Abstract
Heterodimeric αβ soluble guanylyl cyclase (sGC) is a recognized receptor for nitric oxide (NO) and mediates many of its physiological functions. Although it has been clear that the heme moiety coordinated by His-105 of the β subunit is crucial for mediating the activation of the enzyme by NO, it is not understood whether the heme moiety plays any role in the function of the enzyme in the absence of NO. Here we analyze the effects of biochemical and genetic removal of heme and its reconstitution on the activity of the enzyme. Detergent-induced loss of heme from the wild-type αβ enzyme resulted in several-fold activation of the enzyme. This activation was inhibited after hemin reconstitution. A heme-deficient mutant αβCys-105 with Cys substituted for His-105 was constitutively active with specific activity approaching the activity of the wild-type enzyme activated by NO. However, reconstitution of mutant enzyme with heme and/or DTT treatment significantly inhibited the enzyme. Mutant enzyme reconstituted with ferrous heme was activated by NO and CO alone and showed additive effects between gaseous effectors and the allosteric activator 5-cyclopropyl-2-[1-(2-fluoro-benzyl)-1H-pyrazolo[3,4-b]pyridin-3-yl]-pyrimidin-4-ylamine. We propose that the heme moiety through its coordination with His-105 of the β subunit acts as an endogenous inhibitor of sGC. Disruption of the heme-coordinating bond induced by binding of NO releases the restrictions imposed by this bond and allows the formation of an optimally organized catalytic center in the heterodimer.
Soluble guanylyl cyclase (sGC), rhizobial O2-sensing FixL protein, and CooA transcription factor from Rhodospirillum rubrum constitute a unique family of hemoproteins that stand apart from the rest of the heme-containing proteins (1). In contrast to various hemoproteins, which use the heme moiety as a catalyst in redox reactions (e.g., cytochrome P450 and NO synthases) or as an oxygen storage-transporting moiety (hemoglobin and myoglobin), the heme group of these enzymes seems to have a unique role of being receptors specific to various physiologically relevant gaseous activators such as NO for sGC (2), O2 for FixL (3), and CO for CooA transcription factor (4). Binding of gaseous effectors induces conformational changes in the protein, resulting in the transition to an activated state.
sGC [GTP pyrophosphate-lyase (cycling), EC 4.6.1.2] is a key enzyme in the NO/cGMP-dependent pathway, which plays an important role in mammalian physiology. sGC-mediated increases in intracellular cGMP affect smooth muscle relaxation, platelet aggregation, leukocyte adhesion, cell proliferation and migration, neurotransmission, and other effects (5, 6). Mammalian sGC is a heterodimeric protein composed of one α subunit and one β subunit (7). Two isoforms for each subunit (α1, α2, β1, and β2) have been described, but only ubiquitously expressed heterodimer α1β1 and placenta-specific heterodimer α2β1 have been identified thus far in human tissues. Both subunits are required for a fully active sGC (8), although a recent report suggests that the β2 homodimer possesses some residual catalytic activity (9) albeit only with non-physiological concentrations of Mn2+ ion.
NO binds to a heme prosthetic group found in the native enzyme. sGC heterodimer contains only one ferrous pentacoordinated heme with the His-105 residue of the β subunit acting as the axial ligand (10, 11). The binding of NO to the heme results in stimulation of sGC activity (12), which increases the activity of purified enzyme several hundred-fold (13–15).
Various analyses of UV-visible (UV-Vis) (10, 14), EPR (16), and Raman (17, 18) spectra described to a great extent the transformation of the sGC heme prosthetic group after binding of NO. Binding of NO to heme Fe2+ and the formation of nitrosyl heme result in the disruption of the axial coordinating bond and displacement of iron from the protoporphyrin plane. The kinetic and mechanistic aspects of these transformations are currently subjects of investigation and debate (19–22).
Although the connection between the formation of the nitrosyl heme and activation of cGMP synthesis by sGC is well documented and accepted, little is known about the mechanism coupling these two events. Current views of sGC structure define a heme-containing regulatory domain, formed by N-terminal portions of both subunits, and a catalytic domain formed by the C-terminal half of both subunits (23). It is postulated that NO binding induces changes in the regulatory domain (24, 25) that are transmitted through a yet-unknown mechanism to the catalytic site, which results in a significant increase in Vmax of the enzyme and some decrease in the Km for GTP substrate (15, 26, 27).
Although the key role of heme in NO activation is undisputed, there is no clear understanding of the role of the heme group in basal inactivated enzyme. In this report we investigate the role of the heme moiety in the basal state of recombinantly expressed human sGC. We present biochemical data from the analysis of the wild-type enzyme and a constitutively active heme-deficient mutant suggesting that the heme moiety and the heme-coordinating bond have an inhibitory function. On the basis of our data we propose that the heme moiety keeps the regulatory domain in a “restricted” basal conformation. This restricted basal conformation could be released not only after binding the NO ligand to heme but also, to various degrees, by the chemical or genetic removal of the heme moiety from the regulatory domain, which allows the enzyme to achieve the “activated” conformation.
Materials and Methods
Reagents. Hemin, Grace medium, FBS, and imidazole were from Sigma. 3-(2-Hydroxyl-1-methyl-2-nitrosohydrazino)-N-methyl-1-propanamine was from Calbiochem. The 5-cyclopropyl-2-[1-(2-f luoro-benzyl)-1H-pyrazolo[3,4-b]pyridin-3-yl]-pyrimidin-4-ylamine (BAY41-2272) activator was a generous gift from J. P. Stasch (Bayer, Wuppertal, Germany). [α-32P]GTP was from NEN.
cDNAs and Expression Vectors. The design and generation of baculoviruses expressing human sGC α subunit with a His tag, wild-type β subunits, or mutant βCys-105 subunit has been described (15, 28). To express mutant βCys-105 subunit in mammalian cells, the coding region of βCys-105 variant was cloned into EcoRI and NotI sites of the pCDNA3.1(+) vector (Invitrogen).
sGC Expression and Purification. Human wild-type and αβCys-105 mutant sGC were expressed in Sf9 cells as described (28). Sf9 cells were lysed by sonication in buffer A [25 mM triethanolamine (TEA), pH 7.5/10% glycerol/4 mM MgCl2/5 mM DTT/1 mM PMSF/5 mg/ml each of pepstatin A, leupeptin, aprotinin, and chymostatin], and the 100,000 g of supernatant fraction was loaded on a 20-ml DEAE-Sepharose column. Lysate with mutant enzyme was prepared in the absence of DTT. After extensive washes, the enzyme was eluted with a linear 0–100% gradient of buffer B (25 mM TEA, pH 7.5/10% glycerol/500 mM NaCl). Yellow fractions containing sGC were purified further by nickel chromatography as described (15, 28). To remove the imidazole present in sGC fractions collected after nickel chromatography, the sGC sample was loaded on a 2-ml Hi-Trap DEAE-Sepharose column (Amersham Pharmacia Biotech), washed with buffer A, and eluted with buffer A with 250 mM NaCl. The enzyme obtained at this stage (≥95% purity) was used for experiments.
Assay of sGC Activity. Enzyme activity was assayed by formation of [32P]cGMP from [α-32P]GTP at 37°C as described (29). The concentration of DMSO used as a vehicle for BAY41-2272 was not >0.1% and had no effect on sGC activity. Activity of CO-treated enzyme was measured in gas-tight vials by using CO-equilibrated reaction buffer.
UV-Vis Spectroscopy. All measurements were recorded with a dual-beam Cecil 9500 spectrophotometer at 25°C. To monitor the heme reconstitution, wild-type and mutant enzymes in 25 mM TEA, pH 7.5/10% glycerol/250 mM NaCl/4 mM MgCl2 were used. A 5 mM stock solution of hemin prepared in alkaline 50% ethanol was used to make a 350 μM working solution in buffer A without DTT and protease inhibitors. Identical amounts of hemin (0.5–10 μM final concentration) were added to both sample and reference cuvettes, and difference spectra were recorded between 320 and 700 nm.
To obtain heme-reconstituted αβCys-105 enzyme, 1 ml of 4 μM mutant enzyme was supplied with 8 μM hemin, and unbound hemin was removed by filtration through a 5-ml Hi-Trap desalting column (Amersham Biosciences). To reduce the ferric heme moiety, several grains of dithionate or 5 mM DTT were added to reconstituted enzyme. To measure the effects of NO, 50 μM 3-(2-hydroxyl-1-methyl-2-nitrosohydrazino)-N-methyl-1-propanamine was added to both sample and reference cuvettes, and spectra were recorded 15 min later. The reduced sample was also treated with CO for 2 min in a gas-tight vial and transferred anaerobically to a sealed cuvette flashed with CO, and spectra were recorded as described above.
Assay of cGMP Accumulation in Intact Sf9 Cells. Forty-eight hours postinfection, Sf9 cells expressing either wild-type or αβCys-105 mutant sGC were washed twice with Dulbecco's PBS and preincubated for 10 min in PBS with 0.5 mM phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine in a 50-μl final volume of 107 cells per ml. After this, 250 μM spermine NONOate or vehicle was added, and the cells were incubated for an additional 5 min at 37°C. The reaction was terminated by the addition of 50 μl of 1 M perchloric acid, and cGMP was extracted on ice for 1 h. The extract was centrifuged, neutralized with 2 M K2CO3, and used for cGMP determination by an RIA (30). The pellet was dissolved in 0.1 M NaOH and used for a protein assay by the method of Lowry et al. (31).
Generation of Neuroblastoma Cells Stably Transfected with βCys-105. Neuroblastoma cell line BE2 (American Type Culture Collection) was cultured in DMEM/F12K medium (GIBCO) supplemented with 10% FBS (Sigma)/1% penicillin-streptomycin mixture/1mM glutamine/1 mM nonessential amino acids/1 mM sodium pyruvate. pcDNA-βCys-105 plasmid was transfected by using Lipofectamine reagent (Invitrogen) according to manufacturer protocol. Forty-eight hours posttransfection, 800 μg/ml G418 antibiotic (GIBCO) was added to the culture medium. Two weeks later, individual NeoR colonies were collected by using a 0.5-cm-diameter cloning ring (Corning) and expanded in 100-mm tissue-culture dishes. βCys-105-specific 5′ primer (5′-CAGAACCTTGATGCTCTGTGC-3′) and pcDNA-specific 3′ primer (5′-CTGATCAGCGGGTTTAAACGG-3′) were used to perform the RT-PCR on mRNA purified from 20 individual NeoR colonies. Three clones with the highest levels of βCys-105 expression were used for further studies.
Results
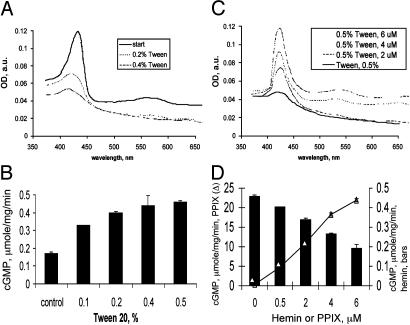
Changes in the Heme Content Affect the Basal Activity of the Wild-Type Enzyme. To test the role of the heme moiety in regulating the basal function of sGC, we investigated the effect of heme removal on the activity of the enzyme. Treatment of the wild-type sGC with increasing concentrations of Tween 20 resulted in the decreased heme content of the enzyme (Fig. 1), which correlated with an increase in the basal activity (Fig. 1). Reconstitution of the partially heme-depleted sGC with exogenously added hemin not only resulted in the increase of the Soret peak but also decreased the activity of the enzyme (Fig. 1). These data suggest that the heme moiety has some inhibitory function in the basal enzyme.
Fig. 1.
Removal and reconstitution of heme regulates the basal activity of wild-type sGC. (A) Representative spectra of 3 μM sGC in 50 mM TEA, pH 7.4/200 mM NaCl treated with 0.1–0.5% Tween 20. a.u., arbitrary units. (B) After each Tween 20 treatment, basal activity was determined. Enzyme (3 μM) in 50 mM TEA, pH 7.4/200 mM NaCl treated with 0.5% Tween 20 was reconstituted with 0.5–6 μM hemin. (C) Difference spectra of obtained enzyme. (D) Basal activity of hemin-reconstituted enzyme (filled bars) was compared with the activity of PPIX-reconstituted sGC (line, open triangles). Data are representative of three independent experiments, with similar results performed in triplicate shown. Values are shown as means ± SD.
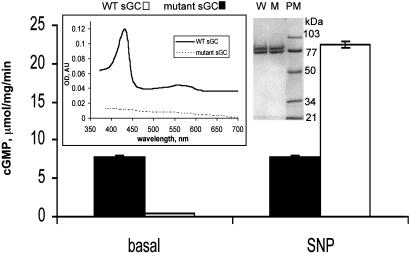
Heme-Deficient αβCys-105 Mutant sGC Has a High Constitutive Basal Activity. To test this hypothesis we analyzed the properties of the mutant sGC enzyme carrying a substitution of the His-105 residue of β subunit by a Cys. Mutant enzyme purified to homogeneity (Fig. 2 Right Inset) was heme-deficient (Fig. 2 Left Inset) but in the absence of any thiols had a remarkably high basal activity of 7.8 ± 0.2 μmol/mg per min, which is ≈70 times higher than the basal activity of the heme-competent wild-type enzyme (Fig. 2). This basal activity of the mutant was not affected by NO but was somewhat less than 22.1 ± 0.12 μmol/mg per min of activity of NO-stimulated wild-type enzyme (Fig. 2).
Fig. 2.
Basal activity of the mutantαβCys-105 enzyme is comparable to the activity of NO-treated sGC. Specific activity of the purified mutant (filled bars) and wild-type (open bars) enzymes in the absence (basal) or presence of 100 μM SNP was determined. Data are shown as means ± SD of three independent experiments performed in triplicate. (Left Inset) UV-Vis spectra of 3 μM purified wild-type and mutant sGC. AU, arbitrary units. (Right) Coomassie blue staining of 5 μg of the wild-type (W) and mutant (M) sGC preparation. The size (in kilodaltons) of the molecular mass markers (PM) is indicated.
In Vivo Activity of the αβCys-105 sGC. To test whether constitutively high basal activity of the mutant enzyme is preserved in vivo, we compared the cGMP accumulation in intact Sf9 cells expressing the αβCys-105 mutant or the wild-type enzymes (Table 1). As expected, addition of the NO donor spermine NONOate to Sf9 cells expressing wild-type enzyme increased the rate of cGMP accumulation (0.4 ± 0.02 vs. 4.9 ± 0.13 nmol/mg per 5 min). However, the rate of cGMP accumulation in cells expressing the αβCys-105 mutant both before and after spermine NONOate treatment was significantly higher (35 nmol/mg per min) than the rates observed for cells with wild-type enzyme (Table 1).
Table 1. Activity of mutant sGC in intact Sf9 cells or neuroblastoma cell lysates.
|
Sf9 with mutant*
|
Sf9 with wild type*
|
BE2/pCDNA3.1 βCys-105
|
BE2/pCDNA3.1 vector
|
|||||
|---|---|---|---|---|---|---|---|---|
| Basal | NO† | Basal | NO† | Basal | NO† | Basal | NO† | |
| cGMP levels, nmol/mg of protein | 35.0 ± 1.59 | 35.8 ± 1.42 | 0.4 ± 0.02 | 4.9 ± 0.13 | ||||
| sGC activity, pmol/min per mg of protein | 60.0 ± 12.1 | 150.2 ± 8.2 | <5‡ | 300.4 ± 15.9 | ||||
Values are means ± SD of three measurements.
cGMP levels 48 h postinfection for αβCys-105 and 72 h postinfection for αβ sGC.
250 μM NO donor spermine NONOate was used for intact Sf9 cells and 100 μM SNP was used for BE2 lysates.
The activity is at or lower than the detection level for this method.
The human neuroblastoma cell line BE2(C) was reported previously to increase intracellular levels of cGMP after exposure to NO. Our tests indicated that the lysate of these cells posses a specific activity of >50 pmol/mg per min (data not shown), which is sufficient to detect sGC activity by measuring directly the conversion of [32P]GTP to [32P]cGMP. This cell line was used as host to stably transfect the control pcDNA3.1 vector and the plasmid constitutively expressing βCys-105 variant of the β sGC subunit. Although basal levels of sGC activity in both BE2 and BE2-pCDNA3.1 cells are at the limit of detection (5 pmol/min per mg of cGMP), basal activity of the BE2-βCys-105 cell lysate was significantly higher (60 pmol/min per mg cGMP) and comparable with sodium nitroprusside (SNP)-induced activity in control lysates (300 pmol/min per mg) (Table 1). Moreover, SNP-induced activity was diminished in BE2-βCys-105 cell lysate to 150 pmol/min per mg of cGMP, suggesting that the mutant subunit effectively competes with the wild-type β subunit for the formation of sGC heterodimer. These measurements indicate that the mutant αβCys-105 sGC in vivo also displays a constitutively high specific activity in the absence of sGC activators.
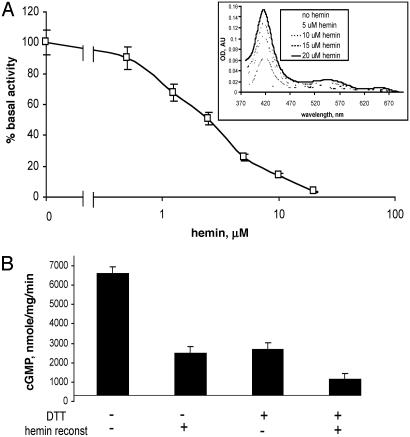
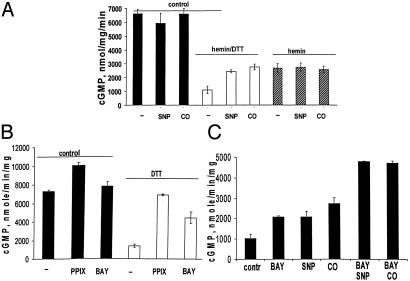
High Basal Activity of the Heme-Deficient Mutant Is Inhibited by DTT and Hemin. As shown in Fig. 3A and reported previously (28), the αβCys-105 mutant is heme-deficient. The addition of hemin to the mutant enzyme decreases the activity of the mutant in a concentration-dependent manner (Fig. 3A). Conversely, the addition of protoporphyrin IX (PPIX) slightly increased the activity to 10 ± 0.3 μmol/mg per min. The addition of 1 mM DTT to the mutant enzyme significantly inhibited the basal activity (Fig. 3B). In fact, storage of the mutant enzyme in 5 mM DTT overnight results in an enzyme with a basal activity only 2- to 3-fold higher than the basal activity of the wild-type sGC (28). DTT and hemin inhibition had an additive effect (Fig. 3B).
Fig. 3.
Hemin-dependent inhibition of the mutant enzyme. (A) αβCys-105 (4 μM) was supplied with 0.5–20 μM hemin, and specific activity at each hemin concentration was determined and normalized to basal 7.6 ± 0.3 μmol/min per mg activity of the mutant specified as 100%. (Inset) Difference spectra of reconstituted enzyme recorded in 50 mM TEA, pH 7.4/200 mM NaCl. AU, arbitrary units. (B) Specific activity of the purified mutant was determined in the reaction buffers containing 1 mM DTT, 2μM hemin, or both. Data are shown as means ± SD of four independent experiments performed in triplicate.
To test whether the mutation affected the properties of the catalytic center of sGC, the Km for Mg2+-GTP substrate was measured. We found that the GTP Km for the αβCys-105 mutant was ≈150 ± 5 μM in the presence and 145 ± 9 in the absence of DTT. These values are well within the 65–450 μM range of GTP Km measured for various recombinant sGC (15, 26, 32, 33) and are higher than the ≈65 and ≈50 μM GTP Km values determined for the human basal and NO-treated sGC, respectively (15). This suggests that the high activity of the αβCys-105 mutant is not due to changes in catalysis but rather to changes in regulation of sGC activity.
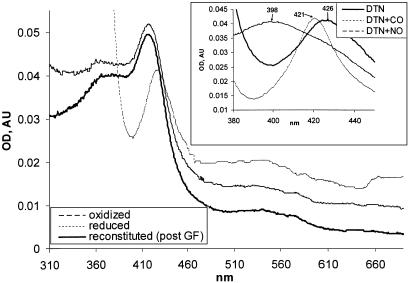
Heme-Reconstituted αβCys-105 Mutant Regains Ability to Bind and Be Activated by Gaseous Ligands. Heme-dependent inhibition and partial activation by PPIX of αβCys-105 enzyme suggested that the mutant sGC can be reconstituted with heme. The results of the reconstitution experiment are presented in Fig. 4A. Recorded difference spectra indicate that increasing concentrations of heme result in the increased Soret peak. When the reconstituted enzyme was subjected to gel filtration, only a portion of the enzyme (≈60%) retained the heme moiety as judged by change in the OD280/OD417 ratio (data not shown). Unlike the Soret peak of the wild-type ferrous sGC (maximum at 431 nm) or oxidized ferric sGC (maximum at 393 nm), the maximum was observed at 417 nm (Fig. 4B and Table 2).
Fig. 4.
Heme reconstitution of αβCys-105 enzyme. (A) αβCys-105 (4 μM) enzyme was supplied with 10 μM hemin and, after 15 min at 24°C, passed through a Hi-Trap desalting column (Amersham Biosciences). The UV-Vis spectra of reconstituted enzyme (solid line) were recorded in 50 mM TEA, pH 7.4, with 200 mM NaCl/10% glycerol. The spectra of reconstituted αβCys-105 oxidized with 5 μM K3[Fe(CN)6] (interrupted line) or reduced by several grains of dithionate (dotted line) are shown. (Inset) Changes in the Soret region of dithionate-reduced hemin-reconstituted αβCys-105 mutant (DTN, solid line) after 15 min of treatment with 50μM 3-(2-hydroxyl-1-methyl-2-nitrosohydrazino)-N-methyl-1-propanamine (interrupted line) or after exposure to CO (dotted line). post GF, post gel filtration; AU, arbitrary units.
Table 2. Absorption spectra of various forms of native and altered hemoproteins.
| n.u.v. | Soret | β | α | Ref. | |
|---|---|---|---|---|---|
| Wild-type sGC | |||||
| Ferric | 393 | 14, 27 | |||
| Ferrous (native) | 431 | 560-590 | 560-590 | 27 | |
| Ferrous + CO | 423 | 541 | 567 | 14 | |
| Ferrous + NO | 399 | 537 | 572 | 14, 27 | |
| Reconstituted αβCys-105 | sGC | ||||
| Ferric | 365 | 417 | 542 | 573 | This study |
| Ferrous | ND | 426 | 540 | This study | |
| Ferrous + CO | ND | 421 | 536 | 567 | This study |
| Ferrous + NO | 399 | 567 | 611 | This study | |
| Cytochrome P450 | |||||
| Ferric | 393 | 645 | 39, 40 | ||
| Ferrous | 411 | 542 | 39, 40 | ||
| Ferrous + CO | 366 | 447 | 39, 40 | ||
| Cytochrome P420 | |||||
| Ferric | 366 | 418 | 540 | 570 | 39, 40 |
| Ferrous | 325 | 428 | 535 | 562 | 39, 40 |
| Ferrous + CO | 341 | 423 | 541 | 570 | 39, 40 |
Values for the αβCys-105 mutant are reported. n.u.v., near-UV band; ND, not detected because of the overlapping dithionate absorption but absent when DTT was used (data not shown).
The addition of potassium ferricyanide did not change the spectra and dithionite shifted the Soret peak to 426 nm (Fig. 4), suggesting that the mutant is reconstituted with a ferric heme; 5 mM DTT has effects similar to dithionate (data not shown).
The αβCys-105 enzyme with reduced heme was able to bind CO. After CO treatment, the 426-nm Soret peak shifted to 421 nm (Fig. 4 and Table 2), whereas the addition of NO donor DEA-NO resulted in the shift of the spectra toward 398 nm (Fig. 4 and Table 2).
Spectral studies of reconstituted mutant correlated well with changes in its catalytic properties. Only the enzyme with reduced heme by at least 1 mM DTT was activated by NO and CO, whereas αβCys-105 mutant with ferric heme was not affected (Fig. 5A). Interestingly, mutant reconstituted with reduced heme was activated to a similar extent by NO or CO (Fig. 5 A and C), which is different than the wild-type enzyme (10). Heme reconstitution did not increase the attenuated response of the αβCys-105 mutant to BAY41-2272 (Fig. 5B) but restored the additive effect of the allosteric regulator and NO (Fig. 5C).
Fig. 5.
Reconstitution of ferrous heme restores sensitivity of the mutant enzyme to NO and CO after DTT treatment. (A) Mutant enzyme treated with 1 mM DTT, 2 μM hemin, or both was assayed in the presence of SNP or under anaerobic conditions with CO-saturated reaction buffer. (B) Specific activity of the mutant enzyme exposed to 2 μM PPIX or 2 μM BAY41-2272 (BAY) was determined in the absence or presence of 1 mM DTT. The data for A and B are shown as means ± SD of three independent experiments performed in triplicate. (C) Heme-reconstituted mutant assayed in the presence of 1 mM DTT with or without 2 μM BAY41-2272 and treated with 100 μM SNP or with CO-saturated buffer. Representative data (means ± SD) of four independent experiments performed in triplicate are shown.
Discussion
Heme Presence and Basal Activity. To analyze the function of heme in the basal state of sGC we varied the heme content in wild-type sGC enzyme by using a previously developed method (35) of Tween 20-dependent heme depletion/reconstitution. Depletion of heme results in up to 3-fold activation of the enzyme (Fig. 1 A and B), whereas reconstitution of heme inhibits sGC activity (Fig. 1 C and D). This correlation of the heme content with the specific activity (Fig. 1D) of the heme-depleted/reconstituted enzyme suggests that the heme moiety of sGC may have an inhibitory function in the basal state of the enzyme. Increase in the activity of the detergent-treated enzyme was also shown in earlier studies in both crude lysates (36) and purified enzyme (35). All these data suggest that the heme group confines the enzyme in a restricted state.
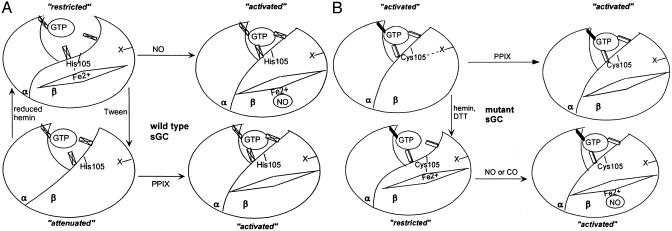
However, removal of the restrictions imposed by the heme-coordinating bond is not sufficient to result in full activation of the enzyme. The presence of PPIX (Fig. 1D and refs. 35 and 37) or uncoordinated nitrosyl heme is necessary to provide changes in the regulatory domain, resulting in a fully activated sGC. Thus, it seems that the heme moiety plays a dual role in the function of sGC. In the basal enzyme heme is coordinated with the His-105 residue, which keeps the enzyme in a restricted conformation (Fig. 6). Removal of the heme by Tween 20 releases the inhibition and produces an enzyme with an “attenuated” conformation, which is permissive to the activating effects of the uncoordinated porphyrin moiety of PPIX or nitrosyl heme (Fig. 6A).
Fig. 6.
Schematic representation of heme-dependent regulation of wild type (A) and αβCys-105 mutant (B). Catalytic center is schematically represented as a GTP molecule interacting with three structural elements. Maximal catalytic activity is achieved when all three elements are firmly interacting with GTP: activated conformation. Insertion of the heme and formation of a coordinating bond with His-105 (wild type) or Cys-105 (mutant) induces changes in the regulatory domain, affects the optimal structure of catalytic center, and results in a restricted or inhibited conformation. Activated conformation can be achieved by binding of NO to the heme moiety and disruption of the heme-coordinating bond. Removal of heme in wild-type enzyme results in an intermediate attenuated conformation, which in theαβCys-105 mutant mimics activated conformation due to a stabilizing effect of a DTT-sensitive modification or interaction designated as “X.” Binding of PPIX to a heme-deficient enzyme (wild type or mutant) also results in activated conformation.
Constitutively Active αβCys-105 Enzyme. sGC mutant enzyme with His-105 residue substituted for Cys-105 provides additional evidence supporting the hypothesis described above. Although the Cys residue is known to coordinate heme in many hemoproteins (e.g., cytochrome 450 or NO synthase enzymes), αβCys-105 enzyme is heme-deficient (Fig. 2). In contrast to the previously described αβPhe-105, which is only slightly more active than the basal wild-type enzyme (35), αβCys-105 enzyme has a specific activity, which is 70-fold higher than the basal activity of the heme-competent wild-type enzyme and is ≈30% of the activity of the NO-activated wild-type sGC (Fig. 2). Heme-deficient wild-type enzyme is significantly activated by PPIX (Fig. 1D and refs. 35 and 37), whereas the PPIX effect on NO-activated heme-competent enzyme is marginal. Similarly, αβCys-105, although heme-deficient, is activated only fractionally by PPIX (Fig. 5), suggesting that αβCys-105 enzyme is already in an activated conformation.
The Mutant Enzyme Is Also Constitutively Active in Intact Cells. Sf9 cells expressing mutant enzyme accumulates in the absence of NO donors' high levels of cGMP, which cannot be degraded efficiently by endogenous phosphodiesterase enzymes (Table 1). In similar conditions, SF9 cells expressing wild-type enzyme have a low level of cGMP, which is increased after treatment with NO donors. Expression of the mutant βCys-105 subunit alone in BE2 neuroblastoma cells also increased the level of basal sGC activity and decreased the response to NO (Table 1), suggesting that the mutant βCys-105 effectively competes with the wild-type β subunit to form a heterodimer and displays a high NO-independent constitutive activity.
This activated state of the mutant enzyme is sensitive to treatment with thiols and/or heme reconstitution. DTT treatment partially inactivated mutant enzyme (Figs. 3–5), making it receptive to the activation by PPIX and partial activation by BAY41-2272 (Fig. 5B). It seems that a thiol-dependent interaction(s) within the mutant enzyme mimics the activation effects of PPIX and stabilizes it in an activated conformation (Fig. 6B). Presumably the thiol environment with DTT present prevents this thiol interaction in the enzyme.
Hemin reconstitution of purified mutant enzyme significantly inactivated αβCys-105 enzyme (Fig. 3), providing additional proof that heme liganding stabilizes the regulatory domain in a restricted conformation, impeding activation of the catalytic center. The DTT-inhibited enzyme was inhibited even further after reconstitution with hemin (Fig. 3B). Hemin-reconstituted, DTT-treated enzyme displayed not only partial activation by NO or allosteric activation by BAY41-2272 but also demonstrated an additive effect of NO and BAY41-2272 (Fig. 5C). Wild-type sGC is synergistically activated by NO and BAY41-2272 at low concentrations of NO, but at high NO concentrations, fully activated sGC is affected only modestly by BAY41-2272 (38). Thus, the mutant enzyme displays properties similar to the wild-type enzyme activated by saturating concentrations of NO. Interestingly, heme-reconstituted mutant enzyme was activated to a similar extent by NO and CO (Fig. 5). Because the binding of NO and CO to sGC requires reduced ferrous heme, measurements were done in the presence of millimolar DTT. In our experience dithionate inhibits sGC and cannot be used in activity assays (E.M., unpublished observation). Due to the presence of inhibiting DTT, NO and CO could only partially activate the heme-reconstituted mutant enzyme.
UV-Vis spectral analysis of the heme-reconstituted enzyme shed some light on events that are affecting the activity of the enzyme. As summarized in Table 2, His → Cys substitution resulted in spectral characteristics of the mutant enzyme different than the wild-type enzyme, presumably due to the formation of thiolate coordination of heme. Heme oxidation with potassium ferricyanide and reduction with dithionate (Fig. 4) or DTT (data not shown) clearly indicate that reconstituted enzyme contained ferric heme. Changes in the spectra of the CO- and NO-treated αβCys-105 mutant with reduced heme are indicative of changes in heme coordination (Fig. 4 and Table 1).
Spectral properties of reconstituted αβCys-105 mutant were compared with the properties of cytochrome P450, a model hemoprotein with thiolate-coordinated heme, or its pressure-induced state, known as cytochrome P420. The positions of the Soret maxima of the ferric and ferrous states of heme in the mutant enzyme differ from the characteristics of cytochrome P450 (39) but are almost identical to cytochrome P420. Most indicative is the absence in the CO-treated mutant of the red shift in the Soret band, characteristic for cytochrome P450 (39). However, the position of the Soret peak after CO treatment is the same as for the pressure-induced cytochrome P420 (Table 2). Extensive EPR, magnetic circular dichroism, and Raman spectra analysis of cytochrome P420 demonstrated that, similar to cytochrome P450, the ferric heme is coordinated by the Cys-derived thiolate anion, although with a weakened iron-sulfur ligation (increased Fe—S stretch) possibly due to protonation of the thiolate anion (40). This comparison suggests that reconstituted αβCys-105 sGC also has a pentacoordinated heme with Cys as the ligand, although with coordination weaker than in cytochrome P450. It should be pointed out that reconstituted αβCys-105 sGC also has a prominent shoulder at 360 nm, which increases after storage of reconstituted mutant αβCys-105 on ice. Appearance of this shoulder may reflect some transition to a mixed low- and high-spin coordinated heme, similar to thermal shift described for the mutant His-93-Cys myoglobin at –60°C (41). Alternatively, heme could form a new bond with a different proximal ligand (e.g., adjacent His-107 or Tyr-114), similar to the ligand switch suggested for the CooA transcription factor (42). Only additional studies will clarify this issue.
It has been suggested that carbonyl heme in cytochrome P420 and His-93-Cys myoglobin is pentacoordinated because of the loss of the weak thiolate coordination (40, 41). Similar assumptions can be made about the αβCys-105 sGC. In this respect CO-induced loss of heme coordination is similar to the effects of NO. Consequently, αβCys-105 mutant is equally stimulated by NO and CO (Fig. 5 A and C).
In summary, our data suggest that the heme prosthetic group of sGC maintains the enzyme in a basal state with the regulatory domain of sGC in an inhibited restricted conformation through its coordination with an axial residue (Fig. 6). Disruption of the coordination and removal of this inhibition permits the transition of the enzyme to an activated state due to the activating role of the porphyrin moiety of nitrosylated heme. Substitution of coordinating His-105 with Cys and the resulting heme deficiency favor the transition of the sGC regulatory domain into an activated conformation. This transition is facilitated or stabilized by some thiol-dependent interaction. Mutant sGC enzyme can be maintained in an activated state without heme and stimulatory ligands, such as NO or allosteric regulators. The constitutively active mutant could be a useful reagent to screen for novel inhibitors and activators of sGC or to analyze cGMP-dependent effects on cellular physiology. Gene delivery of the mutant αβCys-105 sGC could also be beneficial in disorders for which increased cGMP levels are desired.†
Acknowledgments
We thank Dr. Yu-Chen Lee for generating mutant baculovirus, Qui Wu for technical assistance, and Dr. Karen Davis for valuable suggestions. Research support was provided by the John S. Dunn Foundation, the Harold and Leila Y. Mathers Foundation, the Welch Foundation, the National Space Biomedical Research Institute, U.S. Army Medical Research, the National Institutes of Health, and the University of Texas.
Abbreviations: sGC, soluble guanylyl cyclase; UV-Vis, UV-visible; BAY 41-2272, 5-cyclopropyl-2-[1-(2-fluoro-benzyl)-1H-pyrazolo[3,4-b]pyridin-3-yl]-pyrimidin-4-ylamine; TEA, triethanolamine; SNP, sodium nitroprusside; PPIX, protoporphyrin IX.
Footnotes
A patent application for the use of the αβCys105 mutant sGC is pending [Martin, E. & Murad, F. (2003) U.S. Patent Appl. 60/446,427, pending].
References
- 1.Rodgers, K. R. (1999) Curr. Opin. Chem. Biol. 3, 158–167. [DOI] [PubMed] [Google Scholar]
- 2.Murad, F. (1998) Recent Prog. Horm. Res. 53, 43–59. [PubMed] [Google Scholar]
- 3.Pellequer, J. L., Brudler, R. & Getzoff, E. D. (1999) Curr. Biol. 9, R416–R418. [DOI] [PubMed] [Google Scholar]
- 4.Aono, S., Honma, Y., Ohkubo, K., Tawara, T., Kamiya, T. & Nakajima, H. (2000) J. Inorg. Biochem. 82, 51–56. [DOI] [PubMed] [Google Scholar]
- 5.Lucas, K. A., Pitari, G. M., Kazerounian, S., Ruiz-Stewart, I., Park, J., Schulz, S., Chepenik, K. P. & Waldman, S. A. (2000) Pharmacol. Rev. 52, 375–414. [PubMed] [Google Scholar]
- 6.Martin, E., Davis, K., Bian, K., Lee, Y. C. & Murad, F. (2000) Semin. Perinatol. 24, 2–6. [DOI] [PubMed] [Google Scholar]
- 7.Kamisaki, Y., Saheki, S., Nakane, M., Palmieri, J. A., Kuno, T., Chang, B. Y., Waldman, S. A. & Murad, F. (1986) J. Biol. Chem. 261, 7236–7241. [PubMed] [Google Scholar]
- 8.Buechler, W. A., Nakane, M. & Murad, F. (1991) Biochem. Biophys. Res. Commun. 174, 351–357. [DOI] [PubMed] [Google Scholar]
- 9.Koglin, M., Vehse, K., Budaeus, L., Scholz, H. & Behrends, S. (2001) J. Biol. Chem. 276, 30737–30743. [DOI] [PubMed] [Google Scholar]
- 10.Stone, J. R. & Marletta, M. A. (1994) Biochemistry 33, 5636–5640. [DOI] [PubMed] [Google Scholar]
- 11.Wedel, B., Humbert, P., Harteneck, C., Foerster, J., Malkewitz, J., Bohme, E., Schultz, G. & Koesling, D. (1994) Proc. Natl. Acad. Sci. USA 91, 2592–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katsuki, S., Arnold, W., Mittal, C. & Murad, F. (1977) J. Cyclic Nucleotide Res. 3, 23–35. [PubMed] [Google Scholar]
- 13.Humbert, P., Niroomand, F., Fischer, G., Mayer, B., Koesling, D., Hinsch, K. D., Gausepohl, H., Frank, R., Schultz, G. & Bohme, E. (1990) Eur. J. Biochem. 190, 273–278. [DOI] [PubMed] [Google Scholar]
- 14.Stone, J. R. & Marletta, M. A. (1996) Biochemistry 35, 1093–1099. [DOI] [PubMed] [Google Scholar]
- 15.Lee, Y. C., Martin, E. & Murad, F. (2000) Proc. Natl. Acad. Sci. USA 97, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stone, J. R., Sands, R. H., Dunham, W. R. & Marletta, M. A. (1995) Biochem. Biophys. Res. Commun. 207, 572–577. [DOI] [PubMed] [Google Scholar]
- 17.Tomita, T., Ogura, T., Tsuyama, S., Imai, Y. & Kitagawa, T. (1997) Biochemistry 36, 10155–10160. [DOI] [PubMed] [Google Scholar]
- 18.Deinum, G., Stone, J. R., Babcock, G. T. & Marletta, M. A. (1996) Biochemistry 35, 1540–1547. [DOI] [PubMed] [Google Scholar]
- 19.Kharitonov, V. G., Sharma, V. S., Magde, D. & Koesling, D. (1997) Biochemistry 36, 6814–6818. [DOI] [PubMed] [Google Scholar]
- 20.Kharitonov, V. G., Russwurm, M., Magde, D., Sharma, V. S. & Koesling, D. (1997) Biochem. Biophys. Res. Commun. 239, 284–286. [DOI] [PubMed] [Google Scholar]
- 21.Zhao, Y., Brandish, P. E., Ballou, D. P. & Marletta, M. A. (1999) Proc. Natl. Acad. Sci. USA 96, 14753–14758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bellamy, T. C., Wood, J. & Garthwaite, J. (2002) Proc. Natl. Acad. Sci. USA 99, 507–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koesling, D. (1999) Methods 19, 485–493. [DOI] [PubMed] [Google Scholar]
- 24.Koesling, D. & Friebe, A. (1999) Rev. Physiol. Biochem. Pharmacol. 135, 41–65. [DOI] [PubMed] [Google Scholar]
- 25.Sharma, V. S. & Magde, D. (1999) Methods 19, 494–505. [DOI] [PubMed] [Google Scholar]
- 26.Denninger, J. W., Schelvis, J. P., Brandish, P. E., Zhao, Y., Babcock, G. T. & Marletta, M. A. (2000) Biochemistry 39, 4191–4198. [DOI] [PubMed] [Google Scholar]
- 27.Gerzer, R., Bohme, E., Hofmann, F. & Schultz, G. (1981) FEBS Lett. 132, 71–74. [DOI] [PubMed] [Google Scholar]
- 28.Martin, E., Lee, Y. C. & Murad, F. (2001) Proc. Natl. Acad. Sci. USA 98, 12938–12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz, G. & Bohme, E. (1984) Methods of Enzymatic Analysis (Verlag Chemie, Weinheim, Germany).
- 30.Steiner, A. L., Parker, C. W. & Kipnis, D. M. (1972) J. Biol. Chem. 247, 1106–1113. [PubMed] [Google Scholar]
- 31.Lowry, O. H., Rosebrough, N. J., Farr, A. L. & Randall, R. J. (1951) J. Biol. Chem. 193, 265–275. [PubMed] [Google Scholar]
- 32.Nakane, M., Hsieh, G., Miller, L. N., Chang, R., Terranova, M. A., Moreland, R. B., Kolasa, T. & Brioni, J. D. (2002) Int. J. Impot. Res. 14, 121–127. [DOI] [PubMed] [Google Scholar]
- 33.Friebe, A., Russwurm, M., Mergia, E. & Koesling, D. (1999) Biochemistry 38, 15253–15257. [DOI] [PubMed] [Google Scholar]
- 34.Friebe, A. & Koesling, D. (1998) Mol. Pharmacol. 53, 123–127. [DOI] [PubMed] [Google Scholar]
- 35.Foerster, J., Harteneck, C., Malkewitz, J., Schultz, G. & Koesling, D. (1996) Eur. J. Biochem. 240, 380–386. [DOI] [PubMed] [Google Scholar]
- 36.Kimura, H. & Murad, F. (1975) J. Biol. Chem. 250, 4810–4817. [PubMed] [Google Scholar]
- 37.Wolin, M. S., Wood, K. S. & Ignarro, L. J. (1982) J. Biol. Chem. 257, 13312–13320. [PubMed] [Google Scholar]
- 38.Stasch, J. P., Becker, E. M., Alonso-Alija, C., Apeler, H., Dembowsky, K., Feurer, A., Gerzer, R., Minuth, T., Perzborn, E., Pleiss, U., et al. (2001) Nature 410, 212–215. [DOI] [PubMed] [Google Scholar]
- 39.White, R. E. & Coon, M. J. (1982) J. Biol. Chem. 257, 3073–3083. [PubMed] [Google Scholar]
- 40.Martinis, S. A., Blanke, S. R., Hager, L. P., Sligar, S. G., Hoa, G. H., Rux, J. J. & Dawson, J. H. (1996) Biochemistry 35, 14530–14536. [DOI] [PubMed] [Google Scholar]
- 41.Adachi, S., Nagano, S., Ishimori, K., Watanabe, Y., Morishima, I., Egawa, T., Kitagawa, T. & Makino, R. (1993) Biochemistry 32, 241–252. [DOI] [PubMed] [Google Scholar]
- 42.Shelver, D., Thorsteisson, M. V., Kerby, R. L., Chung, S.-Y., Roberts, G. R., Reynolds, M. F., Parks, R. B. & Burstyn, J. N. (1999) Biochemistry 38, 2669–2678. [DOI] [PubMed] [Google Scholar]