Abstract
Lipoxygenase (LOX) enzymes form fatty acid hydroperoxides used in membrane remodeling and cell signaling. Mammalian epidermal LOX type 3 (eLOX3) is distinctive in totally lacking this typical oxygenase activity. Surprisingly, genetic evidence has linked mutations in eLOX3 or a colocalizing enzyme, 12R-LOX, to disruption of the normal permeability barrier of the skin [Jobard, F., Lefèvre, C., Karaduman, A., Blanchet-Bardon, C., Emre, S., Weissenbach, J., Özgüc, M., Lathrop, M., Prud'homme, J. F. & Fischer, J. (2002) Hum. Mol. Genet. 11, 107–113]. Herein we identify a logical link of the biochemistry to the genetics. eLOX3 functions as a hydroperoxide isomerase (epoxyalcohol synthase) by using the product of 12R-LOX as the preferred substrate. 12R-Hydroperoxyeicosatetraenoic acid (12R-HPETE) is converted to 8R-hydroxy-11R,12R-epoxyeicosa-5Z,9E,14Z-trienoic acid, one of the isomers of hepoxilin A3, and to 12-ketoeicosatetraenoic acid in a 2:1 ratio. Other hydroperoxides, including 8R-HPETE, 12S-HPETE, and 15S-HPETE, as well as the 13S- and 13R-hydroperoxides of linoleic acid are converted less efficiently. Mass spectrometric analysis of the epoxyalcohol formed from [18O]15S-HPETE showed that both hydroperoxy oxygens are retained in the product. We propose that the ferrous form of eLOX3 initiates a redox cycle, unprecedented among LOX in being autocatalytic, in which the hydroperoxy substrate is isomerized to the epoxyalcohol or keto product. Our results provide strong biochemical evidence for a functional linkage of 12R-LOX and eLOX3 and clues into skin biochemistry and the etiology of ichthyosiform diseases in humans.
Lipoxygenases (LOXs) are a family of nonheme iron-containing enzymes that oxygenate unsaturated fatty acids such as arachidonic acid to specific hydroperoxide products (1). These may be metabolized further to various bioactive lipid mediators including leukotrienes, lipoxins, hydroxyeicosatetraenoic acids (HETEs), and hepoxilins (2). Although LOX enzymes are catalytically active with free fatty acid substrates, some will also oxygenate esterified substrates such as the phospholipid or cholesterol esters. LOX metabolites play important roles in cell signaling or modification of membrane structures (1, 3).
There are five active LOXs found in human beings: 5-LOX, 12S-LOX, 12R-LOX, 15-LOX-1, and 15-LOX-2. A sixth gene family member, epidermal LOX type 3 (eLOX3, gene symbol ALOXE3) was described first in the mouse (4), and in humans in 2001 (5). The amino acid sequence of human eLOX3 shows the closest similarity to 12R-LOX (54% identity) and 15-LOX-2 (51%). It contains the characteristic well conserved amino acid residues found in all LOXs including the putative iron-binding ligands and additional structure-determining residues. These features clearly indicate that eLOX3 belongs to the LOX gene family. The question of the catalytic activity of eLOX3, nonetheless, has remained elusive. No enzymatic activity has been detected by using linoleic or arachidonic acids, the prototypical C18 and C20 LOX substrates, nor with methyl arachidonate or cholesteryl arachidonate (4). In the first section of Results we confirm and extend these findings.
Studies in humans and the mouse indicate that eLOX3 has a limited scope of tissue expression, being mainly confined to keratinized epithelia such as skin. From PCR evidence it seems to be coexpressed in tissues that express the 12R-LOX (5). A functional relationship to skin pathophysiology is strongly suggested by a recent genetic study reporting that eLOX3 or 12R-LOX are mutated in six families affected by nonbullous congenital ichthyosiform erythroderma (NCIE) (6). NCIE is a major subtype of autosomal recessive congenital ichthyosis characterized by a generalized ichthyosiform (scaly skin) phenotype. Based on these genetic findings in NCIE it was suggested that eLOX3 and 12R-LOX are both involved in skin development and that they may belong to the same metabolic pathway (6).
In this article we report biochemical studies identifying a catalytic activity of eLOX3 in the conversion of HPETE substrates. The products of this reaction are specific epoxyalcohols (hepoxilins or hepoxilin-type products). 12R-HPETE is a particularly good substrate, and it is converted to a product of very distinctive structure. Our results suggest that eLOX3, although named LOX based on its gene sequence, lacks the typical catalytic activity of this enzyme class and instead represents a unique type of epoxyalcohol synthase.
Materials and Methods
Expression and Purification of Human eLOX3. The cDNA for human eLOX3 was cloned by PCR with cDNA prepared from human keratinocytes. To prepare the eLOX3 protein with an N-terminal (His)6 tag, the eLOX3 cDNA was subcloned into the pET3a expression vector (Novagen) with the 5′ sequence encoded as ATG CAT CAC CAT CAC CAT CAC GCA, with the last codon representing the start of the wild-type enzyme. The human eLOX3 was expressed in Escherichia coli BL21 (DE3) cells (Novagen), and the (His)6-tagged protein was purified on nickel-nitrilotriacetic acid agarose (Qiagen, Valencia, CA) according to manufacturer instructions. Fractions of 0.5 ml were collected off the affinity column and assayed by using SDS/PAGE. Fractions containing eLOX3 were pooled and dialyzed against a buffer of 50 mM Tris (pH 7.5) containing 300 mM NaCl to remove the imidazole.
Preparation of Hydroperoxides. 15S-HPETE and [18O]15S-HPETE were prepared from arachidonic acid by using soybean LOX (Sigma type V) (7). 12R-HPETE and 12S-HPETE were prepared from arachidonate methyl ester by the following route: (i) autoxidation of arachidonate methyl ester in the presence of α-tocopherol (8); (ii) isolation of a mixture of HPETE methyl esters with a 70-g open-bed silica column (3.0 × 25 cm) and a solvent system of 5% ethyl acetate in hexane; (iii) isolation and purification of 12R,S-HPETE methyl ester by RP-HPLC [Waters Symmetry C18 7-μm, 1.9 × 30-cm, solvent system of acetonitrile/water (70:30 by volume) and a flow rate of 10 ml/min]; (iv) resolution of 12R- and 12S-HPETE methyl esters with a Chiralpak AD-RH column, eluted with a solvent of methanol/water (88:12 by volume) and a flow rate of 1 ml/min; the R enantiomer eluted at ≈10 min, and the S enantiomer eluted at 15 min; (v) preparation of the free acids by treatment for 30 min with 0.5 M KOH in water/methanol/dichloromethane (1:1:0.05) at room temperature, followed by acidification to pH 6.0 and extraction into dichloromethane; and (vi) final purification of the free acids (12R-HPETE or 12S-HPETE) by straight-phase HPLC (Alltech Associates Econosil silica column, solvent system of hexane/isopropanol/acetic acid (100:1:0.1 by volume) and a flow rate of 2 ml/min).
eLOX3 Activity Assay. Incubations with the purified enzyme were conducted typically in 500 μl of incubation buffer (50 mM Tris/150 mM NaCl, pH 7.5) by using 0.01–0.1 μM enzyme concentration in a 1-cm path length microcuvette. HPETE (5–10 μg) was added and incubated at room temperature for 10 min. eLOX3 activity was monitored by repetitive scanning in the range of 350–200 nm or by monitoring disappearance of the signal at 235 nm in the time-drive mode. To measure the rate of eLOX3 reaction over the substrate concentration range of 5–250 μM, reactions were conducted in a 2-mm path length microcuvette (0.5 ml); the decrease of absorbance at 235 nm was followed, and the rate was calculated from the initial linear part of the curve.
HPLC Analysis. Products of the eLOX3 reactions with HPETE substrates were analyzed initially by RP-HPLC with a Waters Symmetry C18 5-μm column (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with methanol/water/acetic acid (80:20:0.01 by volume) and UV detection at 205, 220, 235, and 270 nm with an Agilent 1100 series diode array detector. The main products were recovered from the reversed-phase solvent by the addition of water and extraction with dichloromethane. Further purification was carried out by straight-phase HPLC with an Alltech Associates Econosil silica column (0.46 × 25 cm), a solvent system of hexane/isopropanol/acetic acid (100:2:0.1 by volume), and a flow rate of 1 ml/min.
GC-MS Analysis. Analysis of the methyl ester trimethylsilyl ether derivatives of the products was carried out in the positive-ion electron impact mode (70 eV) by using either a Hewlett–Packard 5989A mass spectrometer coupled to a Hewlett–Packard 5890 gas chromatograph equipped with an RTX-1701 fused silica capillary column (17 m × 0.25 mm internal diameter) or a ThermoFinnigan Trace DSQ GC-MS system equipped with a Restek (Bellefonte, PA) Rtx-1 fused silica capillary column (15 m × 0.25 mm internal diameter). Samples were injected at 150°C, and after 1 min the temperature was programmed to 300°C at 10 or 20°C/min. For analysis of 18O content of 15-HETE and its epoxyalcohol product, samples were analyzed as the pentafluorobenzyl ester trimethylsilyl ether derivatives in the negative-ion chemical ionization mode. Rapid repetitive scanning was carried out over the mass ranges covering the [M-pentafluorobenzyl] ions of the unlabeled and 18O-labeled species (m/z 388–399 for the HETE derivative, and m/z 405–415 for the epoxyalcohol). Spectra collected during elution of the GC peak (typically ≈20 spectra) were averaged for calculation of the isotopic compositions.
NMR. 1H NMR and 2D (H,H-COSY) NMR spectra were recorded on a Bruker DRX 400-MHz spectrometer. The ppm values are reported relative to residual nondeuterated solvent (δ = 7.24 ppm for C6H6, and δ = 1.92 ppm for CH3CN).
CD Spectroscopy. CD spectra were recorded on a Jasco (Easton, MD) J-700 spectropolarimeter. Hydroxyl chirality of the main product from 12R-HPETE was assigned from the Cotton effects on the benzoate ester derivative (9, 10). The benzoate derivative was selected because its longitudinal transition moment is sufficiently close to that of the adjacent double bond in the fatty acid carbon chain to allow for an efficient coupling of the double bond and benzoate chromophores (11). Due to the relatively low λmax of both chromophores, only the first Cotton effect at ≈227 nm is observed, which, however, is sufficient to determine the absolute configuration of the molecule (11, 12). Details of the derivatization and CD analysis are given in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Results
Lack of LOX Activity of eLOX3. The following substrates were tested and found not to be oxygenated by mouse or human eLOX3: linoleic, arachidonic, and eicosapentaenoic acids, the methyl esters of arachidonic acid and linoleic acid, arachidonyl phosphatidylcholine, anandamide, and the cholesteryl ester of arachidonic acid. Because the related human 12R-LOX was found to have optimal activity at pH 6 (13), incubations were performed at pH 6, 7, and 8 with radiolabeled linoleic and arachidonic acids (50 μM); again, no oxygenated products were formed. These results confirm and extend a previous report of the absence of detectable LOX activity for mouse eLOX3 expressed in HEK293 cells (4).
Reaction of eLOX3 with HPETEs. When hydroperoxy fatty acid substrates (50–75 μM) were incubated in a microcuvette at room temperature with eLOX3 (1–4 μg/ml protein, ≈12–50 nM), the characteristic UV absorbance of the HPETE substrate decreased during the incubation (Fig. 1A). This decreased absorbance at 235 nm was accompanied by a lesser rise in the UV absorbance at 285 nm. Heat pretreatment (60°C, 10 min) of the enzyme impaired the activity, and it was eliminated by boiling.
Fig. 1.
Reaction of eLOX3 with hydroperoxide substrates. (A) Overlay of UV spectra of reaction of eLOX3 (0.05 μM) with 12R-HPETE (40 μM). The sample was scanned from 350 to 200 nm before the addition of enzyme (t = 0 min) and then immediately after mixing (≈15 sec) and at reaction times of 1, 2, 3, 4, and 5 min. The arrows indicate the decreasing absorbance at 235 nm and the increase at 285 nm during the reaction. (B) Rates were measured using 0.025μM enzyme by continuous recording of the decrease in absorbance at 235 nm. An observed value of 100 milliabsorbance units (mAU) per min is equivalent to 2.2 μmol of HPETE consumed per min per mg of protein. Reactions were carried out in a 0.2-cm path length microcuvette to allow measurement of the higher substrate concentrations, and all the data reported here are corrected to absorbance values at a 1-cm path length. The computed kinetic values for 12R-, 12S-, and 15S-HPETE gave Michaelis constants, Km,of46 ± 6, 28 ± 4, and 32 ± 13 μM and Vmax values of 148 ± 7, 39 ± 2, and 22 ± 3 mAU/min, respectively.
Individual HPETE isomers reacted at different rates, with 12R-HPETE being by far the best substrate tested (Fig. 1B). Initial rates for 12R-HPETE metabolism corresponded to approximately five turnovers per second. Other fatty acid hydroperoxides including 12S-HPETE, 15S-HPETE, 8R-HPETE, and the 9S-, 13R-, and 13S-hydroperoxides of linoleic acid were converted at 5- to 10-fold lower rates.
The reaction of 12R-HPETE with eLOX3 was monitored at pH 7.5 or 6. Reactions proceeded similarly, with slightly more ketodiene chromophore appearing at pH 7.5, and, significantly, with similar rates at the two pH values. The lower pH value was tested because it corresponds to the pH optimum of human 12R-LOX in the conversion of arachidonic acid to the 12R hydroperoxide (13). This may have physiological significance, given the evidence for an acidic environment in epidermis where 12R-LOX and eLOX3 are expressed (14).
RP-HPLC Analysis of eLOX3 Reaction Products. The products of eLOX3 reactions were extracted and analyzed by RP-HPLC (Fig. 2). A typical chromatogram from 12R-HPETE incubations (Fig. 2A) is dominated by a main product with retention time of ≈10 min that displayed only end absorbance in the UV (205-nm signal). A second product that eluted near 17 min had the UV spectrum of a conjugated dienone with λmax at 285 nm in the reversed-phase column solvent. Treatment of this product with NaBH4 yielded a product that cochromatographed on RP-HPLC with a 12-HETE standard, which, in accord with the UV spectrum and the mobility on RP-HPLC, points to the original product being 12-ketoeicosa-5Z,8Z,10E,14Z-tetraenoic acid. GC-MS analysis (electron impact mode) of the hydrogenated methyl ester derivative gave a mass spectrum with structurally significant ions at m/z 341 [M + 1]+, 309 [M-OCH3]+, 141 (C12-C20, [COC7H14CH3]+), 227 (C1-C12, [CH3CO2C10H20CO]+), 184 (C1-C10, [CH3CO2C8H16CH]·+), 156 (C11-C20, [CH3COC7H14CH3]+), 242 (C1-C13, [CH3CO2C10H20-COCH3]+), and 98 (C14-C20, [CHC5H10CH3]·+). The spectrum is completely consistent with a structure of methyl 12-ketoeicosanoate; the ions at m/z 141 and 227 indicate cleavage α to the 12-keto group, and the last four fragments are derived from β-cleavage. The mass spectrum showed the predicted shifts in major ion fragments compared with the reported spectrum of methyl 15-ketoeicosanoate (15).
Fig. 2.
RP-HPLC analysis of the products in eLOX3 reactions. (A)12R-HPETE plus eLOX3. (B)12S-HPETE plus eLOX3. (C)15S-HPETE plus eLOX3. The products were analyzed by RP-HPLC with a Waters Symmetry C18 5-μm column (0.46 × 25 cm) eluted at a flow rate of 1 ml/min with methanol/water/acetic acid (80:20:0.01 by volume) and UV detection at 205 nm. In A and B, the 12R-HPETE or 12S-HPETE substrates (retention time, 18 min) were converted completely and do not appear on the chromatograms. KETE, ketoeicosatetraenoic acid.
The RP-HPLC chromatogram from incubation of 12S-HPETE with eLOX3 (Fig. 2B) was more complex, with a series of peaks at retention times of 7–9 min, the major product at 12 min, the second major product at 10.5 min, and a peak of 12-ketoeicosatetraenoic acid (identified as above) near 17 min. The very small peaks at 7- to 8-min retention time had the UV spectra of conjugated trienes, whereas the peaks at 9–12 min, including the two major products, showed only end absorbance in the UV.
The chromatogram from 15S-HPETE incubation (Fig. 2C) showed a main product at ≈10 min with additional earlier peaks at 7.5–9 min, all displaying only end absorbance in the UV. A minor peak at ≈15 min was identified as 15-ketoeicosa-5Z,8Z,11Z,13E-tetraenoic acid by comparison to an authentic standard and by its conversion to 15-HETE after treatment with NaBH4. Some unreacted 15S-HPETE is also seen as the last eluting peak on the chromatogram. The corresponding HETE was not a product in any of the eLOX3 reactions analyzed.
Identification of the Main Product from 12R-HPETE. To prepare sufficient product for NMR analysis, 1.5 mg of 12R-HPETE were incubated in 25 ml of incubation buffer with 0.1 μM eLOX3. After collection of the main product from RP-HPLC, it was repurified by using straight-phase HPLC, where it also chromatographed as a single peak. GC-MS analysis (electron impact mode) gave a mass spectrum of the methyl ester trimethylsilyl ether derivative with structurally significant ions (with assignment and relative abundance in brackets) at m/z 422 (M+, 0.1%), 407 (M-15, 3%), 281 {C8-C20, [HCOSi(CH3)3 C2H2C2H2OC8H15]+, 74%}, and 243 {C1-C8, [CH3CO2 C6H10CHOSi(CH3)3]+, 14%}, the two together indicating a C-8 hydroxyl and a base peak at m/z 73 (100%) (Fig. 7, which is published as supporting information on the PNAS web site). After hydrogenation, the high-mass ions shifted by 6 mass units (m/z 413, M-15, 10% abundance; and m/z 397, M-31, 0.5%), whereas the two α-cleavage ions around C-8 appeared at m/z 285 (281 + 4) (100%), and 245 (243 + 2) (98%) (Fig. 8, which is published as supporting information on the PNAS web site). When considered together with the (lack of) UV spectral characteristics, the structure based on the GC-MS data is compatible with a C20 fatty acid methyl ester containing a C-8 hydroxyl, an epoxide moiety, and three double bonds. The spectrum of the nonhydrogenated product showed the same major cleavage ions as an uncharacterized isomer of 8-hydroxy-11,12-epoxyeicosa-5,9,14-trienoic acid (hepoxilin A3) (16). 1H-NMR (400 MHz, in deuterated benzene) defined the complete covalent structure of the product as a single diastereomer of 8-hydroxy-11R,12R-epoxyeicosa-5,9E,14-trienoic acid (Fig. 3 and Table 1, which is published as supporting information on the PNAS web site). All proton signals were assigned by H,H-COSY analysis. The coupling constant between the epoxide protons H11 and H12 (J ≈ 2 Hz) indicates the trans configuration of the 11,12-epoxide, i.e., 11R,12R-epoxy, assuming, as expected, that the original 12R configuration is retained. The 9,10 double bond is trans (J9,10 = 15.6 Hz); the 5,6 and 14,15 double bonds do not participate in the transformation from 12R-HPETE and should retain the original cis configurations.
Fig. 3.
NMR analysis of the main product of 12R-HPETE reacted with eLOX3. (A) Structure of the main product. (B) Proton spectrum with an expanded view of the geminal hydroxyl and epoxide protons (H8, H11, and H12) above. (C) H,H-COSY analysis with the main couplings indicated.
Based on published NMR data it is difficult to make an assignment of the 8-hydroxyl configuration due to chemical shifts or coupling constants (17, 18). Therefore, the stereoconfiguration of the 8-hydroxyl was determined by CD spectroscopy (9, 10). This method requires the presence of two suitable chromophores at the asymmetric carbon, one being provided by the 9,10 double bond and the other introduced at C-8 by derivatization to the benzoate ester. The through-space coupling of the two chromophores gives rise to Cotton effects in the CD spectrum that allow assignment of the absolute configuration (9). The CD spectrum showed a negative Cotton effect at 227 nm (Δε, –12.5), indicating negative chirality between the two chromophores, and therefore the absolute configuration can be assigned as 8R (Fig. 9, which is published as supporting information on the PNAS web site). Thus, the product of the eLOX3 reaction with 12R-HPETE was identified as 8R-hydroxy-11R,12R-epoxyeicosa-5Z,9E,14Z-trienoic acid (Fig. 4).
Fig. 4.
Structures of the epoxyalcohol products. The products shown are from 12R-HPETE (8R-hydroxy-11R,12R-epoxyeicosa-5Z,9E,14Z-trienoic acid), 12S-HPETE [10R-hydroxy, 11S,12S-epoxyeicosa-5Z,8Z,14Z-trienoic acid (product 1), and 8R-hydroxy-11S,12S-epoxyeicosa-5Z,9E,14Z-trienoic acid (product 2)], and 15S-HPETE (13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid). KETE, ketoeicosatetraenoic acid.
Identification of the Two Main Products from 12S-HPETE. The most prominent of the mixture of products from 12S-HPETE analyzed on RP-HPLC (Fig. 2B, ≈12-min retention time) gave a single peak when further purified on straight-phase HPLC (data not shown). GC-MS analysis of the methyl ester trimethylsilyl ether derivative gave a mass spectrum {m/z 407 [M-CH3]+, 311 [CH3CO2C8H12-CHOSi(CH3)3C2H2O]+, 282 [CH3CO2C8H12CHOSi(CH3)3 CH]+, and 269 [CH3CO2C8H12CHOSi(CH3)3]+} that was very similar to a published spectrum of the same derivative of hepoxilin B3 (10-hydroxy-11,12-epoxyeicosa-5,9,14-trienoic acid) (16). 1H NMR (400 MHz, in deuterated benzene), assigned from the H,H-COSY analysis, defined the covalent structure and provided key details of the stereochemistry (Table 2, which is published as supporting information on the PNAS web site). For the proton signals from the 10-hydroxy-11,12-epoxy moiety, it was clear that the product from the reaction of 12S-HPETE with eLOX3 has a similar visual appearance and similar coupling constants to the 10R diastereomer (19, 20). The coupling constant between H11 and H12 (≈2.2 Hz) indicates the 11,12-trans epoxide configuration, i.e., 11S,12S-epoxy in this case. Assuming that the double bonds not involved in the reaction retain their original cis configuration, we assign the structure as 10R-hydroxy, 11S,12S-epoxyeicosa-5Z,8Z,14Z-trienoic acid (Fig. 4).
GC-MS analysis of the second most prominent product (10.5-min retention time on RP-HPLC, Fig. 2B) as the hydrogenated methyl ester trimethylsilyl ether derivative gave a mass spectrum with structurally significant ions at m/z 413 [M-CH3]+, 285 {C8-C20, [HCOSi(CH3)3C2H4C2H2OC8H17]+}, 245 {C1-C8, [CH3CO2-C6H12CHOSi(CH3)3]+}, and 315 {C1-C12, [CH3CO2C6H12CHOSi(CH3)3C2H4C2H2O]+}, which is consistent with the reported mass spectrum of hydrogenated hepoxilin A3 (16). Although we did not obtain further structural data on this second most prominent product from 12S-HPETE, we can make some likely deductions. Based on its chromatographic properties, it is a diastereomer (not the enantiomer) of the major 12R-HPETE-derived epoxyalcohol; the two separate on RP-HPLC. If it is assumed that the 12S-HPETE product is a trans-11S,12S epoxide, then this would make the predicted structure 8R-hydroxy-11S,12S-epoxyeicosa-5Z,9E,14Z-trienoic acid (Fig. 4).
Identification of the Main Product from 15S-HPETE. By using the same lines of evidence from GC-MS and NMR and comparison to published data (21, 22), the main product from the reaction of 15S-HPETE with eLOX3 was identified as an analog of the main 12S-HPETE product, i.e., 13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid. The mass spectrum of the methyl ester trimethylsilyl ether derivative showed characteristic ions at m/z 407 [M-CH3]+, 309 [CH3CO2C11H22CHOSi(CH3)3]+, and 351 [CH3CO2C11H22CHOSi(CH3)3C2H2O]+ (indicating the C-13 hydroxyl). After hydrogenation, the main structurally significant ions shifted to m/z 413 [M-CH3]+, 315 [CH3CO2C11H16CHOSi(CH3)3]+, and 215 [HCOSi(CH3)3C2H2OC5H11]+. Both spectra are essentially identical to the published spectra (23). The NMR data are summarized in Table 3, which is published as supporting information on the PNAS web site. We also compared this product with the products from the hematin-catalyzed transformation of 15S-HPETE, which we previously characterized by oxidative ozonolysis, GC-MS, and 1H NMR (22). This confirmed that the main product from the eLOX3 reaction with 15S-HPETE is 13R-hydroxy-14S,15S-epoxyeicosa-5Z,8Z,11Z-trienoic acid (Fig. 4).
Reaction of eLOX3 with 18O-Labeled Hydroperoxy Substrate. [18O]15S-HPETE, with the two oxygens of the hydroperoxide labeled with 18O, was reacted with eLOX3, and the epoxyalcohol product was collected from HPLC and analyzed for 18O content by negative-ion chemical ionization GC-MS. A corresponding GC-MS analysis of 15-HETE derived from the starting material indicated that it comprised 92.4% 18O and 7.6% unlabeled molecules; because nearly all the molecules contain either two 16O or two 18O, the 18O-labeled 15-HPETE, with two heavy oxygens, is calculated to have a 92% content of two atoms of 18O. GC-MS analysis of the epoxyalcohol demonstrated that 91.6% of the product molecules contained two atoms of 18O (2% contained one 18O, and 6.4% of the molecules were unlabeled). These results indicate that there was essentially complete retention of the original hydroperoxide oxygens in the epoxyalcohol product. eLOX3, therefore, is a hydroperoxide isomerase. Although other 18O-labeled hydroperoxide substrates were not available, we were able to show by using an oxygen electrode that exogenous molecular oxygen made no significant contribution to the eLOX3-catalyzed transformations of the hydroperoxide substrates.
Effect of Nordihydroguaiaretic Acid (NDGA) on eLOX3 Reaction. As expanded on in Discussion, from our understanding of the catalytic cycle of the eLOX3 reaction with HPETEs we predicted that an agent capable of reducing the eLOX3 active-site iron would accelerate the rate of reaction. As reducing agent we used NDGA. NDGA frequently is used as a nonspecific inhibitor for typical LOXs, although it can also be used as a cosubstrate to support the pseudoperoxidase activity of LOXs (24, 25). Submicromolar concentrations of NDGA caused a dose-dependent increase in reaction rates of eLOX3 with HPETE substrates. The addition of NDGA increased the rate of eLOX3 reaction with 12R-HPETE by 3- to 4-fold (Fig. 5) and with 15S-HPETE by up to 6-fold.
Fig. 5.
Effect of NDGA on eLOX3 rate of reaction. The dose-dependent effects of NDGA are shown for the reaction of 12R-HPETE with eLOX3. Rates were determined by following the decrease in absorbance at 235 nm.
Discussion
Genetic evidence links mutations in the coding sequence of the human eLOX3 gene to the development of an inherited skin disease, NCIE (6). NCIE represents the first example in which improper LOX expression has been shown to have direct pathophysiological consequences. The disease is characterized by hyperkeratosis and epidermal dysfunction, leading to a white, flaky skin with transepidermal water loss. A second group of families with NCIE showed somatic mutations in the gene of a related LOX, 12R-LOX. The finding that mutations in either 12R-LOX or eLOX3 resulted in the same disease phenotype led the authors to speculate that both enzymes might participate in the same metabolic pathway, and that this pathway would have a crucial role in the normal functioning of human skin (6). From a biochemical point of view, at first glance this hypothesis seemed surprising. eLOX3 and 12R-LOX are both LOX enzymes theoretically catalyzing the same type of reaction on polyunsaturated fatty acid substrates. Furthermore, recombinant eLOX3 has been shown to be lacking any demonstrable catalytic activity at all (4).
Here we provide biochemical evidence that demonstrates a specific catalytic activity for eLOX3 and potentially provides a functional link with 12R-LOX. eLOX3 exhibits potent enzymatic activity toward the transformation of the 12R-LOX-derived product, 12R-HPETE, into a specific epoxy alcohol product, 8R-hydroxy-11R,12R-epoxyeicosa-5Z,9E,14Z-trienoic acid. We also have shown that the LOX products 12S-HPETE and 15S-HPETE are converted to specific epoxyalcohol products of related structure, albeit with lower catalytic efficiency. The very fact of enzymatic conversion and especially the preference of eLOX3 for 12R-HPETE support the hypothesis suggested by Jobard et al. (6) on the existence of a specific pathway involving the two LOXs. An additional supporting finding is that eLOX3 and 12R-LOX have similar tissue expression patterns in humans, making the conversion of 12R-HPETE into the specific epoxyalcohol a reaction likely to occur in vivo (5). The platelet 12-LOX and the two 15-LOXs are also expressed in human skin, potentially providing 12S-HPETE and 15S-HPETE as alternative substrates.
NCIE is associated with three different mutations in the ORF of the eLOX3 gene (6). One of these, a truncation mutant, is certain to eliminate the catalytic activity of the eLOX3 protein. In preliminary experiments we prepared the other two point mutations and attempted to express the cDNAs in our usual E. coli system. In contrast to the wild-type enzyme, no protein was recovered at the nickel affinity column step, suggesting to us that the mutated proteins are unstable and fail to accumulate in the bacteria.
The term “hepoxilin” is generally used to refer to groups of 12-HPETE-derived epoxyalcohol fatty acids that have been detected in skin and other tissues (26). Hepoxilin A3 is used for any stereoisomer with an 8-hydroxy-11,12-epoxy structure, whereas hepoxilin B3 refers to the isomers with a 10-hydroxy-11,12-epoxyeicosatrienoic acid structure. Thus far, an enzyme with hepoxilin synthase activity in human skin has not been characterized, but there are reports about the detection of this activity in rat and human skin and appearance of the products in human psoriatic lesions (27–29). The recombinant human eLOX3 forms one particular stereoisomer of hepoxilin A3 from 12R-HPETE and mainly one isomer of hepoxilin B3 from the substrate 12S-HPETE. It remains to be elucidated the extent to which the eLOX3 activity is responsible for the formation of hepoxilin isomers that have been detected in human skin or whether additional activities or nonenzymatic reactions have to be considered.
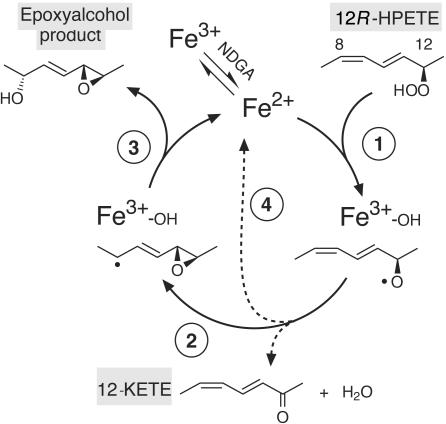
In terms of mechanism, the facets of eLOX3 reactivity that make it stand out are the complete absence of typical LOX (oxygenase) activity under any of the variety of conditions that have been explored and also the unusual autocatalytic nature of the reaction with HPETE substrates. Equivalent hydroperoxide rearrangements by other LOXs are limited to single turnovers unless promoted by a reducing cofactor (25, 30). In the conventional Fe2+/Fe3+ redox cycle of normal LOX catalysis, the Fe3+ enzyme is the active form that performs the hydrogen abstraction from the bis-allylic methylene of the fatty acid substrate. Oxygenation of the fatty acid radical and reduction of the peroxyl radical to the fatty acid hydroperoxide completes the cycle. By contrast, in the reaction of eLOX3 with HPETE substrates it can be assumed that the Fe2+ enzyme is the active species. Based on this, we propose a mechanism for epoxyalcohol formation by eLOX3 as depicted in Fig. 6. The Fe2+ enzyme initiates a homolytic cleavage of the hydroperoxide O–O bond (Fig. 6, step 1), the resulting alkoxyl radical cyclizes to an epoxyallylic carbon radical (step 2) while the other oxygen of the original hydroperoxide is retained in a Fe3+–OH complex. The cycle is completed by an oxygen-rebound type of reaction that forms the epoxyalcohol product (step 3) while the iron is restored to the active Fe2+ form. The corresponding ketoeicosatetraenoic acid is formed as a by-product in some of the catalytic cycles (step 4). Other potential rearrangement products such as aldehydes resulting from chain cleavage are not formed. The reducing factor NDGA speeds up catalysis by converting the pool of enzyme molecules from predominantly ferric (Fe3+) to the active ferrous state.
Fig. 6.
Proposed mechanism for eLOX3 catalysis.
Because the reaction mechanism in Fig. 6 could be proposed “on paper” for any LOX, why is such a self-sufficient catalytic cycling observed only with eLOX3? We speculate that the redox state of eLOX3 may make the ferric enzyme incapable of performing a hydrogen abstraction from a typical LOX substrate, and thus the protein is incapable of oxygenating a polyunsaturated fatty acid. This shift in balance of redox potential, in turn, may favor the reduction reaction that constitutes the basis of the catalytic activity we describe with HPETE substrates.
Although eLOX3 is clearly a member of the LOX gene family, it does not have the LOX activity predicted from proteomic analysis. In an unprecedented reaction for LOX enzymes, eLOX3 converts the product of a typical epidermal LOX, 12R-LOX, into a specific epoxyalcohol isomer. Taking our findings together with the genetic evidence from NCIE, we can surmise that 12R-LOX and eLOX3 participate in the formation of a product with a role in keratinocyte differentiation. The hepoxilin isomer formed by eLOX3 from 12R-HPETE has not been characterized previously, and its biological activity remains to be assessed. It may act as a signaling molecule or be transformed to an active product. The bioactive principal of one of the endothelial-derived relaxing factors, for example, is a trihydroxy eicosanoid of the type formed by hydrolysis of the eLOX3-derived hepoxilin (31). Because an increase in intracellular calcium is one of the most important factors in inducing keratinocyte differentiation (32), and such an activity has been reported for other hepoxilin isomers (33), this provides a potential basis for the participation of eLOX3 and 12R-LOX in skin development.
Supplementary Material
Acknowledgments
We thank Dr. David Hachey and Ms. Betty Fox for help with the GC-MS analyses. This work was supported by National Institutes of Health Grants AR-45943 (to A.R.B.) and CA-89450 (to L.J.M.) and Core Laboratories of the Vanderbilt Skin Disease Research Center Grant AR-41943.
Abbreviations: LOX, lipoxygenase; H(P)ETE, hydro(pero)xyeicosatetraenoic acid; eLOX3, epidermal LOX type 3; NCIE, nonbullous congenital ichthyosiform erythroderma; NDGA, nordihydroguaiaretic acid.
References
- 1.Brash, A. R. (1999) J. Biol. Chem. 274, 23679–23682. [DOI] [PubMed] [Google Scholar]
- 2.Pace-Asciak, C. R. & Asotra, S. (1989) Free Radical Biol. Med. 7, 409–433. [DOI] [PubMed] [Google Scholar]
- 3.Funk, C. D. (2001) Science 294, 1871–1875. [DOI] [PubMed] [Google Scholar]
- 4.Kinzig, A., Heidt, M., Fürstenberger, G., Marks, F. & Krieg, P. (1999) Genomics 58, 158–164. [DOI] [PubMed] [Google Scholar]
- 5.Krieg, P., Marks, F. & Fürstenberger, G. (2001) Genomics 73, 323–300. [DOI] [PubMed] [Google Scholar]
- 6.Jobard, F., Lefèvre, C., Karaduman, A., Blanchet-Bardon, C., Emre, S., Weissenbach, J., Özgüc, M., Lathrop, M., Prud'homme, J. F. & Fischer, J. (2002) Hum. Mol. Genet. 11, 107–113. [DOI] [PubMed] [Google Scholar]
- 7.Brash, A. R. & Song, W.-C. (1996) Methods Enzymol. 272, 250–259. [DOI] [PubMed] [Google Scholar]
- 8.Peers, K. F. & Coxon, D. T. (1983) Chem. Phys. Lipids 32, 49–56. [Google Scholar]
- 9.Schneider, C., Schreier, P. & Humpf, H.-U. (1997) Chirality 9, 563–567. [Google Scholar]
- 10.Schneider, C., Tallman, K. A., Porter, N. A. & Brash, A. R. (2001) J. Biol. Chem. 276, 20831–20838. [DOI] [PubMed] [Google Scholar]
- 11.Humpf, H.-U., Berova, N., Nakanishi, K., Jarstfer, M. B. & Poulter, C. D. (1995) J. Org. Chem. 60, 3539–3542. [Google Scholar]
- 12.Gonnella, N. C., Nakanishi, K., Martin, V. S. & Sharpless, B. K. (1982) J. Am. Chem. Soc. 104, 3775–3776. [Google Scholar]
- 13.Schneider, C., Keeney, D. S., Boeglin, W. E. & Brash, A. R. (2001) Arch. Biochem. Biophys. 386, 268–274. [DOI] [PubMed] [Google Scholar]
- 14.Rippke, F., Schreiner, V. & Schwanitz, H. J. (2002) Am. J. Clin. Dermatol. 3, 261–272. [DOI] [PubMed] [Google Scholar]
- 15.Hamberg, M. & Samuelsson, B. (1967) J. Biol. Chem. 242, 5344–5354. [PubMed] [Google Scholar]
- 16.Pace-Asciak, C. R., Granstrom, E. & Samuelsson, B. (1983) J. Biol. Chem. 258, 6835–6840. [PubMed] [Google Scholar]
- 17.Corey, E. J. & Su, W.-G. (1984) Tetrahedron Lett., 5119–5122.
- 18.Lumin, S., Falck, J. R., Capdevila, J. H. & Karara, A. (1992) Tetrahedron Lett., 2091–2094.
- 19.Vasiljeva, L. L., Manukina, T. A., Demin, P. M., Lapitskaja, M. A. & Pivnitsky, K. K. (1993) Tetrahedron 49, 4099–4106. [Google Scholar]
- 20.Bernart, M. W. & Gerwick, W. H. (1994) Phytochemistry 36, 1233–1240. [Google Scholar]
- 21.Corey, E. J. & Mehrotra, M. M. (1983) Tetrahedron Lett., 4921–4922.
- 22.Chang, M. S., Boeglin, W. E., Guengerich, F. P. & Brash, A. R. (1996) Biochemistry 35, 464–471. [DOI] [PubMed] [Google Scholar]
- 23.Narumiya, S., Salmon, J. A., Cottee, F. H., Weatherley, B. C. & Flower, R. J. (1981) J. Biol. Chem. 256, 9583–9592. [PubMed] [Google Scholar]
- 24.Kemal, C., Louis-Flamberg, P., Krupinski-Olsen, R. & Shorter, A. L. (1987) Biochemistry 26, 7064–7072. [DOI] [PubMed] [Google Scholar]
- 25.Riendeau, D., Falgueyret, J. P., Guay, J., Ueda, N. & Yamamoto, S. (1991) Biochem. J. 274, 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pace-Asciak, C. R., Reynaud, D. & Demin, P. M. (1995) Lipids 30, 107–114. [DOI] [PubMed] [Google Scholar]
- 27.Nugteren, D. H., Christ-Hazelhof, E., van der Beek, A. & Houtsmuller, U. M. (1985) Biochim. Biophys. Acta 834, 429–436. [DOI] [PubMed] [Google Scholar]
- 28.Anton, R., Puig, L., Esgleyes, T., de Moragas, J. M. & Vila, L. (1998) J. Invest. Dermatol. 110, 303–310. [DOI] [PubMed] [Google Scholar]
- 29.Antón, R. & Vila, L. (2000) J. Invest. Dermatol. 114, 554–559. [DOI] [PubMed] [Google Scholar]
- 30.Garssen, G. J., Veldink, G. A., Vliegenthart, J. F. G. & Boldingh, J. (1976) Eur. J. Biochem. 62, 33–36. [DOI] [PubMed] [Google Scholar]
- 31.Pfister, S. L., Spitzbarth, N., Nithipatikom, K., Edgemond, W. S., Falck, J. R. & Campbell, W. B. (1998) J. Biol. Chem. 273, 30879–30887. [DOI] [PubMed] [Google Scholar]
- 32.Yuspa, S. H., Hennings, H., Tucker, R. W., Jaken, S., Kilkenny, A. E. & Roop, D. R. (1988) Ann. N.Y. Acad. Sci. 548, 191–196. [DOI] [PubMed] [Google Scholar]
- 33.Reynaud, D., Demin, P. M., Sutherland, M., Nigam, S. & Pace-Asciak, C. R. (1999) FEBS Lett. 446, 236–238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.