Abstract
High-pressure liquid chromatography–tandem mass spectrometry was used to obtain a protein profile of Escherichia coli strain MG1655 grown in minimal medium with glycerol as the carbon source. By using cell lysate from only 3 × 108 cells, at least four different tryptic peptides were detected for each of 404 proteins in a short 4-h experiment. At least one peptide with a high reliability score was detected for 986 proteins. Because membrane proteins were underrepresented, a second experiment was performed with a preparation enriched in membranes. An additional 161 proteins were detected, of which from half to two-thirds were membrane proteins. Overall, 1,147 different E. coli proteins were identified, almost 4 times as many as had been identified previously by using other tools. The protein list was compared with the transcription profile obtained on Affymetrix GeneChips. Expression of 1,113 (97%) of the genes whose protein products were found was detected at the mRNA level. The arithmetic mean mRNA signal intensity for these genes was 3-fold higher than that for all 4,300 protein-coding genes of E. coli. Thus, GeneChip data confirmed the high reliability of the protein list, which contains about one-fourth of the proteins of E. coli. Detection of even those membrane proteins and proteins of undefined function that are encoded by the same operons (transcriptional units) encoding proteins on the list remained low.
Neidhardt and colleagues pioneered the use of 2D gel electrophoresis to determine the protein composition of the bacterium Escherichia coli (1), an approach that has been intensively pursued by others (2–5). When coupled with Edman degradation (3), electrospray ionization (3), or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (2, 4, 5) and the availability of a complete genome sequence, this tool has allowed identification of up to 300 gene products. Genomewide studies of transcription have also been initiated. Those performed with Affymetrix GeneChips not only allow comparison of mRNA levels under different conditions but also provide a statistical estimate of which genes are transcribed under a single condition: the global transcription profile (6–9).
For an organism whose genome sequence has been determined, analysis of complex mixtures of tryptic peptides by HPLC–tandem mass spectrometry (MS/MS) provides a powerful means of determining its protein composition (10, 11). This can be achieved rapidly by using small amounts of cell extract, and the data can be analyzed automatically by using the sequest algorithm (12). We have applied these tools to E. coli strain MG1655 grown on a minimal medium with glycerol as the carbon source and have identified more than 1,100 proteins, a quarter of those coded in its genome. We have compared this protein profile to the transcription profile of the organism under the same conditions and have assessed it in terms of the operon organization of E. coli genes (i.e., the pattern of linked genes that are cotranscribed).
Materials and Methods
Growth of E. coli MG1655 and Preparation of Cell Extracts. E. coli strain MG1655 (CGSC 6300; 100 ml in a 500-ml flask) was grown with agitation at 37°C in minimal medium N–C– with 0.2% glycerol as the carbon source and 10 mM ammonium chloride as the nitrogen source (25). At midexponential phase (OD600 of 0.4), cells were chilled on ice and harvested by centrifugation at 8,000 × g for 5 min at 4°C. The cell pellet was frozen on dry ice and stored at –80°C. For preparation of a crude cell extract, the pellet was defrosted on ice and suspended in 1 ml of ice-cold breakage buffer (0.1 mM ammonium bicarbonate/1 mM dithiothreitol). Cells were disrupted at 4°C by passage through a French pressure cell twice at 12,000 lb/in2 (83 MPa). Magnesium chloride was added to the lysate to a final concentration of 10 mM, and nucleic acids were digested for 15 min on ice with 20 units of DNase I (Sigma) and 20 units of RNase mixture (Ambion, Austin, TX). EDTA was then added to a final concentration of 1 mM and the lysate was cleared by centrifugation at 12,000 × g for 15 min at 4°C. The sample was frozen on dry ice and stored at –80°C. The protein concentration was usually ≈4–5 mg/ml and the total volume of extract was ≈1 ml.
For preparation of a sample enriched in membranes, the above protocol was modified as follows: the cell pellet from a 2-liter culture was suspended in 40 ml of ice-cold breakage buffer supplemented with 0.3 M sodium chloride. After the steps described above, the cleared lysate was divided into four portions of 10 ml, which were then subjected to centrifugation at 150,000 × g for 60 min at 4°C. The pellets were not washed and were frozen dry on dry ice and stored at –80°C. The amount of protein in each pellet was ≈5 mg.
Tryptic Digestion and HPLC-MS/MS Analysis. E. coli proteins from ≈3 × 109 cells (≈200 μg in 50 μl) were added to 50 μl of ammonium bicarbonate (100 mM, pH 8.5), reduced with 400 mM dithiothreitol (35 μl) at 51°C for 1 h, carboxyamidomethylated with 800 mM iodoacetamide (50 μl) in the dark at room temperature for 1 h, and digested with modified trypsin (5 μg, 20 μl; Promega) at 37°C for 17 h. Proteolysis was terminated by acidification of the reaction mixture with glacial acetic acid. Tryptic peptides in 64 μl of the final reaction mixture (≈109 cell equivalents) were desalted, concentrated, and then fractionated on a strong cation exchange column [polysulfoethyl aspartamide (PolyLC, Columbia, MD, 360 × 100 μm, 8 cm of 5-μm particles; see supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html, and refs. 13–15)]. Peptides were eluted stepwise with solutions containing 2, 5, 10, 15, 25, 50, 75, 100, and 500 mM KCl in 5 mM potassium phosphate buffer (pH 3) containing 5% acetonitrile.
Samples, corresponding to 10% (≈108 cell equivalents) of each of the above ion exchange fractions, were loaded onto nano-HPLC precolumns, which were washed and connected to analytical columns (13–15). Samples were then analyzed by a combination of a nano-HPLC/microelectrospray ionization on a LCQ Deca mass spectrometer (ThermoFinnigan, San Jose, CA) as described previously (14). The HPLC gradient (A = 100 mM acetic acid in water, B = 70% acetonitrile/100 mM acetic acid in water) was 0–2% B in 4 min, 2–10% B in 24 min, 10–16% B in 44 min, 16–22% B in 48 min, 22–30%Bin48min,30–50%Bin52min,50–70%Bin10min, 70–100% B in 4 min, 100% B for 2 min, 100–0% B in 2 min, and 0% B for 5 min. Full-scan mass spectra were acquired over the m/z range 400–1,500.
A membrane pellet from ≈1010 cells (≈90 μg of protein) was placed in 0.2% SDS (30 μl), and the resulting suspension was then vortexed and sonicated. To remove lipids (16), the above solution was treated with methanol (90 μl), mixed briefly, treated with chloroform (30 μl), and stirred to yield a single phase. Addition of water (60 μl) with vigorous mixing afforded two phases that were separated by centrifugation (10,000 × g for 2 min) into two layers, with a precipitate at the interface. The bulk of the upper aqueous methanol phase was removed and the pellet was dried under vacuum on a Speed Vac with the tube inverted. The resulting protein pellet was dissolved in 0.2% SDS (40 μl), reduced with 50 mM dithiothreitol (5 μl) at 37°C for 1 h, and carboxyamidomethylated with 500 mM iodoacetamide (5 μl) in the dark at room temperature for 1 h. For proteolysis, the sample was diluted with 100 mM ammonium bicarbonate (30 μl) and digested with trypsin (3.5 μg, 7 μl; Promega) at 37°C for 13 h at pH 8.5 and then for an additional 22 h at 37°C after treatment with a second aliquot of trypsin (3.5 μg, 7 μl). Proteolysis was terminated by acidifying the reaction mixture with glacial acetic acid (10 μl).
Half of the above digest (≈5 × 109 cell equivalents, ≈45 μg of protein) was desalted, concentrated, and fractionated on a cation exchange column (Poros 20 HS, 360 × 200 μm, 8-cm packing, PerSeptive Biosystems, Framingham, MA). After washing the HS column with 100 μl of 0.1% acetic acid, peptides were eluted stepwise with 100 μl of 0, 5, 25, 50, 100, and 500 mM KCl in 5 mM potassium phosphate buffer (pH 3) containing 5% acetonitrile.
Twenty percent of each fraction (≈109 cell equivalents) was diluted 3-fold with 0.1% acetic acid to reduce the acetonitrile concentration and then loaded for HPLC as described above for the cell extract. The HPLC gradient (A and B as above) was 0–16% B in 28 min, 16–22% B in 48 min, 22–30% B in 48 min, 30–50% B in 52 min, 50–70% B in 10 min, 70–100% B in 4 min, 100% B for 2 min, 100–0% B in 2 min, and 0% B for 5 min. Spectra were acquired as described above.
Criteria for Proteins in the Short, Long, Membrane Sample, and Total Lists. A protein was assigned as present on the short list if the software program sequest matched MS/MS spectra with an Xcorr ≥2.4 (ref. 12) to 4 or more different tryptic peptides from the same protein in the E. coli database (17). A protein was assigned as present on the long list or membrane sample list if the software program sequest matched an MS/MS spectrum to at least one peptide from a protein in the E. coli database using the following parameters: Del Mass < 1, Xcorr > 2.4, Del cn > 0.1, Sp > 500, Rsp < 10, Ion Ratio > 0.6, and at least one end of the peptide resulted from cleavage C-terminal to Lys or Arg. The total list was formed by compiling the proteins in the long and membrane sample lists. When the total list was compiled, the value for the number of tryptic peptides detected for a protein found in both the long and membrane sample lists was the larger of the two values.
Definition of Membrane Proteins. Three criteria were used to compile lists of known and predicted membrane proteins. (i) if the gene description [taken from the E. coli Entry Point (http://coli.berkeley.edu/cgi-bin/ecoli/coli_entry.pl)] indicated that the protein was a membrane protein, it was designated a known membrane protein (156 proteins); (ii) if the protein was listed as a membrane protein in the GenProtEC database (http://genprotec.mbl.edu), it was considered a known membrane protein (634 proteins); (iii) if a protein was predicted to contain at least two transmembrane helices by the phd algorithm (18), it was designated a predicted membrane protein (821 proteins). Our low estimate of membrane proteins contained those present on at least two lists (532 proteins), whereas our high estimate contained those present on any one list (1,017 proteins). These values are given as the range of low estimate to high estimate. For purposes of calculations no distinction was made between known and predicted membrane proteins.
Affymetrix DNA Microarrays. E. coli strain MG1655 was grown essentially as described above, except that the glycerol concentration was 0.4%. Three independent assessments of gene expression (more accurately, mRNA levels) were made on Affymetrix E. coli Antisense GeneChip arrays as described (25). In each case, total RNA (100–300 μg) was isolated from 25 ml of cells at OD600 = 0.4–0.5; ≈15 μg was used for synthesis of cDNA and 1–1.5 μg of cDNA was used per chip (http://nature.berkeley.edu/~opaliy/papers/Proteomics.html). Raw data files were analyzed by the statistical algorithm (6, 7) in the Microarray Analysis Suite (MAS) 5.0 (Affymetrix) by using the default parameters. Experiments were scaled globally to the same target intensity of 1,500 by using only the probe sets for E. coli genes and not for intergenic regions. In each experiment, expression of ≈3% of all protein-coding genes was called “marginal” (marginally expressed), whereas expression of all others was called “present” or “absent.” Reproducibility between experiments was assessed by calculating the pairwise concordance of presence calls, which was 85–90%, and by computing the pairwise correlation coefficient of log-transformed signal intensities (average of 0.91). Consensus presence calls for gene expression were made as follows: “absent” if absent in at least two experiments or absent in one experiment and marginal in the other two, and otherwise “present.” The mRNA signal intensity for each gene, whether present or absent, was calculated as a mean of values in the three experiments.
Proteome–Transcriptome Comparison. A spreadsheet file was constructed which included b number, gene name, strand orientation, protein length, mRNA signal intensity and presence call, number of tryptic peptides predicted, proteins detected and number of tryptic peptides detected (total list, short list, long list, membrane sample), and assignment as membrane protein. Gene description, functional category assignment (19), and operon organization (17) were also included. Unless otherwise noted, all data not from this study were taken from the E. coli Entry Point. For various lists or groups of proteins and their corresponding genes we calculated arithmetic means for the number of tryptic peptides predicted or detected per protein and for the mRNA signal intensity. For mRNA signal intensity we also calculated geometric means. {x̄G = antilog[(Σ log Xi)/n]}. Mean values for peptides were rounded to one decimal place and those for mRNA signal intensity to the nearest 10.
Results
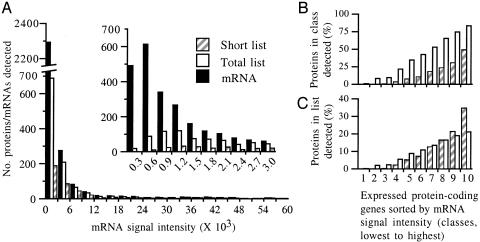
Initial List of 404 Proteins (Short List). To define the protein profile of E. coli MG1655 grown in minimal medium with glycerol as the carbon source, we began with a list of 404 proteins in a cell extract for which at least four different tryptic peptides per protein were found (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html; Table 1, column 1). These represent ≈1/10 of the proteins coded by the E. coli genome (4,290 proteins; ref. 17). The genes coding for 401 (99%) of the 404 proteins were considered expressed at the mRNA level on Affymetrix GeneChips (i.e., called “present”; see Materials and Methods). They were a subset of the 2,826 protein-coding genes considered expressed, which we have not analyzed separately in this study. The mRNA signal intensities (approximations of mRNA levels; see Discussion) for the 401 genes corresponding to proteins on the short list differed by at least two orders of magnitude (≈100–60,000 arbitrary units; Fig. 1A). However, few genes had mRNA signal intensities >9,000 and hence there were few proteins detected in this range. Most proteins detected had mRNA signal intensities between 600 and 9,000. Below a signal intensity of 600, few proteins were detected despite the fact that the number of genes expressed remained high (Fig. 1A Inset). To look at the data in another way, we sorted the 2,826 expressed protein-coding genes by mRNA signal intensity from lowest to highest and divided them into 10 equal classes of 283 each. Note that the range of mRNA signal intensity differs greatly from class to class (Fig. 1 legend). We then determined the percentage of proteins detected in each class (Fig. 1B) and the proportion of all proteins on the short list present in each class (Fig. 1C). Both were progressively higher with higher mRNA signal intensity. Neither the average length of all E. coli proteins nor the average number of predicted tryptic peptides per protein increased with mRNA signal intensity (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html), and hence the relationship between protein detection and signal intensity was not a trivial consequence of these other relationships.
Table 1. Analysis of the protein lists and comparison to mRNA signal intensities on Affymetrix GeneChips.
| Profile element | Short list | Long list | Membrane sample* | Total list | Protein-coding genes expressed on Affymetrix arrays† |
|---|---|---|---|---|---|
| Proteins | 404 | 986 | 287 | 1,147 | — |
| Corresponding mRNA detected | 401 | 964 | 275 | 1,113 | 2,826 |
| Mean mRNA signal (arithmetic) | 7,990 | 5,670 | 8,080 | 5,330 | 2,850 |
| Mean mRNA signal (geometric) | 3,830 | 2,450 | 2,940 | 2,270 | 1,020 |
| Mean number of tryptic peptides detected | 8.0‡ | 4.2 | 2.4 | 4.0§ | — |
| Mean number of tryptic peptides predicted | 21.0 | 16.1 | 16.5 | 15.9 | 13.4 |
| Membrane proteins (low estimate to high estimate) | 7-15 | 22-56 | 94-138 | 99-160 | 317-592 |
| Proteins of unknown function¶ | 14 | 163 | 49 | 199 | 803 |
If we consider only the membrane proteins in the sample, which was prepared without washing, the values are as follows (low estimate to high estimate): proteins, 94-138; corresponding mRNA detected, 87-130; mean mRNA signal (arithmetic), 6,530-5,840; mean mRNA signal (geometric), 2,240-2,130; mean number of tryptic peptides detected, 2.8-2.7; mean number of tryptic peptides predicted, 15.1-15.9; proteins of unknown function, 3-17. If we consider the proteins identified only in the membrane sample, the values are as follows: proteins, 161; corresponding mRNA detected, 149; mean mRNA signal (arithmetic), 3,240; mean mRNA signal (geometric), 1,410; mean number of tryptic peptides detected, 2.0; mean number of tryptic peptides predicted, 15.0; membrane proteins, 77-104; proteins of unknown function, 36.
E. coli has a total of 4,291 protein-coding genes. Values for these were as follows: mean mRNA signal (arithmetic), 1,950; mean mRNA signal (geometric), 460; mean number of tryptic peptides predicted, 13.1; membrane proteins, 532-1,017; proteins of unknown function, 1,409.
With the criteria employed for the total list (see Materials and Methods), this value was 7.8.
Only one tryptic peptide was detected for 381 of the proteins.
This is the same as open reading frames (19). The overlap between these and membrane proteins is as follows: short list, 0-0; long list, 1-4; membrane sample, 3-17; total list, 3-18; all expressed protein-coding genes, 11-134.
Fig. 1.
E. coli proteins detected as a function of mRNA signal intensity. (A) The number of proteins detected in the short (hatched bars) or total (open bars) lists or the number of mRNAs (filled bars) detected for protein-coding genes on Affymetrix arrays was plotted as a function of mRNA signal intensity. (A Inset) An expansion of the data for proteins/mRNAs with signal intensities (×103) ≤3. (B) To get the x axis, the 2,826 protein-coding genes expressed on Affymetrix arrays were first sorted from lowest to highest mRNA signal intensity. They were then divided into 10 equal classes (283 each). Class 1 contained the 10% of genes with the lowest intensities, and so on. The proportion (percent) of the 283 proteins in each class that was detected was plotted on the y axis (short list or total list). (C) The x axis is as for B. The proportion (percent) of all proteins in a list (short or total) that corresponded to genes in each class was plotted on the y axis. The ranges of mRNA signal intensities for classes 1–10 were as follows: 1, 51–227; 2, 227–327; 3, 327–446; 4, 447–625; 5, 625–878; 6, 879–1,179; 7, 1,181–1,781; 8, 1,782–2,828; 9, 2,830–5,467; 10, 5,468–58,952.
Results in Fig. 1 A Inset, B, and C reflected the fact that the arithmetic and geometric mean mRNA signal intensities for genes corresponding to proteins on the short list were higher than those for all 2,826 protein-coding genes that were expressed (2.8- and 3.8-fold, respectively; Table 1). On average, the 404 proteins in the short list were considerably longer than the average protein product of an expressed gene or the average E. coli protein and had more predicted tryptic peptides. The short list contained only 2–4% membrane proteins (low estimate to high estimate; see Materials and Methods), whereas the percentage of membrane proteins among expressed or total protein-coding genes was 5- or 6-fold higher. Likewise, the short list contained a much lower fraction of proteins of unknown function than the fraction among expressed or total genes. [Note, however, that our estimates of proteins of unknown function are high because we have used the original annotations (17).]
The genes coding for proteins in the short list (404) were contained in 335 operons coding for 681 proteins (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html). Thus we had not detected ≈40% of the proteins coded by these operons (Table 2, column 1). Although most of the genes corresponding to the missing proteins were expressed at the mRNA level, their arithmetic and geometric mean mRNA signal intensities were lower than those for genes corresponding to the proteins that were detected (Tables 2 versus 1, column 1). In addition, the 277 undetected proteins were much shorter (11.2 versus 21.0 predicted tryptic peptides). The list of undetected proteins included a higher proportion of membrane proteins (23–30%) and a somewhat higher proportion of proteins of unknown function (11%) than the short list.
Table 2. Characteristics of proteins missing in expanded operon lists.
| Profile element | Expanded short list | Expanded total list |
|---|---|---|
| Proteins not detected* | 277 | 471 |
| Corresponding mRNA detected | 263† | 383‡ |
| Mean mRNA signal (arithmetic)§ | 5,560 | 1,840 |
| Mean mRNA signal (geometric) | 2,240 | 740 |
| Mean number of tryptic peptides predicted | 11.2 | 11.5 |
| Membrane proteins (low estimate to high estimate) | 64-82 | 92-143 |
| Proteins of unknown function¶ | 30 | 134 |
The genes coding for proteins in the short list, total list, and list of all E. coli proteins were contained in 335, 868, and 2,583 operons composed of 681, 1,618, and 4,286 protein-coding genes, respectively. (Five protein-coding genes had no operon assignment.) The mean numbers of genes in the operons were 2.03, 1.86, and 1.66, respectively, and the numbers of these operons composed of a single gene were 188, 520, and 1,748, respectively.
For data on the 404 and 1,147 proteins detected in the short and total lists, see Table 1, columns 1 and 4, respectively.
One gene was not represented on Affymetrix GeneChips.
Four genes were not represented on Affymetrix GeneChips.
When membrane proteins and proteins of unknown function were not considered, the corresponding values for arithmetic and geometric mean mRNA signal intensities for the remaining missing proteins were as follows: short list, 6,390 and 2,620, respectively; total list, 2,110 and 870, respectively.
This is the same as open reading frames (19). The overlap between these and membrane proteins is as follows: expanded short list, 0-2; expanded total list, 1-19.
Long List of Proteins (986), Proteins in Membrane Sample (287), and Total Proteins (1,147). We next expanded the short list in two ways: (i) we included all proteins for which a single tryptic peptide was identified with high reliability criteria (see Materials and Methods) (long list; 986 proteins); (ii) we analyzed a preparation enriched in membranes (membrane sample; 287 proteins). From the long list, which included all of the proteins in the short list, and the membrane sample we compiled a list of 1,147 different proteins detected (total list; supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html) (Table 1). The total list contains about one-fourth of the proteins of E. coli. We focus here on the membrane sample and the total list.
Of the proteins identified in the membrane sample 33–48% were indeed membrane proteins (Table 1, column 3), and the majority of these were identified only in this sample (i.e., not found in the long list). The percentage of membrane proteins in the total list was 4- to 5-fold higher than in the short list. Of the 74 proteins annotated in the GenProtEC database as outer membrane proteins, 21 (28%) were detected in the membrane sample.
Expression of 97% of the genes coding for proteins in the total list was detected at the mRNA level (Table 1, column 4). As was true for proteins in the short list, the arithmetic and geometric mean mRNA signal intensities for these genes were higher than for all expressed protein-coding genes of E. coli (Fig. 1 and Table 1). However, the differences for proteins in the total list were smaller. With respect to the short list, higher percentages of proteins in the total list (and the membrane sample) were proteins of unknown function. They were nevertheless underrepresented with respect to all expressed protein-coding genes.
The total list of 1,147 proteins contained 45% (126) of those proteins missing when the short list was expanded to operons. Genes corresponding to the proteins that were found had a 3-fold higher arithmetic mean mRNA signal intensity than those corresponding to proteins that remained missing. Eight of the proteins that remained missing had only one tryptic peptide and one had no tryptic peptides. Four of these were short regulatory peptides called leader peptides.
Expansion of the total list to operons indicated that the 1,147 proteins were contained in 868 operons coding for 1,618 proteins (Table 2 legend). Thus, we had not detected ≈30% of the protein products of these operons. Genes corresponding to 82% of the missing proteins were expressed at the mRNA level. Again, the mean mRNA signal intensity for the genes corresponding to the 471 undetected proteins was 3-fold lower than that for genes corresponding to the 1,147 proteins detected. This discrepancy was twice as large as that for the short list. As was the case for the short list, the undetected proteins were smaller than those detected (11.5 versus 15.9 mean predicted tryptic peptides, respectively), and there was some enrichment (1.6-fold) for proteins of unknown function. Although we considered the possibility that genes coding for proteins that were not detected had particular positions in operons, we did not find evidence for this (O.P., unpublished work).
Categories of Proteins in the Total List. We have analyzed the total list of 1,147 proteins with respect to the functional categories defined by Riley and Labedan (19) (Table 3). There are a few striking findings. In general, the fraction of proteins detected in a category was a function of the mean mRNA signal intensity for genes or expressed genes in that category (Table 3). In comparison to other categories, a low fraction of the proteins involved in cell processes was detected, presumably because more than half the proteins in this category (58%, high estimate) are membrane proteins. Low fractions of the proteins in the large categories “miscellaneous” and “open reading frames” (proteins of putative or unknown function) were detected. Proteins in the open reading frames category are unusually short (9.8 mean predicted tryptic peptides). The mean mRNA signal intensity for genes in these two categories was low (<800) and detection of mRNAs was also somewhat low. Despite the fact that proteins in the “structural elements” category are also short (11.6 mean predicted tryptic peptides), they were well represented in the protein list, likely because the mean mRNA signal intensity for the corresponding genes was exceptionally high. Detection of proteins involved in macromolecule metabolism was highest of any category (50%). Proteins in this category, which have the largest average number of domains per protein (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html), are very long (19.4 mean predicted tryptic peptides) and their mRNA signals were strong.
Table 3. Analysis of the total protein list with respect to functional categories.
|
Proteins detected
|
Membrane proteins
detected*
|
mRNA detected
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | Genes/proteins | Membrane proteins* | Mean no. of tryptic peptides predicted† | Mean mRNA signal (arithmetic)† | Mean mRNA signal (geometric)† | No. | % | No. | % | No. | % |
| 1 Cell processes | 608 | 355 | 12.6 | 2,000 | 550 | 171 | 28.1 | 63 | 17.7 | 431 | 70.9 |
| 2 Extrachromosomal | 92 | 12 | 14.2 | 1,270‡ | 210‡ | 8 | 8.8 | 4 | 33.3 | 25 | 48.1‡ |
| 3 Global functions | 55 | 12 | 15.8 | 2,610 | 1,050 | 22 | 40.7 | 3 | 25.0 | 47 | 85.5 |
| 4 Macromolecule metabolism | 315 | 43 | 19.4 | 3,100 | 1,090 | 156 | 49.7 | 15 | 34.9 | 283 | 89.8 |
| 5 Metabolism of small molecules | 881 | 111 | 15.6 | 2,630 | 820 | 385 | 43.7 | 25 | 22.5 | 673 | 76.4 |
| 6 Miscellaneous | 742 | 180 | 14.1 | 780§ | 280§ | 117 | 15.8 | 14 | 7.8 | 421 | 57.4§ |
| 7 Open reading frames¶ | 1,409 | 258 | 9.8 | 780∥ | 280∥ | 199 | 14.1 | 18 | 7.0 | 803 | 57.7∥ |
| 8 Structural elements | 215 | 46 | 11.6 | 17,000 | 2,160 | 89 | 46.8** | 18 | 39.1 | 1,369 | 78.6 |
| 9 tRNA | 86 | — | — | 7,800 | 2,150 | — | — | — | — | 71 | 82.6 |
| 10 Sum or average†† | 4,403 | 1,017 | 13.1 | 2,470‡‡ | 490‡‡ | 1,147 | 26.7** | 160 | 15.7 | 2,923 | 67.4‡‡ |
High estimate.
Data are for all proteins/genes in category. The values for arithmetic means for expressed genes in each category were as follows: 1, 2,770; 2, 2,530; 3, 3,020; 4, 3,420; 5, 3,400; 6, 1,260; 7, 1,260; 8, 21,590; 9, 9,430; 10, 3,600. The values for geometric means for expressed genes were as follows: 1, 1,060; 2, 830; 3, 1,530; 4, 1,360; 5, 1,470; 6, 620; 7, 680; 8, 5,000; 9, 4,810; 10, 1,090.
Forty genes were not represented on Affymetrix GeneChips.
Nine genes were not represented on Affymetrix GeneChips.
Seventeen genes were not represented on Affymetrix GeneChips.
Stable RNAs were excluded from calculations.
As appropriate.
Sixty-six genes were not represented on Affymetrix GeneChips.
Discussion
In a crude cell extract of E. coli and a sample enriched in membranes we detected about one-fourth of all E. coli proteins in short HPLC-MS/MS experiments. Expression of most of the corresponding genes was detected at the mRNA level on Affymetrix GeneChips, and genes corresponding to the proteins detected had relatively higher mRNA signal intensities than did all protein-coding genes that were expressed. Signal intensities probably provide an approximation of transcript abundance (Affymetrix, technical note, 2001) and therefore the high signal intensities observed provide independent evidence that the protein list contains few false positives. The proteins detected include most of those identified on 2D gels from cells grown under a variety of conditions (refs. 1–5; O.P., unpublished data) and increase by almost 4-fold the number so identified. They include many more membrane proteins than were detected previously (>14 times as many as deposited in SWISS-2DPAGE as of August 2002). Particularly under the steady-state growth conditions we used, it is proteins that determine the phenotype of an organism, and hence protein profiling provides an important complement to monitoring gene expression at the mRNA level. Given the small amounts of material required for HPLCMS/MS and the rapidity of the analyses, these methods will be exceptionally valuable not only for E. coli and other well studied microorganisms but also for microbes that cannot be cultivated and for mixtures of organisms, as long as their genome sequences are known.
The operons coding for the 1,147 proteins we detected coded for ≈1600 proteins overall, more than one-third of all E. coli proteins. To the degree that the operon annotations are correct, we can infer that all 1,600 proteins are present and we should be able to detect them directly. Although use of a sample enriched in membranes helped with detection of membrane proteins, the missing proteins remained high in membrane proteins. The mean mRNA signal intensity for genes corresponding to the missing proteins was markedly lower than that for genes corresponding to proteins detected. This indicates that detection of the missing proteins will require the use of more material, additional fractionation steps, use of longer, flatter gradients for HPLC/MS experiments, and/or instrumentation with a greater dynamic range. The assessment that an additional 1,200 genes are expressed at the mRNA level (giving a total of 2,800 genes expressed on Affymetrix arrays) remains to be evaluated (see below for problems of interpretation in any case). Even though we have used only a single growth condition, we note in passing that low detection of open reading frames and the low mRNA signal intensities for genes corresponding to them may help to explain why the functions of these proteins were not known when the E. coli genome sequence was published.
Given our detection limit, we estimate that we should have identified reliably proteins that are present at ≥100 copies per cell (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html). We have considered a number of examples in this context (O.P., unpublished work) and here discuss detection of proteins of low abundance. Many E. coli proteins must be expressed at some low level under all growth conditions to allow increases in their expression under appropriate conditions. For example, this is true for products of the lactose catabolic operon, which we did not detect. These proteins are present at only a few copies per cell when E. coli is grown on glycerol but at much higher levels when it is grown on lactose or when expression of the operon is induced with the gratuitous inducer isopropyl β-d-thiogalactopyranoside. Hence, the deduction that these proteins are functionally absent in cells grown on glycerol is valid. By contrast, some Fts proteins, which are required for septation and cell division, are never present in more than a few copies per cell but are essential under all growth conditions (20). Hence the deduction that the Fts proteins we failed to detect (7 of 12, O.P., unpublished work) are functionally absent is not correct. These examples highlight problems of interpretation of negative results. Biological interpretation will be greatly facilitated by the development of methods for determining quantitative differences in the amounts of particular proteins under different growth conditions (21).
Finally, we note that the HPLC-MS/MS protocol we used is expected to detect the most abundant proteins most readily (supplemental data, http://nature.berkeley.edu/~opaliy/papers/Proteomics.html) and that, as noted above, mRNA signal intensity on Affymetrix GeneChips probably provides an approximation of transcript abundance (Affymetrix, technical note, 2001). If these assumptions are correct, our data indicate that there is a positive relationship between protein abundance and transcript abundance during exponential growth of E. coli. Although this initially appears trivial, the explanation(s) are not obvious. Two extreme possibilities are (i) if, in general, all mRNA species are translated with the same frequency, which is known not to be true in individual cases, more abundant transcripts will give rise to more protein; (ii) conversely, if transcripts that are translated most frequently are more resistant to decay, they will be of increased abundance. The observation itself, which is global, should not be confused with the well known fact that E. coli controls the amounts of many of its individual proteins by controlling the amounts of their transcripts (e.g., see example of the lactose catabolic enzymes above). Previous conclusions differ as to whether there is a positive relationship between protein and transcript abundance in other organisms, but these studies included at most several hundred proteins (22–24). Our speculation that there is such a global relationship in E. coli under steady-state growth conditions remains to be tested.
Acknowledgments
We thank Adam Breier for asking about the relationship between protein detection and mRNA signal intensity and David Weiss for information on Fts proteins. This work was supported by National Institutes of Health Grants GM38361 (to S.K.) and GM37537 (to D.F.H.).
Abbreviation: MS/MS, tandem MS.
References
- 1.VanBogelen, R. A., Abshire, K. Z., Pertsemlidis, A., Clark, R. L. & Neidhardt, F. C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaecter, A. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), Vol. 2, pp. 2067–2117. [Google Scholar]
- 2.Champion, K. M., Nishihara, J. C., Joly, J. C. & Arnott, D. (2001) Proteomics 1, 1133–1148. [DOI] [PubMed] [Google Scholar]
- 3.Link, A. J., Robison, K. & Church, G. M. (1997) Electrophoresis 18, 1259–1313. [DOI] [PubMed] [Google Scholar]
- 4.Loo, R. R., Cavalcoli, J. D., VanBogelen, R. A., Mitchell, C., Loo, J. A., Moldover, B. & Andrews, P. C. (2001) Anal. Chem. 73, 4063–4070. [DOI] [PubMed] [Google Scholar]
- 5.Tonella, L., Hoogland, C., Binz, P. A., Appel, R. D., Hochstrasser, D. F. & Sanchez, J. C. (2001) Proteomics 1, 409–423. [DOI] [PubMed] [Google Scholar]
- 6.Hubbell, E., Liu, W.-M. & Mei, R. (2002) Bioinformatics 18, 1585–1592. [DOI] [PubMed] [Google Scholar]
- 7.Liu, W.-m., Mei, R., Di, X., Ryder, T. B., Hubbell, E., Dee, S., Webster, T. A., Harrington, C. A., Ho, M.-h., Baid, J., et al. (2002) Bioinformatics 18, 1593–1599. [DOI] [PubMed] [Google Scholar]
- 8.Selinger, D. W., Cheung, K. J., Mei, R., Johansson, E. M., Richmond, C. S., Blattner, F. R., Lockhart, D. J. & Church, G. M. (2000) Nat. Biotechnol. 18, 1262–1268. [DOI] [PubMed] [Google Scholar]
- 9.Wassarman, K. M., Repoila, F., Rosenow, C., Storz, G. & Gottesman, S. (2001) Genes Dev. 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Florens, L., Washburn, M. P., Raine, J. D., Anthony, R. M., Grainger, M., Haynes, J. D., Moch, J. K., Muster, N., Sacci, J. B., Tabb, D. L., et al. (2002) Nature 419, 520–526. [DOI] [PubMed] [Google Scholar]
- 11.Lipton, M. S., Pasa-Tolic, L., Anderson, G. A., Anderson, D. J., Auberry, D. L., Battista, J. R., Daly, M. J., Fredrickson, J., Hixson, K. K., Kostandarithes, H., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 11049–11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eng, J. K., McCormack, A. L. & Yates, J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989. [DOI] [PubMed] [Google Scholar]
- 13.Basrur, V., Yang, F., Kushimoto, T., Higashimoto, Y., Yasumoto, K., Valencia, J., Muller, J., Vieira, W. D., Watabe, H., Shabanowitz, J., et al. (2003) J. Proteome Res. 2, 69–79. [DOI] [PubMed] [Google Scholar]
- 14.Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., Hunt, D. F. & White, F. M. (2002) Nat. Biotechnol. 20, 301–305. [DOI] [PubMed] [Google Scholar]
- 15.Martin, S. E., Shabanowitz, J., Hunt, D. F. & Marto, J. A. (2000) Anal. Chem. 72, 4266–4274. [DOI] [PubMed] [Google Scholar]
- 16.le Coutre, J., Whitelegge, J. P., Gross, A., Turk, E., Wright, E. M., Kaback, H. R. & Faull, K. F. (2000) Biochemistry 39, 4237–4242. [DOI] [PubMed] [Google Scholar]
- 17.Blattner, F. R., Plunketti, G. I., Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 18.Rost, B., Casadio, R., Fariselli, P. & Sander, C. (1995) Protein Sci. 4, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riley, M. & Labedan, B. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F., Curtiss, R., III, Ingraham, J., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), 2nd Ed., pp. 2118–2202.
- 20.Errington, J., Daniel, R. A. & Scheffers, D. J. (2003) Microbiol. Mol. Biol. Rev. 67, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gygi, S. P., Rist, B., Gerber, S. A., Turecek, F., Gelb, M. H. & Aebersold, R. (1999) Nat. Biotechnol. 17, 994–999. [DOI] [PubMed] [Google Scholar]
- 22.Anderson, L. & Seilhamer, J. (1997) Electrophoresis 18, 533–537. [DOI] [PubMed] [Google Scholar]
- 23.Futcher, B., Latter, G. I., Monardo, P., McLaughlin, C. S. & Garrels, J. I. (1999) Mol. Cell. Biol. 19, 7357–7368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, T. J., Gygi, S. P., Ideker, T., Rist, B., Eng, J., Hood, L. & Aebersold, R. (2002) Mol. Cell. Proteomics 1, 323–333. [DOI] [PubMed] [Google Scholar]
- 25.Soupene, E., van Heeswijk, W. C., Plumbridge, J., Stewart, V., Bertenthal, D., Lee, H., Gyaneshwar, P., Paliy, O., Charernnoppakul, P. & Kustu, S. (2003) J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]