Abstract
Late blight, caused by the oomycete pathogen Phytophthora infestans, is the most devastating potato disease in the world. Control of late blight in the United States and other developed countries relies extensively on fungicide application. We previously demonstrated that the wild diploid potato species Solanum bulbocastanum is highly resistant to all known races of P. infestans. Potato germplasm derived from S. bulbocastanum has shown durable and effective resistance in the field. Here we report the cloning of the major resistance gene RB in S. bulbocastanum by using a map-based approach in combination with a long-range (LR)-PCR strategy. A cluster of four resistance genes of the CC-NBS-LRR (coiled coil–nucleotide binding site–Leu-rich repeat) class was found within the genetically mapped RB region. Transgenic plants containing a LR-PCR product of one of these four genes displayed broad spectrum late blight resistance. The cloned RB gene provides a new resource for developing late blight-resistant potato varieties. Our results also demonstrate that LR-PCR is a valuable approach to isolate genes that cannot be maintained in the bacterial artificial chromosome system.
Potato late blight, a disease caused by the oomycete pathogen Phytophthora infestans, is one of the world's most devastating plant diseases. Late blight was responsible for the European potato famine in the 19th century, which caused the starvation deaths of more than one million people in Ireland alone. Despite its historic significance, none of the currently grown potato cultivars in the United States have adequate late blight resistance. Currently, late blight is responsible for multibillion-dollar losses annually in both potato and tomato production (1). Furthermore, in developing countries, where funds for purchasing fungicides are limited, late blight can completely eliminate the potato crop.
A number of wild potato species, such as Solanum demissum (2n = 6x = 72), coevolved with P. infestans, and have provided the primary germplasm for breeding late blight resistance in cultivated potato. At least 11 resistance (R) genes that originated from S. demissum have been incorporated into various potato cultivars (2). All of these 11 R genes confer race-specific hypersensitive resistance. Potato cultivars possessing such R genes are not resistant to all races of the pathogen. These race-specific R genes provide only short-lived resistance in the field as new virulent races of the pathogen rapidly overcome the resistance encoded by single race-specific resistance genes (3, 4).
A wild diploid potato species, Solanum bulbocastanum (2n = 2x = 24), is highly resistant to all known races of P. infestans, even under intense disease pressure (5). Because S. bulbocastanum is sexually incompatible with potato, somatic hybrids between potato and this wild species were developed with the long-term goal of capturing this resistance for use in potato cultivars. The somatic hybrids and a number of backcrossed progenies consistently displayed late blight resistance similar to the parental S. bulbocastanum clone PT29 (5, 6). Unlike potato varieties containing the R genes derived from S. demissum, no obvious necrotic lesions, which are characteristic of the classical hypersensitive response, were observed. In fact, the pathogen sometimes sporulates on PT29-derived resistant materials. The resistance of the PT29-derived plants is manifested as a slow progression of lesion development that substantially decreases the rate of disease development in the plants. This phenotype of general suppression but not elimination of symptom development has been consistently observed in field tests at various locations in the United States and in Toluca, Mexico, between 1995 and 2002. The late blight resistance associated with the PT29-derived materials could be considered rate-reducing or, in some terms, partial resistance, and can be effectively used in resistance breeding programs (7).
Several populations have been developed from a fertile somatic hybrid between potato and S. bulbocastanum clone PT29 and its backcrossed progenies. A major resistance locus, RB, was mapped to a specific location on chromosome 8 of S. bulbocastanum (8). A bacterial artificial chromosome (BAC) contig spanning the RB gene was constructed by using a reiterative approach of BAC walking and high-resolution genetic mapping (9). In this paper, we report the cloning of the RB gene by a map-based approach used in combination with long-range (LR)-PCR. The cloned RB gene will provide an important resource to develop late blight-resistant potato cultivars and further understand disease resistance mechanisms in plants.
Materials and Methods
DNA Sequencing and Analysis. The DNA sequences of the S. bulbocastanum BAC clones 177O13 and CB3A14 were determined by using a shotgun sequencing strategy as described by Yuan et al. (10). The two BACs were deposited in the PLN division of GenBank [accession nos. AY303171 (177O13) and AY303170 (CB3A14)]. Multiple sequence alignments were conducted by using clustalx 1.81 software (11). Diversifying selections were investigated by using paml (12, 13).
Long-Range PCR. Primers were designed for each candidate RB gene (from the resistant RB haplotype) based on the sequence information of BAC 177O13 (the susceptible rb haplotype). Individual candidate RB genes were amplified by LR-PCR, using genomic DNA of S. bulbocastanum clone PT29 as a template. Eight-base pair sequences (CGGGATCC) were introduced into all LR-PCR primers (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org) to create a BamHI restriction site to facilitate cloning of the LR-PCR products. LR-PCR reactions were carried out in a total volume of 50 μl containing 2.5 units of Takara LA Taq (PanVera, Madison, WI), 1× reaction buffer, 400 μM dNTP, 100 ng of template DNA, and 0.2 μM each primer. The PCR conditions were 1 min at 94°C followed by 35 cycles of 20 sec at 94°C, 20 sec at 60°C, and 15 min at 68°C. The LR-PCR products from three independent reactions were cloned into the pGEM-T or pCR-XL-TOPO vectors. The haplotype origin (RB or rb haplotype) of the cloned LR-PCR fragments was determined by cleaved amplified polymorphic sequence markers (Table 4, which is published as supporting information on the PNAS web site) developed for fine mapping (9). The final LR-PCR products from the RB haplotype were recloned into the BamHI site of the binary cosmid vector pCLD04541 (14) for use in potato transformation.
Complementation Analysis. The pCLD04541 constructs containing putative RB genes derived from LR-PCR and BAC CB3A14 were introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Potato (Solanum tuberosum cv. Katahdin) transformation was performed essentially as described by Ziegelhoffer et al. (15). Kanamycin-resistant transgenic plants were tested by PCR, using both gene-specific primers and CAPS markers. Plants with confirmed insertion of the putative genes were clonally propagated in vitro and screened for late blight resistance. The greenhouse resistance assay was carried out in triplicate by inoculating plants with sporangial suspensions of different isolates of P. infestans (Table 1) according to Naess et al. (8). Foliage blight scores were recorded at 3, 4, 5, 7, and 10 days after inoculation. The ratings and ranges of percentage infections associated with the scores were as follows: 9, no visible infection; 8, <10%; 7, 11–25%; 6, 26–40%; 5, 41–60%; 4, 61–70%; 3, 71–80%; 2, 81–90%; 1, >90%; 0, 100%.
Table 1. P. infestans isolates used in late blight resistance evaluation.
| Isolate | Genotype | Mating type | Race | Source |
|---|---|---|---|---|
| US930287 | US-8 | A2 | ND | William Fry, Cornell University, Ithaca, NY |
| 126C18 | US-8 | A2 | 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 | Neil Gudmestad, North Dakota State University |
| US940480 | US-8 | A2 | 0, 1, 2, 3, 4, 5, 6, 7, 9, 10, 11 | William Fry, Cornell University |
| MSU-96 | US-8 | A2 | 0, 1, 2, 3, 4, 5, 6, 7, 8, 10, 11 | Ken Deahl, U.S. Department of Agriculture—Agricultural Research Service, Beltsville, MD |
| US940501 | US-1 | A1 | 0, (6)* | William Fry, Cornell University |
| US980008 | US-11 | A1 | ND | William Fry, Cornell University |
ND, not determined.
Parentheses denote variable results; i.e., both compatible and incompatible reactions were observed in different experiments (William Fry, personal communication).
Rapid Amplification of cDNA Ends (RACE). Three-week-old S. bulbocastanum PT29 plants were inoculated with P. infestans isolate US930287 and maintained in greenhouses. Equal amounts of challenged and unchallenged leaves at 12 h and 1, 2, 3, 4, and 5 days after inoculation were collected and pooled. Total RNA was isolated from the combined materials by using TRIzol (Invitrogen). Poly(A)+ RNA was isolated by using PolyATract mRNA Isolation Systems (Promega). The 5′ and 3′ ends of the cDNA were determined by RACE, using the GeneRacer kit (Invitrogen). The RACE primers (Table 5, which is published as supporting information on the PNAS web site) are identical between the resistant and susceptible haplotypes.
Results
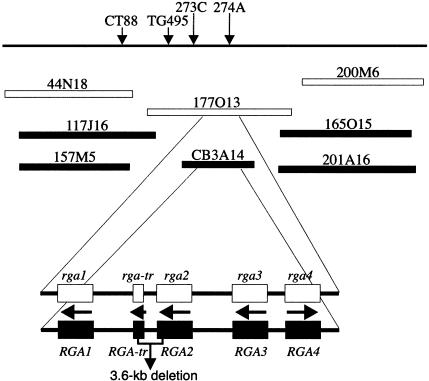
Cloning of the RB Gene by LR-PCR. The RB locus was previously mapped to chromosome 8 of S. bulbocastanum (8). The RB locus is heterozygous (RB/rb) in the original S. bulbocastanum clone PT29 used in the development of genetic mapping populations and BAC libraries. Surprisingly, all 11 BACs associated with the RB locus were derived from the rb haplotype (9). One of these 11 BACs, 177O13, was fully sequenced. BAC 177O13 contains 163,635 bp, and includes one truncated and four complete CC-NBS-LRR (coiled coil–nucleotide binding site–Leu-rich repeat)-class R-gene analogs (RGAs) (Fig. 1). The one truncated and four complete RGAs in 177O13 are named rga-tr, rga1, rga2, rga3, and rga4, respectively.
Fig. 1.
Genetic and physical maps of the genomic region containing RB. The RB locus is closely linked to markers CT88, TG495, 273C, and 274A (8, 9). BAC clones from the RB haplotype are represented as filled boxes, and BAC clones from the rb haplotype are represented as open boxes. Both 177O13 and CB3A14 contain one truncated and four complete RGAs. The direction of transcription of each gene is indicated by an arrow. The 3.6-kb deletion region between RGA2 and RGA-tr ismarked. This 3.6-kb fragment is not present in BAC CB3A14 but is drawn for the purpose of comparison with BAC 177O13.
We used a LR-PCR-based approach to clone the RGAs from the RB haplotype. Four primer sets (Table 3) were designed based on the sequences in BAC 177O13 to amplify each of the four complete RGAs. DNA isolated from S. bulbocastanum clone PT29 was used as the LR-PCR template. Four LR-PCR products with expected sizes of 13.0, 8.6, 11.8, and 7.9 kb were successfully amplified from S. bulbocastanum and cloned into the pGEM-T or pCR-XL-TOPO vectors. End sequencing and CAPS markers (Table 4) were used to determine the rb or RB origin of each cloned LR-PCR fragment. To avoid potential PCR artifacts, we cloned three independent LR-PCR products for each of the four RGAs. All four RGAs derived from both rb and RB haplotypes were recovered from the LR-PCR experiments. The four RGAs derived from the RB haplotype are denoted RGA1-PCR, RGA2-PCR, RGA3-PCR, and RGA4-PCR, and were cloned into the BamHI site of the binary vector pCLD04541 for complementation studies.
Complementation Analysis of the LR-PCR-Derived RGA Clones. The four RGA clones amplified from the RB haplotype were transferred into A. tumefaciens strain LBA4404. Katahdin, a potato variety susceptible to late blight, was used for Agrobacterium-mediated transformation. Transgenic Katahdin plants (Table 2) developed from the four RGA constructs were inoculated with isolate US930287 of P. infestans. Katahdin plants containing RGA1-PCR, RGA3-PCR, and RGA4-PCR, as well as untransformed controls, were susceptible to late blight (Fig. 2 and Table 2). Leaves of all susceptible plants displayed typical spreading lesions, with water-soaked areas and extensive rotting developed at later time points. In contrast, all 14 plants derived from independent transformations by using the RGA2-PCR construct displayed resistance to the pathogen. P. infestans infection of these plants was significantly delayed and limited to the lower leaves. Small lesions that developed on the lower leaves were restricted to the infected area.
Table 2. Late blight screening of transgenic plants by using isolate US930287.
| Construct in transgenic Katahdin | Number of plants tested* | Number of plants showing resistance |
|---|---|---|
| RGA1-PCR | 13 | 0 |
| RGA2-PCR (RB) | 14 | 14† |
| RGA3-PCR | 21 | 0 |
| RGA4-PCR | 18 | 0 |
| RGA1-BAC | 13 | 0 |
| RGA2-BAC | 11 | 0 |
| RGA3-BAC | 20 | 0 |
| RGA4-BAC | 26 | 0 |
| Katahdin control | 8 | 0 |
| S. bulbocastanum (PT29) control | 6 | 6 |
Plants were inoculated with P. infestans US930287 and scored for symptoms 7 days after inoculation. Plants were scored as resistant (R) if the resistance score was ≥7.0 (≤25% infection); plants were scored as susceptible (S) if the resistance scores was ≤6.9 (>25% infection).
These correspond to independent transformation events.
Of the 14 resistant plants, nine plants had a score of 7 and five plants had a score of 8.
Fig. 2.
Complementation analysis of putative RB genes. Transgenic Katahdin and control plants were inoculated with the US930287 isolate of P. infestans. Disease symptoms were recorded 7 days after inoculation. (A–C) Transgenic Katahdin plants containing constructs RGA1-PCR, RGA2-PCR, and RGA4-PCR, respectively. (D) Control Katahdin plant. (E) Katahdin plant that was not inoculated. (F–I) Transgenic Katahdin plants containing constructs RGA1-BAC, RGA2-BAC, RGA3-BAC, and RGA4-BAC, respectively.
One of the RGA2-PCR transgenic lines, SP922, was subsequently tested by using five additional isolates of P. infestans (Table 1). The SP922 plants showed remarkable resistance to all five isolates, including 126C18, a “super race” that overcomes all 11 major R genes identified in S. demissum (16). Untransformed Katahdin plants were all susceptible (data not shown). SP922 was susceptible to early blight when it was inoculated with isolate AS98-100 of the early blight pathogen Alternaria solani (data not shown). This result is consistent with independent segregation of early blight resistance and the RB locus observed in BC1 populations derived from the potato–PT29 somatic hybrids (J.P.H., unpublished work). The complementation results demonstrated that RGA2 represents the functional RB gene.
Attempts to Clone RB from BAC. Sequence analysis of BAC 177O13 revealed that a 102-kb region, which spans rb, is devoid of BamHI restriction sites. Because approximately two-thirds of the PT29 BAC library was developed from partial BamHI digestion (9, 17), the lack of BAC clones containing the RB allele may be due to the lack of BamHI sites in this region. We constructed a BAC library, consisting of 8,448 clones, by complete BamHI digestion of PT29 DNA. Four BAC clones were identified by using probes located in the RB region. Three of the four clones were derived from the rb haplotype. The fourth BAC, CB3A14, was derived from the RB haplotype.
Complete sequencing of CB3A14 revealed four complete RGAs. The truncated RGA found in 177O13 was not observed in CB3A14 (Fig. 1). The four RGAs in CB3A14 were designated RGA1-BAC, RGA2-BAC, RGA3-BAC, and RGA4-BAC. A cosmid library was developed from CB3A14 by using the binary vector pCLD04541. Cosmid clones containing each of the four RGAs were selected by PCR, using two sets of primers designed for each RGA (Table 6, which is published as supporting information on the PNAS web site), and were used to transform Katahdin. Surprisingly, transgenic Katahdin plants containing all four constructs, including RGA2-BAC, did not show increased resistance compared with the control Katahdin plants (Table 2 and Fig. 2). These results indicate that RGA2-PCR and RGA2-BAC do not represent the same genomic DNA sequences. Sequence analyses of RGA2-PCR and RGA2-BAC, as well as BACs 177O13 and CB3A14, revealed that a 3.6-kb deletion occurred in CB3A14 between the 3′ region of RGA2 and the middle of RGA-tr (Fig. 1).
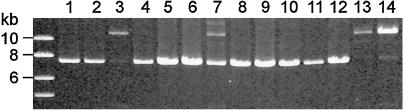
A pair of primers (Table 3) spanning the putative 3.6-kb deletion were designed based on the CB3A14 sequence. An expected 7-kb fragment was amplified from CB3A14, whereas a 10-kb fragment was amplified from both PT 29 genomic DNA and BAC177O13 (Fig. 3). We then conducted PCR analysis of 12 independent clones from the original well containing BAC CB3A14. A 7-kb band was amplified from 10 of the 12 clones. One clone (Fig. 3, lane 7) produced both 7- and 10-kb products, indicating that it is likely a mixture of the deletion derivatives and intact fragments. A second clone (Fig. 3, lane 3) produced a major 10-kb band and a few faint smaller bands, similar to the pattern produced from PT29 genomic DNA. We were unable to rescue this clone by retransforming the BAC plasmid into different E. coli strains because a similar deletion pattern was again observed. These results confirmed that the original BAC CB3A14 cannot be stably maintained in E. coli.
Fig. 3.
Stability analysis of CB3A14. Culture from the original well of BAC CB3A14 was streaked, and plasmid DNA from 12 independent clones (lanes 1–12) was isolated and used as a template for PCR analysis with primers GAP-1a and GAP-1b (Table 3) that span the 3.6-kb deletion (Fig. 1). In the experiment represented by lane 13, genomic DNA from S. bulbocastanum clone PT29 was used as a template. In the experiment represented by lane 14, DNA from BAC 177O13 was used as a template.
Transcript Analysis of the RGAs. The structures of the four RGAs were determined by comparing the genomic DNA sequences derived from BAC CB3A14 and LR-PCR products with 5′ and 3′ RACE products. Multiple RACE products from each experiment were sequenced to avoid PCR errors. Sequences of the 5′ and 3′ RACE products for each RGA overlapped and provided complete coverage of the transcribed region. RACE analysis revealed that RGA1, RGA2 (RB), RGA3, and RGA4 are all expressed and have a similar structure with a single intron (Fig. 4A). RB was transcribed in leaves of S. bulbocastanum plants in the absence of pathogen challenge, suggesting that it is constitutively expressed.
Fig. 4.
Structure of the RB gene and the deduced RB protein. (A) Physical structure of the RB gene. Two exons are indicated by open rectangles, and one intron is indicated by lines angled downward. (B) Predicted RB protein sequences. The potential leucine zipper motif and a heptad repeat motif are underlined. The three predicted kinase motifs of the NBS domain are shown in blue. Conserved motifs for plant resistance genes are shown in green. The two amino acid changes (E420-K and K662-M) caused by LR-PCR misincorporation are indicated in bold and underlined. The start point of the 3.6-kb deletion in RGA2-BAC is double underlined. The LRRs are aligned according to the consensus sequence LXXLXXLXXLXLXXN/CXXLXXLXX, where X represents any amino acid. The first L and the last two Ls are not highly conserved in different LRRs. Aliphatic residues L, I, M, V, and F are red; conversed N, C, and T residues are brown.
Sequences of 5′ and 3′ RACE products of RGA2 (RB) revealed two complete primary transcripts. One corresponded to rga2 (rb) in BAC 177O13 and the other matched with the RGA2-PCR sequence except for a 679-bp intron (positions 430–1,108) and three nucleotide mismatches. The three mismatches were caused by LR-PCR, which was confirmed by partial sequencing of the corresponding region of two additional independent LR-PCR products. The 5′ transcript was identical to only the first 2,295 bp of the 5′ sequence of RGA2-BAC, excluding an identical intron region observed in RGA2-PCR. However, none of the three 3′ transcripts matched the RGA2-BAC sequence, confirming that the 3.6-kb deletion starts from the 3′ coding region of RGA2 (RB).
Structure and Evolution of the RB Gene Family. The RB transcript was determined to be 3,319 bp and to contain 130- and 276-bp UTR at the 5′ and 3′ ends, respectively. The RB gene encodes a predicted polypeptide of 970 aa with a molecular weight of 110.3 kDa (Fig. 4B). The predicted RB protein belongs to the NBS-LRR class of R proteins (18). Its putative NBS domain consists of three motifs: kinase 1a or P-loop (positions 182–190), kinase 2 (positions 255–264), and kinase 3a (positions 288–293) (Fig. 4B). Downstream of the kinase motifs is a domain with unknown function conserved among resistance genes: QLPL, CFAY, and MHD motifs (19). The deduced RB protein contains one putative five-heptad leucine zipper motif near the N terminus (positions 10–45). Another region containing four-heptad repeats (positions 588–609) was observed within the LRR domain (Fig. 4B). The LRR domain consists of 21 LRR repeats, several of them imperfect.
The RB gene family includes one truncated and four complete genes in both the resistant and susceptible haplotypes (Fig. 1). In the susceptible haplotype, gene rb had a point mutation resulting in a premature stop codon (at the 454th codon). Both rga1 and rga3 had a 1-bp frame-shift deletion. Therefore, rb, rga1, and rga3 are likely to be pseudogenes. The four complete genes in both haplotypes are similar in length (2,895–2,979 bp) and have conserved intron–exon structures. The resistant and susceptible haplotypes were highly conserved, with 98.8% overall nucleotide identity in a 39-kb region. Regions flanking these genes were also highly conserved between the two haplotypes, but differed between different sites within each haplotype. The RB and rb sequences exhibited 99.8% nucleotide identity, with only three synonymous mutations, one nonsynonymous mutation, and one 18-bp deletion. High nucleotide identity (>98.8%) was also observed between the other four pairs of homologs (Fig. 1). In contrast, the paralogs exhibited only 79.6–85.6% nucleotide identity. No obvious sequence exchange between paralogs was found. Therefore, there is an obvious orthologous relationship between members located at the same position in resistant and susceptible haplotypes.
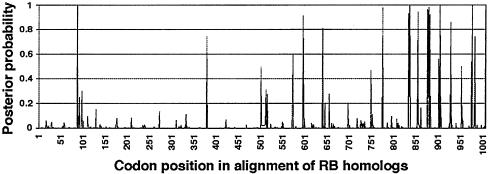
Diversifying selections have been detected in many different resistance gene systems (20). Selections on each site of the RB homologs were investigated by using model M2 in the program codeml of paml (12, 13). Eight sites were found to be under diversifying selection, with posterior probability >0.95 (Fig. 5), seven of them located at the regions potentially encoding solvent-exposed residues within the LXXLXXLXLXXC/NXX motif of the LRR repeats, consistent with previous studies.
Fig. 5.
The posterior probability of diversifying selection at each site in RB homologs. The x axis denotes codon position in the alignment of RB homologs. A site with posterior probability >0.95 is considered to be under significant diversifying selection.
Discussion
We have demonstrated that transgenic Katahdin plants containing the RB gene show resistance to all tested isolates, including a “super race” that can overcome all 11 known R genes in potato. The RB gene encodes a polypeptide of 970 aa and belongs to the CC-NBS-LRR class of plant resistance genes (21). RB is more closely related to the I2 protein of tomato (30% identity, 47% similarity over 1,070 aa) than it is to any other known R protein (22). RB has limited similarity with the protein of R1 (22% identity, 49% similarity over 902 aa), a gene derived from S. demissum that confers hypersensitive resistance to potato late blight (23). Many R genes are organized in clusters (24). Similarly, RB is a member of a four-gene family located within a 40-kb region on chromosome 8. RB transcript was detected in unchallenged plants, indicating that RB is expressed in the absence of the corresponding Avr-expressing pathogen, similar to other R genes that function in pathogen surveillance (25).
Sequence comparison between RB and its susceptible allele rb revealed a C1362 to G point mutation that creates a stop codon in the second exon at Tyr-454. Other than this stop codon within rb, the amino acid sequences deduced from RB and rb are highly similar, with only three synonymous point mutations (C28 to T, T2635 to C, and A2745 to G), a point mutation of T65 to C that changes valine to alanine, and a deletion of an 18-bp sequence that resulted in a loss of six amino acids (KIQLCC) in the 18th LRR repeat. It is likely that the premature stop codon in the rb sequence resulted in the loss of function, but the effects of other mutations, particularly the 18-bp deletion, remain unclear. RGA2-BAC, a chimera of RB and RGA-tr generated during BAC propagation, failed to complement RB function, suggesting that the last 151 aa in the LRR domain are essential for the function of RB resistance.
Both RB and rb contain 21 LRR repeats, whereas RGA1, RGA3, and RGA4 contain 22 LRR repeats (Fig. 6, which is published as supporting information on the PNAS web site). The variation of LRR repeats may play a role in determining the recognition specificity of the RB protein. It has been demonstrated that expansion and contraction of LRR repeats are responsible for loss of function or recognition specificities of plant disease resistance genes. In flax, inactivation of the rust resistance gene M was associated with the loss of a single repeated unit within the LRR coding region (26). Sequence analysis of mutant RPP5 alleles identified four duplicated LRR repeats in comparison to the wild-type RPP5 gene (27). Recently, domain swapping and gene shuffling of tomato proteins Cf-4 and Cf-9 also demonstrated that variation in LRR copy number plays a major role in determining recognition specificity in these proteins (28).
R proteins may perceive the presence of more than one Avr proteins (29). Two recently identified R genes, RPW8.1 and RPW8.2, which consist of only a coiled-coil domain and a single N-terminal transmembrane domain, confer resistance to all tested isolates of four species of powdery mildew of Arabidopsis, indicating that RPW8-mediated resistance may not involve a gene-for-gene interaction (30). Dual recognition has been demonstrated in several cases. For example, RPM1 recognizes two nonhomologous P. syringae avr genes (31). Similarly, the tomato Mi gene confers resistance both to the root-knot nematode and to potato aphid (32). The broad spectrum resistance against multiple races of P. infestans suggests that the RB protein may recognize conserved molecules from different races of the pathogen.
Gene RB shows an evolutionary pattern typical to Type II resistance genes (H.K., E. Nevo, and R. W. Michelmore, unpublished work). Gene RB, like the Type II RGC2 resistance genes in lettuce, might be highly conserved in different geno-types or closely related species and present at high frequencies in natural populations. This is consistent with the observation that many accessions of S. bulbocastanum are highly resistant to all races of P. infestans. There might be purifying selection on RB orthologs, even at the hypervariable sites in the LRR motif, as observed in K orthologs in lettuce (H.K., E. Nevo, and R. W. Michelmore, unpublished work). This hypothesis can be tested when more RB orthologs become available.
Resistance of potato germplasm developed from S. bulbocastanum clone PT29 is effective against all known races of the late blight pathogen. There have been no reports that this resistance has been overcome by any of the P. infestans pathotypes. Transgenic Katahdin plants with the RB gene developed limited lesions on the lower leaves consistent with symptoms noted in field evaluations of the RB-containing progenies derived from S. bulbocastanum clone PT29 (5). The RB locus on chromosome 8 explains 62% of the genetic variation in late blight resistance in the progenies derived from S. bulbocastanum (8). Quantitative trait loci (QTL)-associated late blight resistances, which have been identified in various potato populations (33–35), may also contribute to the resistance of S. bulbocastanum clone PT29. Thus, RB may represent a substantial part, but not all, of the resistance contained within the genome of S. bulbocastanum clone PT29.
Currently, none of the major potato varieties grown in the United States contain resistance to US-8, the most prevalent genotype of P. infestans. Late blight-resistant germplasm has been developed from potato–S. bulbocastanum somatic hybrids (6). These materials can be valuable for breeding new late blight-resistant cultivars through marker-assisted selection. However, because the potato genome is tetraploid and highly heterogeneous, the production of late blight-resistant cultivars acceptable to industry may not be efficiently realized through repeated backcrosses of the S. bulbocastanum-derived germplasm to modern cultivars. Replacing old and disease-susceptible potato varieties has been an extremely slow process because these current varieties are well adapted to the processing industry. Russet Burbank, a late blight-susceptible variety released more than 100 years ago, still accounts for almost half of the potato acreage in the United States. With the deployment of the cloned RB gene it is now possible, through genetic engineering, to render the current popular potato varieties late blight-resistant.
Supplementary Material
Acknowledgments
We acknowledge the work of Institute for Genomic Research sequencing facility personnel in the sequencing of the two BAC clones. We thank Sandy Miller for her excellent technical assistance. This work was supported by National Science Foundation (NSF) Plant Genome Grant DBI-9975866. H.K. was supported by NSF Plant Genome Grant DBI-9975971 (to R. W. Michelmore).
Abbreviations: BAC, bacterial artificial chromosome; LR-PCR, long-range PCR; R, resistance; RGA, R-gene analog.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY303171 and AY303170).
References
- 1.Kamoun, S. (2001) Curr. Opin. Plant Biol. 4, 295–300. [DOI] [PubMed] [Google Scholar]
- 2.Umaerus, V. & Umaerus, M. (1994) Potato Genetics (CAB International, Wallingford, U.K.), pp. 365–401.
- 3.Wastie, R. L. (1991) in Phytophthora infestans: The Cause of Late Blight of Potato, Advances in Plant Pathology, eds. Ingram, D. S. & Williams, P. H. (Academic, London), Vol. 7, pp. 193–223. [Google Scholar]
- 4.Fry, W. E. & Goodwin, S. B. (1997) Bioscience 47, 363–371. [Google Scholar]
- 5.Helgeson, J. P., Pohlman, J. D., Austin, S., Haberlach, G. T., Wielgus, S. M., Ronis, D., Zambolim, L., Tooley, P., McGrath, J. M., James, R. V., et al. (1998) Theor. Appl. Genet. 96, 738–742. [Google Scholar]
- 6.Naess, S. K., Bradeen, J. M., Wielgus, S. M., Haberlach, G. T., McGrath, J. M. & Helgeson, J. P. (2001) Mol. Genet. Genomics 265, 694–704. [DOI] [PubMed] [Google Scholar]
- 7.Dorance, A. E., Inglis, D. A., Helgeson, J. P. & Brown, C. R. (2001) Am. J. Potato Res. 78, 9–17. [Google Scholar]
- 8.Naess, S. K., Bradeen, J. M., Wielgus, S. M., Haberlach, G. T., McGrath, J. M. & Helgeson, J. P. (2000) Theor. Appl. Genet. 101, 697–704. [Google Scholar]
- 9.Bradeen, J. M., Naess, S. K., Song, J., Haberlach, G. T., Wielgus, S. M., Buell, C. R., Jiang, J. & Helgeson, J. P. (2003) Mol. Genet. Genomics, in press. [DOI] [PubMed]
- 10.Yuan, Q., Hill, J., Hsiao, J., Moffat, K., Ouyang, S., Cheng, Z., Jiang, J. & Buell, C. R. (2002) Mol. Genet. Genomics 267, 713–720. [DOI] [PubMed] [Google Scholar]
- 11.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 13.Yang, Z., Nielsen, R., Goldman, N. & Pedersen, A. M. K. (2000) Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, D. A., Thomas, C. M., Hammond-Kosack. K. E., Balint-Kurti, P. J. & Jones, D. J. G. (1992) Transgenic Res. 1, 285–297.1338696 [Google Scholar]
- 15.Ziegelhoffer, T., Will, J. & Austin-Phillips, S. (1999) Mol. Breed. 5, 309–318. [Google Scholar]
- 16.Samen, A.-E., Secor, G. A. & Gudmestad, N. C. (2003) Phytopathology 93, 293–304. [DOI] [PubMed] [Google Scholar]
- 17.Song, J., Dong, F. & Jiang, J. (2000) Genome 43, 199–204. [PubMed] [Google Scholar]
- 18.Jones, D. A. & Jones, J. D. G. (1997) Adv. Bot. Res. 24, 90–167. [Google Scholar]
- 19.van der Biezen, E. A. & Jones, J. D. G. (1998) Curr. Biol. 8, 226–227. [DOI] [PubMed] [Google Scholar]
- 20.Hulbert, S. H., Webb, C. A., Smith, S. M. & Sun, Q. (2001) Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- 21.Baker, B., Zambryski, P., Staskawicz, B. & Dinesh-Kumar, S. P. (1997) Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- 22.Simons, G., Groenendijk, J., Wijbrandi, J., Reijans, M., Groenen, J., Diergaarde, P., Van der Lee, T., Bleeker, M., Onstenk, J., de Both, M., et al. (1998) Plant Cell 10, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballvora, A., Ercolano, M. R., Weiss, J., Meksem, K., Bormann, C. A., Oberhagemann, P., Salamini, F. & Gebhardt, C. (2002) Plant J. 30, 361–371. [DOI] [PubMed] [Google Scholar]
- 24.Michelmore, R. W. & Meyers, B. C. (1998) Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- 25.Hammond-Kosack, K. & Jones, J. D. (1997) Ann. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. [DOI] [PubMed] [Google Scholar]
- 26.Anderson, P. A., Lawrence, G. J., Morrish, B. C., Ayliffe, M. A., Finnegan, E. J. & Ellis, J. G. (1997) Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker, J. E., Coleman, M., Szabo, V., Frost, L. N., Schmidt, R., van der Biezen, E. A., Moores, T., Dean, C., Daniels, M. J. & Jones, J. D. G. (1997) Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wulff, B. B. H., Thomas, C. M., Smoker, M., Grant, M. & Jones, J. D. G. (2001) Plant Cell 13, 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dangl, J. L. & Jones, J. D. G. (2001) Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- 30.Xiao, S., Ellwood, S., Calis, O., Patrick, E., Li, T., Coleman, M. & Turner, J. G. (2001) Science 291, 118–120. [DOI] [PubMed] [Google Scholar]
- 31.Grant, M. R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R. W. & Dangl, J. L. (1996) Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- 32.Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., Frijters, A., Groenendijk, J., Diergaarde, P., Reijans, M., et al. (1998) Nat. Biotechnol. 16, 1365–1369. [DOI] [PubMed] [Google Scholar]
- 33.Leonards-Schippers, C., Gieffers, W., Schafer-Pregl, R., Ritter, E., Knapp, S. J., Salamini, F. & Gebhardt, C. (1994) Genetics 137, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyer, R. C., Milbourne, D., Hackett, C. A., Bradshaw, J. E., McNichol, J. W. & Waugh, R. (1998) Mol. Gen. Genet. 259, 150–160. [DOI] [PubMed] [Google Scholar]
- 35.Trognitz, F., Manosalva, P., Gysin, R., Niño-Liu, D., Simon, R., Herrera, M. R., Trognitz, B., Ghislain, M. & Nelson, R. (2002) Mol. Plant–Microbe Interact. 15, 587–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.