Abstract
We report a strategy for the reagentless transduction of DNA hybridization into a readily detectable electrochemical signal by means of a conformational change analogous to the optical molecular beacon approach. The strategy involves an electroactive, ferrocene-tagged DNA stem–loop structure that self-assembles onto a gold electrode by means of facile gold-thiol chemistry. Hybridization induces a large conformational change in this surface-confined DNA structure, which in turn significantly alters the electron-transfer tunneling distance between the electrode and the redoxable label. The resulting change in electron transfer efficiency is readily measured by cyclic voltammetry at target DNA concentrations as low as 10 pM. In contrast to existing optical approaches, an electrochemical DNA (E-DNA) sensor built on this strategy can detect femtomoles of target DNA without employing cumbersome and expensive optics, light sources, or photodetectors. In contrast to previously reported electrochemical approaches, the E-DNA sensor achieves this impressive sensitivity without the use of exogenous reagents and without sacrificing selectivity or reusability. The E-DNA sensor thus offers the promise of convenient, reusable detection of picomolar DNA.
The detection of DNA hybridization is of significant scientific and technological importance, as manifested in, for example, the growing interest in chip-based characterization of gene expression patterns and the detection of pathogens in both clinical and civil defense settings (1). Consequently, a variety of optical (2–5), acoustic (6, 7), and electronic (8, 9) “gene detection” approaches has been reported. Among these, a solution-phase optical approach termed molecular beacons has attracted significant interest (10) because of its applicability to approaches ranging from in vitro genotyping (11) to in vivo studies within single cells (12). Molecular beacons are composed of a hairpin-like DNA stem–loop structure, with a fluorescent moiety and a fluorescence quencher attached to either terminus. In the absence of target, the molecular beacon is in the folded configuration in which its termini are held in close proximity, and fluorescence emission is thus suppressed. On hybridization with a complementary target sequence, the stem–loop is converted into a rigid, linear double helix, removing the fluorophore from proximity to the quencher and greatly enhancing emission. Previous studies demonstrate that molecular beacons can discriminate even single-base mismatches (13). More recently, a reagentless solid-state version of the optical molecular beacon has been described that may prove suitable for chip-based optical detection (14, 15).
Whereas optical detection methods have historically dominated state-of-the-art real-time or near real-time genosensors (1, 16, 17), the application of electrochemical methods to the sensing of biologically related species may provide very significant advantages (18–20). Specifically, the advantages of bioelectronic approaches include the speed, sensitivity, and low cost/mass/power requirements of electrochemical detection (21); the relatively high stability and environmental insensitivity of electroactive labels; and the availability of electroactive labels with nonoverlapping redox potentials for multicolor labeling and the simultaneous detection of multiple analytes (22).
DNA is electrochemically silent at moderate applied voltages (23) and thus, whereas Armistead and Thorp (24) have electrocatalytically oxidized guanine by means of inorganic metal complexes, electrochemical detection of hybridization typically requires the use of exogenous reporter groups. The first sequence-selective electrochemical method for DNA detection was based on the electrochemical interrogation of exogenous, redox-active intercalators that bind preferably to dsDNA (25–27). Barton and coworkers (28) have improved the sensitivity of this approach by employing exogenous electrocatalytic species for amplification. They also report that the current flow through the double helix is sensitive even to single-base mismatches, paving the way for the direct electrochemical identification of point mutations (9). In an attempt to reduce high backgrounds arising from the inappropriate binding of hybridization indicators to unhybridized DNA, sandwich-type detectors have also been developed and commercialized (29–31). Sandwich approaches employ a surface-confined, electrochemically silent probe sequence to bring target DNA strands to the electrode surface. A signal is generated when this bound target strand is hybridized with an exogenous, redox-labeled signaling sequence. Finally, Mirkin and coworkers (8) have developed an electronic DNA detection approach with high sensitivity and selectivity. In this resistance-based method, the probe-captured target undergoes a second hybridization with exogenous, gold nanoparticle-labeled DNA strands. Subsequent catalytic deposition of exogenously added silver onto the gold nanoparticles produces electrical contact between a closely spaced electrode pair.
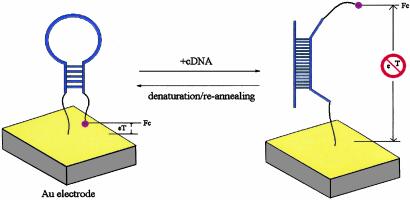
Despite these advances, there has been relatively little progress toward the important goal of creating electrochemical DNA detection methods that are simultaneously sensitive, selective, and reagentless (32); all of the above-described electrochemical hybridization detectors require posthybridization treatment with either hybridization indicators or other exogenous signaling molecules. Here, we report the development of a sensitive, reagentless, reusable electrochemical DNA sensor that combines the significant advantages of electrochemical detection with the versatility of reagentless, reusable, surface-attached molecular beacons. This electrochemical DNA (E-DNA) sensor employs an electrode-attached, molecular beacon-like DNA stem–loop labeled with an electroactive reporter as the hybridization sensing element (Scheme 1). On hybridization, the distance between the label and the electrode is significantly altered, leading to a large, readily measurable signal change. The E-DNA sensor thus provides a ready means for the reagentless, reusable detection of hybridization.
Scheme 1.
A stem–loop oligonucleotide possessing terminal thiol and a ferrocene group is immobilized at a gold electrode through self-assembly. In the absence of target, the stem–loop structure holds the ferrocene tag into close proximity with the electrode surface, thus ensuring rapid electron transfer and efficient redox of the ferrocene label. On hybridization with the target sequence, a large change in redox currents is observed, presumably because the ferrocene label is separated from the electrode surface.
Materials and Methods
Materials. Oligonucleotides were obtained from Synthegen (Houston). The sensor oligonucleotide, 5′-NH2-(CH2)6-GCGAGGTAAAACGACGGCCAGTCCTCGC-(CH2)6-SH-3′ (oligo 1), contains a 5′ hexamethylene amine and a 3′ hexamethylene thiol. A ferrocene tag was conjugated to oligo 1 through coupling the succinimide ester of ferrocene carboxylic acid with the 5′ amine of oligo 1 (33). The final product (oligo 1-ferrocene) was purified by HPLC on a C18 column and confirmed by electrospray mass spectroscopy. The sequences of the target and control DNA oligos were 5′-TTTTTACTGGCCGTCGTTTTACTCTTT-3′ (oligo 2) and 5′-CGTATCATTGGACTGGCCATTTAT-3′ (oligo 3), respectively. The control oligo is a random sequence unrelated to the probe sequence.
Cyclic voltammetry (CV) was performed at room temperature by using a CHI 603 workstation (CH Instruments, Austin, TX). Polycrystalline gold disks (1.6-mm diameter; BAS, West Lafayette, IN) were used as working electrodes. Although potentials are reported versus the normal hydrogen electrode, in actuality we used a platinum electrode as a quasi-reference electrode at an assigned potential of 0.495 V relative to the normal hydrogen electrode (calibrated with Ag/AgCl, 3 M NaCl reference electrode from BAS in 1 M NaClO4 with 1 mM ferricyanide). In all experiments, 1 M NaClO4, which is a weak nucleophile, was used as the electrolyte to avoid the instability of ferrocenium (the oxidized form of ferrocene; ref. 34). Some of the reported CV curves were background-subtracted by using origin 7.0 (Microcal Software, Northampton, MA) through extrapolation to the baseline in regions far from the peak (35).
Construction of E-DNA Sensor. The E-DNA sensor was constructed by assembling the ferrocene-labeled DNA stem–loop at a bioelectronic interface. This surface assembly was achieved by a self-assembly process given the fact that DNA-thiol has a strong tendency to diffuse from diluted aqueous solutions to clean gold surfaces (36). The stem–loop unit (oligo 1) has been designed such that it has five complementary bases at its 5′ and 3′ ends (four of them are G–C pairs), in the hope that the DNA strand will be closed by the thermostable G–C pairs and thus form a stem–loop with either end close to the gold surface. We expect that this folding localizes the ferrocene unit at the 5′ end to proximity of the gold surface.
To construct the sensor as demonstrated in Scheme 1, a 1-μM solution of the stem–loop oligo 1-ferrocene was self-assembled on an extensively cleaned gold surface (37). The prepared surface was subsequently passivated with excess 2-mercaptoethanol at 1 mM. This process has been reported to “cure” relatively disordered self-assembled monolayers by gradually displacing nonspecifically adsorbed oligonucleotides (36). This oligonucleotide-containing, passivated surface does not interact significantly with noncognate DNA sequences, as reported (38) and independently confirmed in our laboratory by monitoring quartz crystal microbalance measurements (data not shown). The modified electrode was thoroughly rinsed, dried, and then incubated in 1 M NaClO4 before use.
Results
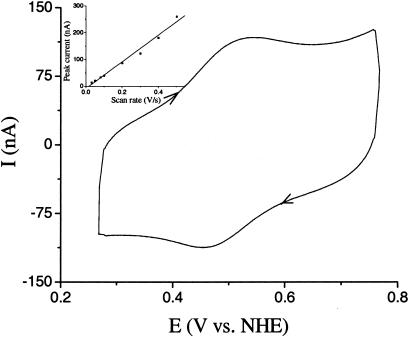
Characterization of the E-DNA-Modified Electrode. In the absence of target DNA, a ferrocene redox peak pair is observed with the E-DNA-modified gold electrodes (Fig. 1). Bare gold electrodes and gold modified with either 2-mercaptoethanol or 2-mercaptoethanol/ferrocene-free oligo 1, in contrast, do not produce cyclic voltammetry peaks in the relevant potential range (data not shown). The apparent formal potential (E°′) of the electroactive label is 0.492 V, as estimated from E1/2 = (Ered + Eox)/2. This value falls within the typical redox potential range of ferrocene (22, 30). We thus ascribe this peak pair to the redox conversion of ferrocene labels in close proximity to the gold electrode. Because of electrostatic repulsion between negatively charged DNA strands, high ionic strength is required for the formation of the stem–loop structure (38). The observation that freshly modified electrodes do not produce significant redox peaks without prior incubation in 1 M NaClO4 (data not shown) thus provides strong evidence that the formation of the stem–loop structure is required for efficient electron transfer. This result also suggests that the use of uncharged peptide nucleic acids in place of the sensor DNA might allow hybridization to occur at lower ionic strengths.
Fig. 1.
A cyclic voltammogram for a gold electrode modified with the ferrocene-tagged, stem–loop oligonucleotide in the absence of target DNA (scan rate 0.1 V/s). The electrolyte is 1 M NaClO4.(Inset) The linear relationship between peak currents and scan rates confirms that the redox species is confined to the electrode surface (21).
Modulating the scan rate of the cyclic voltammetries provides further evidence that ferrocene is confined at the electrode surface by the formation of the stem–loop structure. Peak currents of the ferrocene redox reaction (Ip) are directly proportional to scan rates (Fig. 1 Inset), consistent with a surface-confined electrochemical reaction (in contrast to Ip being proportional to the square-root of the scan rate characteristic of diffusion-controlled electrochemical reactions; ref. 21).
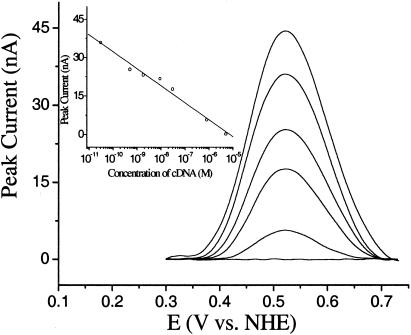
Target Detection. Hybridization of the stem–loop structure with a target sequence complementary to the 17-base loop region competes with the less stable stem structure, moving the ferrocene away from the electrode surface (Scheme 1). Thus, incubating a stem–loop-modified electrode with 5 μM cDNA (oligo 2) in 1 M NaClO4 eliminates the ferrocene reduction and oxidation peaks within ≈30 min (Fig. 2). At lower target concentrations, only partial loss of signal is observed after 30 min (Fig. 2). For example, after a 30-min incubation at 500 pM target, the electrochemical signal attenuates to ≈70% of its initial value (Fig. 3).
Fig. 2.
Background-subtracted (35, 37) voltammograms (anodic scan) for the E-DNA sensor in the presence of complementary DNA at 0 M, 30 pM, 500 pM, 30 nM, 800 nM, and 5 μM (from top to bottom). The hybridization time was fixed at 30 min. (Inset) A calibration curve demonstrating peak height versus target concentration; E-DNA sensor response is logarithmically related to target concentration over at least six orders of magnitude.
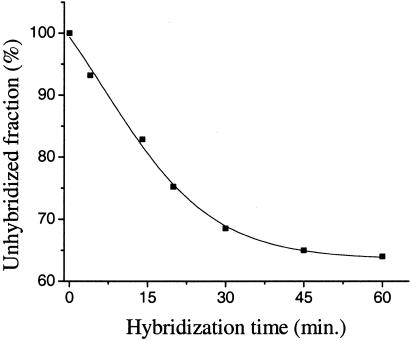
Fig. 3.
The E-DNA signal develops in minutes. At a target concentration of 500 pM, the signal change observed after1hof hybridization implies that 35% of the probe DNA has hybridized with target molecules. At 5 μM target, in contrast, the peak current is entirely abolished within 30 min.
Employing a fixed 30-min incubation time, we have tested the sensitivity and selectivity of the E-DNA sensor. Under these conditions, we observe easily measurable decreases in peak current at target DNA concentrations as low as 10 pM (Fig. 2). We currently employ a sample volume of 500 μl, equating to an absolute detection limit of 5 fmol. The exceptional signal-to-noise of the approach (Fig. 2) suggests, however, that large improvements in electrode size (and thus sample volume and absolute sensitivity) will be straightforward. In contrast, no significant signal change is observed for electrodes incubated in DNA-free hybridization buffer or in the presence of the highest noncognate DNA concentrations we have investigated (10 μM oligo 3). Thus, the selectivity of the sensor is in excess of 106. The E-DNA sensor also exhibits exceptional dynamic range; peak currents are logarithmically related to target concentration across the almost 6-decade range we have investigated (Fig. 2 Inset). Whereas similar logarithmic signal-versus-concentration relationships have been reported for other solid-state DNA sensors (32, 39), the mechanism underlying the relationship has not been determined.
Sensor Regeneration. The electrochemical DNA sensor is readily reusable. We have successfully recovered up to ≈80% of the original signal by washing the electrode with 1 M NaClO4 at 95°C and rechallenging with the target sequence. We believe the minor signal loss during recovery arises because of the relative instability of ferrocene in aqueous solution at high temperature.
Discussion
The E-DNA sensor is thus a reusable, solid-state, electrochemical genosensor. Unlike all previous sequence-specific electronic genosensors, however, the E-DNA sensor is reagentless and thus ideally suited for the continuous, rather than batch, monitoring of a flow of analyte. Moreover, the E-DNA sensor achieves the significant advantages of reagentless, reusable operation without compromising sensitivity or selectivity; the ≈10 pM sensitivity and greater than million-fold selectivity of the E-DNA sensor is competitive with the very best current electrochemical DNA sensors. For example, E-DNA's 10-pM detection limit (≈5 fmol in 0.5 ml) matches both sandwich approaches (J. F. Kayyem, personal communication) and a recently proposed enzyme-amplified electrochemical detection method (32, 39), and is only surpassed by the 0.5 pM detection limit achieved by the reagent-intensive, nonreusable catalytic amplification approach of Mirkin and coworkers (8).
The E-DNA sensor also offers significant advantages over optically detected molecular beacons. For example, whereas the most highly optimized optical approaches can detect a femtomolar target in the laboratory, the picomolar sensitivity of the E-DNA sensor is comparable to or significantly better than the fluorescence-based techniques used in the “real world” (i.e., with lower-power light sources and off-the-shelf detectors; refs. 4 and 40). For example, the sensitivity of the E-DNA sensor exceeds that of typical, charge-coupled device-based fluorescent detectors by at least an order of magnitude (4). The E-DNA sensor also vastly surpasses solid-state optical molecular beacons, for which an ≈1 nM detection limit is reported (14). However, we note that the current E-DNA sensor, like all other state-of-the-art electrochemical or optical DNA sensors, does not meet the high-end requirements of many real-world gene-targeting applications; whereas picomolar sensitivity is orders of magnitude more sensitive than necessary for the detection of amplified targets (and offers the very real possibility of replacing cumbersome gel- or optical-based detection schemes in this role), picomolar sensitivity is not sufficient for the unamplified detection of most pathogens. By employing electrochemical amplification methods, such as coupling with electrocatalysis (9) or extreme catalysis (39), it may be possible to increase the E-DNA signal sufficiently to enable direct pathogen detection.
The E-DNA sensor is a “signal-off” device in that hybridization abolishes the redox current. Signal-off sensors, unfortunately, are susceptible to false-positive responses if, for example, the DNA is degraded by sample contaminants. The availability of “multicolor” redoxable labels (22), however, provides a means of detecting false positives by simultaneously monitoring both sensor elements and control sequences on a single electrode. In contrast, the signal-off nature of the E-DNA sensor provides a significant advantage with respect to false positives arising from the binding of electroactive contaminants; unlike hybridization, contaminants that are redoxable at the ferrocene potentials will increase, rather than decrease, peak currents. The E-DNA approach thus offers a platform for the reagentless detection of DNA hybridization in which both false positives and false negatives are readily detected in real time.
The preparation of the E-DNA sensor is quite straightforward. The key sensing element, the electroactive stem–loop, is compatible with normal solid-state synthesis of oligonucleotides, and the surface assembly chemistry is facile. Because the entire setup can be conveniently prepared and generalized to be consistent with chip-based electrode arrays, the reagentless detection described here seems to provide a promising alternative to traditional, fluorescence-based gene arrays.
Although the signal generation mechanism of the E-DNA sensor has not been determined in detail, our experimental results provide strong support for the claim that the signal change arises from the on-and-off states provided by the DNA stem–loop (Scheme 1). When in the “on” state the ferrocene label is localized to the electrode surface by means of hybridization of the 5-bp stem region of the sensor DNA. Presumably, this spatial proximity allows facile electron transfer between the electroactive label and the gold. In the presence of a complementary target, the stem–loop is disrupted in favor of the thermodynamically more stable, rigid rod-like (41) target-sensor duplex, thereby separating the ferrocene from the electrode and effectively abolishing the exponentially distance-dependent electron transfer process (42, 43). The separation of the electroactive label from the electrode surface may be facilitated by repulsion between the negatively charged DNA and the negative dipole of the 2-mercaptoethanol hydroxyl group (36, 44). Previous neutron reflectivity studies (36) demonstrate that both ssDNA and dsDNA project away from surfaces covered with similar self-assembled monolayers. This agrees with our observation that stem–loop formation is required to generate electrochemical signals in the E-DNA sensor.
A critical aspect of the described E-DNA sensor is the electrochemical detection of a target-induced conformational change of a biopolymer. This suggests that the E-DNA approach may be generalizable to other sensor designs in which significant protein (45, 46) or aptamer (33, 47, 48) conformational changes occur on target binding.
Acknowledgments
This work was supported by National Science Foundation Grant NSF-DMR-0099843, Office of Naval Research Grant N0014-1-1-0239, and National Institutes of Health Grant GM 62958-01.
Abbreviation: E-DNA sensor, electrochemical DNA sensor.
References
- 1.Heller, M. J. (2002) Annu. Rev. Biomed. Eng. 4, 129–153. [DOI] [PubMed] [Google Scholar]
- 2.Xu, X. H. & Bard, A. J. (1995) J. Am. Chem. Soc. 117, 2627–2631. [Google Scholar]
- 3.Cao, Y. W. C., Jin, R. C. & Mirkin, C. A. (2002) Science 297, 1536–1540. [DOI] [PubMed] [Google Scholar]
- 4.Gaylord, B. S., Heeger, A. J. & Bazan, G. C. (2002) Proc. Natl. Acad. Sci. USA 99, 10954–10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peterson, A. W., Wolf, L. K. & Georgiadis, R. M. (2002) J. Am. Chem. Soc. 124, 14601–14607. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, M. A., Dultsev, F. N., Minson, T., Ostanin, V. P., Abell, C. & Klenerman, D. (2001) Nat. Biotechnol. 19, 833–837. [DOI] [PubMed] [Google Scholar]
- 7.Hook, F., Ray, A., Norden, B. & Kasemo, B. (2001) Langmuir 17, 8305–8312. [Google Scholar]
- 8.Park, S. J., Taton, T. A. & Mirkin, C. A. (2002) Science 295, 1503–1506. [DOI] [PubMed] [Google Scholar]
- 9.Boon, E. M., Ceres, D. M., Drummond, T. G., Hill, M. G. & Barton, J. K. (2000) Nat. Biotechnol. 18, 1096–1100. [DOI] [PubMed] [Google Scholar]
- 10.Tyagi, S. & Kramer, F. R. (1996) Nat. Biotechnol. 14, 303–308. [DOI] [PubMed] [Google Scholar]
- 11.Kostrikis, L. G., Tyagi, S., Mhlanga, M. M., Ho, D. D. & Kramer, F. R. (1998) Science 279, 1228–1229. [DOI] [PubMed] [Google Scholar]
- 12.Fang, X., Mi, Y., Li, J. J., Beck, T., Schuster, S. & Tan, W. (2002) Cell Biochem. Biophys. 37, 71–81. [DOI] [PubMed] [Google Scholar]
- 13.Tyagi, S. & Kramer, F. R. (1999) Proc. Natl. Acad. Sci. USA 96, 6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, X. & Tan, W. (1999) Anal. Chem. 71, 5054–5059. [DOI] [PubMed] [Google Scholar]
- 15.Du, H., Disney, M. D., Miller, B. L. & Krauss, T. D. (2003) J. Am. Chem. Soc. 125, 4012–4013. [DOI] [PubMed] [Google Scholar]
- 16.Bowtell, D. D. L. (1999) Nat. Genet. 21, 25–32. [DOI] [PubMed] [Google Scholar]
- 17.Winzeler, E. A., Schena, M. & Davis, R. W. (1999) Methods Enzymol. 306, 3–18. [DOI] [PubMed] [Google Scholar]
- 18.Kuhr, W. G. (2000) Nat. Biotechnol. 18, 1042–1043. [DOI] [PubMed] [Google Scholar]
- 19.Willner, I. (2002) Science 298, 2407–2408. [DOI] [PubMed] [Google Scholar]
- 20.Fritz, J., Cooper, E. B., Gaudet, S., Sorger, P. K. & Manalis, S. R. (2002) Proc. Natl. Acad. Sci. USA 99, 14142–14146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bard, A. J. & Faulkner, L. R. (2001) Electrochemical Methods (Wiley, New York).
- 22.Brazill, S. A., Kim, P. H. & Kuhr, W. G. (2001) Anal. Chem. 73, 4882–4890. [DOI] [PubMed] [Google Scholar]
- 23.Palecek, E. & Jelen, F. (2002) Crit. Rev. Anal. Chem. 32, 261–270. [Google Scholar]
- 24.Armistead, P. M. & Thorp, H. H. (2000) Anal. Chem. 76, 3764–3770. [DOI] [PubMed] [Google Scholar]
- 25.Carter, M. T. & Bard, A. J. (1987) J. Am. Chem. Soc. 109, 7528–7530. [Google Scholar]
- 26.Rodriguez, M. & Bard, A. J. (1990) Anal. Chem. 62, 2658–2662. [DOI] [PubMed] [Google Scholar]
- 27.Millan, K. M. & Mikkelsen, S. R. (1993) Anal. Chem. 65, 2317–2323. [DOI] [PubMed] [Google Scholar]
- 28.Kelley, S. O., Boon, E. M., Barton, J. K., Jackson, N. M. & Hill, M. G. (1999) Nucleic Acids Res. 27, 4830–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ihara, T., Maruo, Y., Takenaka, S. & Takagi, M. (1996) Nucleic Acids Res. 24, 4273–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, C. J., Wan, Y. J., Yowanto, H., Li, J., Tao, C. L., James, M. D., Tan, C. L., Blackburn, G. F. & Meade, T. J. (2001) J. Am. Chem. Soc. 123, 11155–11161. [DOI] [PubMed] [Google Scholar]
- 31.Umek, R. M., Lin, S. W., Vielmetter, J., Terbrueggen, R. H., Irvine, B., Yu, C. J., Kayyem, J. F., Yowanto, H., Blackburn, G. F., Farkas, D. H. & Chen, Y. P. (2001) J. Mol. Diag. 3, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korri-Youssoufi, H., Garnier, F., Srivastava, P., Godillot, P. & Yassar, A. (1997) J. Am. Chem. Soc. 119, 7388–7389. [Google Scholar]
- 33.Fahlman, R. P. & Sen, D. (2002) J. Am. Chem. Soc. 124, 4610–4616. [DOI] [PubMed] [Google Scholar]
- 34.Han, S. W., Seo, H., Chung, Y. K. & Kim, K. (2000) Langmuir 16, 9493–9500. [Google Scholar]
- 35.Hirst, J., Duff, J. L. C., Jameson, G. N. L., Kemper, M. A., Burgess, B. K. & Armstrong, F. A. (1998) J. Am. Chem. Soc. 120, 7085–7094. [Google Scholar]
- 36.Levicky, R., Herne, T. M., Tarlov, M. J. & Satija, S. K. (1998) J. Am. Chem. Soc. 120, 9787–9792. [Google Scholar]
- 37.Fan, C., Gillespie, B., Wang, G., Heeger, A. J. & Plaxco, K. W. (2002) J. Phys. Chem. 106, 11375–11383. [Google Scholar]
- 38.Herne, T. M. & Tarlov, M. J. (1997) J. Am. Chem. Soc. 119, 8916–8920. [Google Scholar]
- 39.Patolsky, F., Lichtenstein, A. & Willner, I. (2001) Nat. Biotechnol. 19, 253–257. [DOI] [PubMed] [Google Scholar]
- 40.Epstein, J. R., Biran, I. & Walt, D. R. (2002) Anal. Chim. Acta 469, 3–36. [Google Scholar]
- 41.Bednar, J., Furrer, P., Katritch, V., Stasiak, A. Z., Dubochet, J. & Stasiak, A. (1995) J. Mol. Biol. 254, 579–594. [DOI] [PubMed] [Google Scholar]
- 42.Chang, I.-J., Lee, J. C., Winkler, J. R. & Gray, H. B. (2003) Proc. Natl. Acad. Sci. USA 100, 3838–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tezcan, F. A., Crane, B. R., Winkler, J. R. & Gray, H. B. (2001) Proc. Natl. Acad. Sci. USA 98, 5002–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gooding, J. J. (2002) Electroanalysis 14, 1149–1156. [Google Scholar]
- 45.Dyson, H. J. & Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12, 54–60. [DOI] [PubMed] [Google Scholar]
- 46.Plaxco, K. W. & Gross, M. (1997) Nature 386, 657. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, M. P. & Ellington, A. (1999) Nat. Biotechnol. 17, 62–66. [DOI] [PubMed] [Google Scholar]
- 48.Pavski, V. & Le, X. C. (2003) Curr. Opin. Biotechnol. 14, 65–73. [DOI] [PubMed] [Google Scholar]