation of all GPCRs. A schematic diagram of the counterion switch in rhodopsin is shown in Fig. 2 B–E. Here, we explore the global issues raised by this interesting mechanism.
Fig. 2.
(A) The second extracellular loop, EII, extends into the chromophore binding site in the form of two β sheets (β3 and β4) (coordinates from ref. 1). Note that the extracellular surface is up, opposite of the orientation used in Fig. 1. Glutamic acid residues 113 and 181 are also shown in A, and the protonation states of these two residues as a function of photobleaching intermediate is schematically shown in B–E. The arrows in B and D indicate the key conformational or chemical changes associated with transformation of rhodopsin (to bathorhodopsin) and Lumirhodopsin (to Meta I). The chromophore geometry in bathorhodopsin remains controversial, but recent theoretical studies suggest it may be inverted from the model shown here (9). The yellow brackets in C are intended to indicate both distortion and conformational uncertainty. The relative location of the residues is approximate and does not reflect possible movement during the thermal relaxations (see text).
The first question to ask is whether the counterion switch shown in Fig. 2 shares mechanistic commonality with the counterion changes observed during the photocycle of bacteriorhodopsin (BR). Such commonality would be a problem because it would suggest that the counterion switch is a general feature of retinal proteins and has a common mechanistic purpose, to force forward directionality on the thermal reactions. Although BR is not a GPCR, it has significant structural similarities to GPCRs and has served for many years as a modeling template for GPCRs because high-resolution structural data have been available since 1998 (17). Also, there is striking similarity between the sequence of intermediates in the early portion of the photocycle of BR [BR (570 nm) → K (590 nm) → L (550 nm) → M (412 nm) →...] and the early photobleaching sequence of rhodopsin [Rho (498 nm) → Batho (529 nm) → Lumi (492 nm) → Meta I (478 nm) →...] (18). The counterion switch in rhodopsin takes place in the Lumi to Meta I reaction. Similarly, BR undergoes a major change in the counterion environment in the L to M reaction with the protonation of the counterion (D85). But this change in the counterion environment is not comparable to the counterion switch observed in rhodopsin. In BR, the D85 residue receives its proton from the chromophore, not another nearby residue. And when the chromophore is reprotonated later in the photocycle, the source is an aspartic acid residue (D96) that is >10 Å away from the chromophore. Thus, a counterion switch does not take place in BR. This comparison suggests that the counterion switch observed in rhodopsin is unique to the visual pigments, and that the proposal by Yan et al. (16) that the counterion switch is an integral component of the activation mechanism deserves serious examination.
Additional evidence that a counterion switch is an important element of visual pigment activation can be found in an examination of the photobleaching sequence of UV pigments. Humans do not have UV cones, but many birds, small animals, insects, and fish rely on UV pigments for friend–foe identification, food harvesting, and mating (4, 19, 20). UV pigments have unprotonated Schiff base chromophores, which yield absorption maxima around 360 nm (21). The nearby E113 counterion is neutral, and the binding site does not appear to have any charged residues near the chromophore (21). Remarkably, the photobleaching sequence of the UV pigments has evolved to include a counterion switch. The primary event involves an 11-cis isomerization of the unprotonated Schiff base retinal chromophore, but the all-trans chromophore picks up a proton in the Lumi state (21). Recent site-directed mutagenesis and spectroscopic studies in our laboratories indicate that a counterion switch occurs from E108 (E113 in rhodopsin) to E176 (E181) during the Lumi to Meta I transition, in close analogy to rhodopsin. The formation of Lumi can be completely blocked by replacing the primary counterion with asparagine (E108Q), but Meta I still forms, although slowly, through donation of a proton from E176. This observation provides support for the importance of the counterion switch to the activation of visual pigments in general. Simply put, why would such a convoluted sequence of events evolve if it were not a key contributor to function? From another perspective, the counterion switch mechanism makes it possible for UV vision. The counterion switch mechanism provides a comprehensive activation strategy that will work for both protonated and unprotonated chromophores.
The basic model of activation proposed by Yan et al. (16) is based on helix translocation during the dark reactions to form Meta I such that the protonated Schiff base moves toward E181 (16). The activated state of rhodopsin is Meta II, which is in equilibrium with Meta I. Meta I has a lower enthalpy than Meta II, and the formation of Meta II is entropy-driven (22, 23). Previous studies have shown that there is significant motion as well as rotation of the transmembrane helices in Meta II, and, in particular, movement of HVII, to which the chromophore is attached (Figs. 1 and 2), is critical for coupling of the activated rhodopsin with transducin (24–26). The counterion switch may serve as the electrostatic component of the driving force that moves HVII. Alternatively, the motion of the helices may induce the counterion switch. Further studies will be necessary to answer this question. Molecular modeling of the latter intermediates of the mouse UV pigment, however, indicate that a counterion switch is energetically favored for an isomerized chromophore without significant helix motion. The presence of two tyrosine residues, Y192 and Y268, in hydrogen bonding proximity to E181 provides significant stabilization of the negative charge on E181, as schematically shown in Fig. 2E.
We conclude by noting that the counterion switch mechanism as proposed by Yan et al. (16) will require some modification to function as part of the photobleaching sequence in many of the medium and long wavelength (M/LWS group) cones. Homology modeling indicates that a histidine residue rather than a glutamic acid residue occupies the E181 site (see Fig. 1B). One possibility is that the H197 (E181; rho) residue is positively charged and donates its labile proton to E129 (E113; rho) during the Lumi to Meta I transition in the M/LWS cones. Simple molecular orbital calculations to test this possibility, however, indicate that this scenario is energetically unfavorable. A more attractive alternative is that the counterion switch involves E102, a residue that replaces methionine in rhodopsin (M86, see Fig. 1). A homology model of the human red cone binding site indicates that the carboxylate oxygen on E129 is separated from the carboxyl alcohol oxygen of the E102 by 7.5 Å, a distance only slightly larger than the 6.4-Å distance separating the corresponding oxygen atoms on E113 and E118 in rhodopsin. Thus, the binding sites of the red cones provide a glutamic acid pair consistent with a counterion switch from E129 to E102. Further work will be required to determine whether the E129 to E102 counterion switch occurs and to answer the interesting question of why the M/LWS cones have evolved a modified mechanism and topology for the counterion switch.
See companion article on page 9262.
References
- 1.Okada, T., Le Trong, I., Fox, B., Behnke, C., Stenkamp, R. & Palczewski, K. (2000) J. Struct. Biol. 130, 73–80. [DOI] [PubMed] [Google Scholar]
- 2.Palczewski, K., Kumasaka, T., Hori, T., Behnke, C. A., Motoshima, H., Fox, B. A., Le Trong, I., Teller, D. C., Okada, T., Stenkamp, R. E., et al. (2000) Science 289, 739–745. [DOI] [PubMed] [Google Scholar]
- 3.Okada, T., Ernst, O., Palczewski, K. & Hofmann, K. (2001) Trends Biochem. Sci. 26, 318–324. [DOI] [PubMed] [Google Scholar]
- 4.Ebrey, T. & Koutalos, Y. (2001) Progr. Retinal Eye Res. 20, 49–94. [DOI] [PubMed] [Google Scholar]
- 5.Wald, W. & Brown, P. (1958) Science 127, 222–226. [DOI] [PubMed] [Google Scholar]
- 6.Schick, G. A., Cooper, T. M., Holloway, R. A., Murray, L. P. & Birge, R. R. (1987) Biochemistry 26, 2556–2562. [DOI] [PubMed] [Google Scholar]
- 7.Schoenlein, R. W., Peteanu, L. A., Mathies, R. A. & Shank, C. V. (1991) Science 254, 412–415. [DOI] [PubMed] [Google Scholar]
- 8.Lewis, J. & Kliger, D. (2000) Methods Enzymol. 315, 164–178. [DOI] [PubMed] [Google Scholar]
- 9.Saam, J., Tajkhorshid, E., Hayashi, S. & Schulten, K. (2002) Biophys. J. 83, 3097–3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honig, B., Dinur, U., Nakanishi, K., Balogh-Nair, V., Gawinowicz, M. A., Arnaboldi, M. & Motto, M. G. (1979) J. Am. Chem. Soc. 101, 7084–7086. [Google Scholar]
- 11.Birge, R. R., Murray, L. P., Pierce, B. M., Akita, H., Balogh-Nair, V., Findsen, L. A. & Nakanishi, K. (1985) Proc. Natl. Acad. Sci. USA 82, 4117–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han, M. & Smith, S. O. (1995) Biophys. Chem. 56, 23–29. [DOI] [PubMed] [Google Scholar]
- 13.Sakmar, T. P., Franke, R. R. & Khorana, H. G. (1989) Proc. Natl. Acad. Sci. USA 86, 8309–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhukovsky, E. A. & Oprian, D. D. (1989) Science 246, 928–931. [DOI] [PubMed] [Google Scholar]
- 15.Kusnetzow, A., Dukkipati, A., Babu, K. R., Singh, D., Vought, B. W., Knox, B. E. & Birge, R. R. (2001) Biochemistry 40, 7832–7844. [DOI] [PubMed] [Google Scholar]
- 16.Yan, E. C. Y., Kazmi, M. A., Ganim, Z., Hou, J.-M., Pan, D., Chang, B. S. W., Sakmar, T. P. & Mathies, R. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9262–9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luecke, H., Richter, H. T. & Lanyi, J. K. (1998) Science 280, 1934–1937. [DOI] [PubMed] [Google Scholar]
- 18.Stuart, J. A. & Birge, R. R. (1996) in Biomembranes, ed. Lee, A. G. (JAI Press, London), Vol. 2A, pp. 33–140. [Google Scholar]
- 19.Gaertner, W. (2000) in Handbook of Biological Physics, eds. Stavenga, D. G., DeGrip, W. J. & Pugh, E. N., Jr. (Elsevier, New York), Vol. 3.
- 20.Yokoyama, S. & Shi, Y. (2000) FEBS Lett. 486, 167–172. [DOI] [PubMed] [Google Scholar]
- 21.Dukkipati, A., Kusnetzow, A., Babu, K. R., Ramos, L., Singh, D., Knox, B. E. & Birge, R. R. (2002) Biochemistry 41, 9842–9851. [DOI] [PubMed] [Google Scholar]
- 22.Birge, R. R. & Vought, B. W. (2000) Methods Enzymol. 315, 143–163. [DOI] [PubMed] [Google Scholar]
- 23.Cooper, A. (1981) FEBS Lett. 123, 324–326. [DOI] [PubMed] [Google Scholar]
- 24.Farrens, D. L., Altenbach, C., Yang, K., Hubbell, W. L. & Khorana, H. G. (1996) Science 274, 768–770. [DOI] [PubMed] [Google Scholar]
- 25.Hamm, H. (2001) Proc. Natl. Acad. Sci. USA 98, 4819–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubbell, W. L., Altenbach, C., Hubbell, C. M. & Khorana, H. G. (2003) Adv. Protein Chem. 63, 243–290. [DOI] [PubMed] [Google Scholar]