Abstract
During the biosynthesis of the vancomycin-class antibiotic chloroeremomycin, TDP-epi-vancosaminyltransferase GtfA catalyzes the attachment of 4-epi-vancosamine from a TDP donor to the β-OHTyr-6 of the aglycone cosubstrate. Glycosyltransferases from this pathway are potential tools for the combinatorial design of new antibiotics that are effective against vancomycin-resistant bacterial strains. These enzymes are members of the GT-B glycosyltransferase superfamily, which share a homologous bidomain topology. We present the 2.8-Å crystal structures of GtfA complexes with vancomycin and the natural monoglycosylated peptide substrate, representing the first direct observation of acceptor substrate binding among closely related glycosyltransferases. The acceptor substrates bind to the N-terminal domain such that the aglycone substrate's reactive hydroxyl group hydrogen bonds to the side chains of Ser-10 and Asp-13, thus identifying these as residues of potential catalytic importance. As well as an open form of the enzyme, the crystal structures have revealed a closed form in which a TDP ligand is bound at a donor substrate site in the interdomain cleft, thereby illustrating not only binding interactions, but the conformational changes in the enzyme that accompany substrate binding.
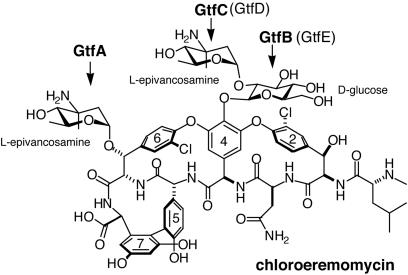
The glycopeptide antibiotics of the vancomycin family are clinically important for the treatment of Gram-positive bacterial infections. These natural products are derived from an oxidatively crosslinked heptapeptide scaffold, which is differentially glycosylated (Fig. 1). The number, identity, and position of these sugar moieties influence bioactivity, both by increasing solubility and by mediating important binding interactions. Glycosylation of the aglycone core is performed in the final stages of biosynthesis by a series of structurally homologous, dedicated glycosyltransferase (Gtf) tailoring enzymes, using an NDP-hexose substrate as a sugar donor (1).
Fig. 1.
Structure and glycosylation pattern of chloroeremomycin. The Gtfs catalyzing sugar attachments are shown in bold; in parentheses are the corresponding Gtfs from the biosynthesis of vancomycin, which has only the disaccharide attachment at residue 4.
The emergence of vancomycin resistance has created an urgent need for novel antibiotics active against resistant bacterial strains. Modification of the carbohydrate moieties on vancomycin has been shown to overcome resistance (2, 3), possibly through different modes of action (3). Because of the complex chemistry involved, vancomycin analogs are prohibitively difficult to produce synthetically. However, the Gtfs from the natural biosynthetic pathways are promising tools for the combinatorial design of new antibiotics with enhanced potency or novel activity by reprogramming them to accept alternative aglycone scaffolds or NDP-sugar donors. Previous studies have demonstrated that some Gtfs from the vancomycin and chloroeremomycin pathways can effectively use nonnatural substrates to generate new compounds (1, 4, 5). Moreover, if the structural determinants of substrate specificity can be understood, the directed genetic modification of these enzymes could be exploited to yield a broader diversity of products.
We previously reported the x-ray crystal structure of the first of these enzymes, GtfB (6). This structure established that the antibiotic Gtfs are members of the structurally homologous GT-B glycosyltransferase superfamily. Only two other GT-B Gtfs are known: UDP-N-acetylglucosaminyltransferase MurG from Escherichia coli, which transfers N-acetyl glucosamine to the peptidoglycan precursor during cell wall biosynthesis (7, 8), and the more distantly related β-glucosyltransferase from T4 phage (BGT), which glucosylates hydroxymethyl cytosine of DNA (9, 10). The GT-B topology is highly conserved between all three GT-B Gtfs, despite having almost no similarity in amino acid sequence. The bidomain architecture of these enzymes, in which the N- and C-terminal domains appear to be independently responsible for the recognition and binding of the acceptor and donor substrates, respectively, suggests that the development of chimeric enzymes may be yet an additional approach to yield new antibiotic products.
Here we report the structural analysis of a second enzyme from this antibiotic Gtf subfamily, the TDP-4-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway, which transfers 4-epi-vancosamine from TDP-epi-vancosamine to the β-OH group of residue 6 of the heptapeptide core (Fig. 1). The structure of the GtfA was determined as a complex with either vancomycin or the monoglycosylated cosubstrate desvancosaminyl vancomycin (DVV) and provides the first direct observations of acceptor substrate binding in the GT-B superfamily. The enzyme was observed in both an open conformation with an empty NDP-sugar binding site, as well as in a closed conformation with the site occupied by an unanticipated nucleotide diphosphate ligand.
Materials and Methods
Protein Expression and Purification. The GtfA gene from the chloroeremomycin producer Amycolatopsis orientalis A82846 was cloned into a pET22b expression vector, adding a C-terminal LEHHHHHH tag, and transformed into E. coli expression strain BL21(DE3). Protein from the lysed cells was purified by using His·Bind (Novagen), Q-Sepharose ion exchange, and gel filtration columns. The protein was concentrated to 10 mg/ml in a storage buffer of 25 mM Tris·HCl buffer, pH 8, with 150 mM NaCl. Selenomethionine (Se-Met)-labeled GtfA was overexpressed as described (11) and purified by elution from a Qiagen (Valencia, CA) Ni-NTA column, followed by anion exchange chromatography using a High-trap Q column, with 2 mM 2-mercaptoethanol included in all buffers. The GtfA from the balhimycin producer Amycolatopsis mediterranei DSM5908 (12), bGtfA, was subcloned, overproduced, and purified as described in the purification of GtfA.
GtfA Assays. In a total volume of 40 μl, 0.5 mM pure TDP-l-4-epi-vancosamine prepared via epivancosamine biosynthesis pathway (13) and either 0.5 mM vancomycin aglycone, vancomycin pseudoaglycone, or vancomycin were incubated with 75 mM Tricine, pH 9/2.5 mM TCEP/2.5 mM MgCl2/1 mg/ml BSA, and either with 5 μM bGtfA or no enzyme. The samples were incubated overnight at 30°C and quenched at specified times with 9 volumes of cold methanol. The samples were analyzed by HPLC, and product peaks were analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry.
Crystallization and Data Collection. Crystals of GtfA were grown in the presence of either ≈1 mM DVV or vancomycin, by using the hanging drop method; the drop was formed by mixing equal parts protein solution and a reservoir solution of 1.3 M sodium potassium phosphate, pH 6.1. The hexagonal rods belong to space group P3221 (a = b = 152.5 Å, c = 98.7 Å) and have two GtfA molecules in the asymmetric unit (≈70% solvent content). Isomorphous crystals of Se-Met-labeled GtfA were grown in the presence of vancomycin without modification of native enzyme protocols. Crystals were transferred in a slow, step-wise manner into a solution of 1.5 M sodium potassium phosphate, pH 6.1, with 30% glycerol added as a cryo-protectant, and then flash-frozen in liquid propane for data collection. Multiwavelength anomalous x-ray diffraction data to 3.6-Å resolution were collected from Se-Met-labeled GtfA crystals at the Structural Biology Center beamline 19-ID at the Advanced Photon Source, Argonne National Laboratory. Data sets were measured at the selenium K-edge peak (0.9793 Å), inflection point (0.9795 Å), and remote (0.9464 Å) energy wavelengths. For native GtfA complexed with vancomycin, data at 2.7-Å resolution were collected the Advanced Photon Source beamline 5-ID (DuPont–Northwestern–Dow Collaborative Access Team). For GtfA complexed with DVV, data at 2.8-Å resolution were collected at beamline X-25 at the National Synchrotron Light Source, Brookhaven National Laboratory. All data were processed and scaled with HKL/HKL2000 and scalepack software (14).
Structure Solution and Refinement. The GtfA structure was solved by Se-Met multiwavelength anomalous dispersion methods. solve/resolve 2.0 (15) was used to determine all 14 selenium positions for two protein molecules in the asymmetric unit, producing initial phases at 3.8-Å resolution having an overall figure of merit of 0.68. The program dm (16) from the CCP4 crystallographic program suite (17) was used to improve the phases by solvent flattening and histogram mapping techniques, which yielded a final figure of merit of 0.84. By using the structure of the homologous GtfB enzyme as a guide, models were built for the two independent GtfA molecules, using chain interactive graphics software (18). The model was partially refined at 3.6-Å resolution against the remote wavelength data set with cnssolve 1.1a (19). The resulting atomic model of Se-GtfA was then used to analyze the higher resolution data sets of native GtfA complexes with DVV and vancomycin. The latter structures were similarly refined by simulated annealing in early stages, followed by positional refinement. As refinement progressed, individual B factors were refined, and water molecules were gradually added to the models. Statistics of data collection and structure refinement for the DVV and vancomycin complexes are summarized in Table 1.
Table 1. Statistics of crystallographic analysis.
| DVV | Vancomycin | |
|---|---|---|
| Resolution, Å | 30.0-2.8 | 30.0-2.8 |
| Completeness, % | 99.2 | 98.3 |
| Average I/σ(I) | 15.1 | 12.6 |
| Rsym* (highest shell) | 8.1 (29.5) | 7.2 (23.6) |
| Reflections | 31,308 | 30,058 |
| Protein atoms | 5,648 | 5,593 |
| Ligand atoms | 207 | 126 |
| Waters | 133 | 49 |
| Rcryst† | 21.3 | 22.8 |
| Rfree‡ | 25.0 | 27.4 |
| 〈B〉, Å2 | 44.2 | 64.2 |
| rmsd bonds, Å | 0.008 | 0.007 |
| rmsd angles, ° | 1.39 | 1.40 |
Rsym = Σ|I- 〈I〉|/ΣI, where I is the observed intensity of a measured reflection, and 〈I〉 is the mean intensity of that reflection.
Rcryst = Σ|Fob - Fcalc|/Σ|Fob|.
Rfree = R factor calculated for 5% of data omitted from refinement calculations.
Results and Discussion
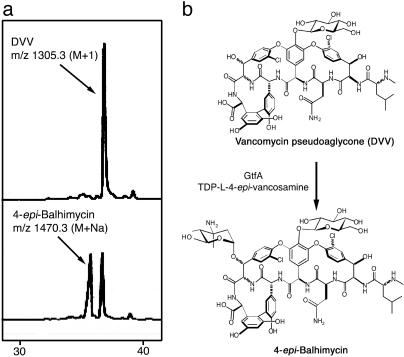
Enzyme Activity. Previously, GtfB was established as the first tailoring enzyme in chloroeremomycin biosynthesis; however, the order of the remaining two glycosylation reactions was unknown. bGtfA, sharing 71% sequence identity with chloroeremomycin GtfA, was used to catalyze the transfer of l-4-epi-vancosamine to the β-OH of Tyr-6 of the aglycone substrate. Incubation of bGtfA with DVV and enzymatically derived TDP-β-l-4-epi-vancosamine (13) resulted in the production of a new peak visible by HPLC after an overnight incubation (Fig. 2). The observed mass for this peak ([M+ Na+] = 1,470.3) correlated with the mass predicted for the compound 4-epi-balhimycin ([M+ H+] = 1,448.4). No activity toward vancomycin was detected, suggesting that GtfA is the second tailoring enzyme in the series and acts specifically on the monoglucosylated intermediate, DVV. The addition of GtfC completed reconstitution of chloroeremomycin biosynthesis by GtfA, B, and C from the vancomycin aglycone (W.L., D. Kahne, and C.T.W., unpublished results).
Fig. 2.
GtfA catalyzes the addition of 4-epi-vancosamine to the DVV substrate. (a) HPLC traces show the production of 4-epi-balhimycin only in the presence of bGtfA. The assay condition is described in Materials and Methods. (b) Schematic representation of the conversion of DVV to 4-epi-balhimycin.
Overall Structure. The atomic structure of GtfA complexed with either DVV or vancomycin is nearly identical. The refined models include residues 1–391, with a disordered segment, residues 316–322, in one of two molecules found in the crystallographic asymmetric unit. As expected, GtfA (Fig. 3) displays the same fold observed previously for GtfB and the other GT-B glycosyltransferases. The N- and C-terminal domains are comprised predominantly of a Rossmann fold motif, a core structure of six parallel β-sheets alternating with connecting α-helices, which is commonly associated with di- and mononucleotide binding. The interface between the two domains forms a deep cleft and creates the active site.
Fig. 3.
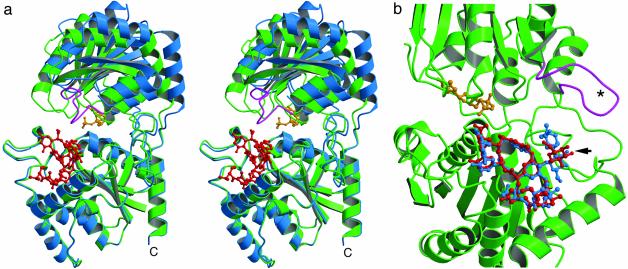
(a) Stereoview comparing the open (blue) and closed (green) forms of GtfA, with bound TDP (gold) and DVV (red) as observed in closed conformation. The flexible loop, unobserved in open form, is shown in magenta. (b) An expanded view looking into cleft of closed form (molecule B). Positions of bound DVV (red) and vancomycin (blue) are superimposed, highlighting the altered binding at the glycosylation site (arrow). The figure was prepared by using molscript (27) and raster3d (28).
Within the asymmetric unit, the two independent GtfA molecules reveal different conformational and ligand-binding states. Molecule A displays a more open conformation (Fig. 3a) and has a well ordered DVV ligand bound on the surface of the N-terminal domain. Molecule B adopts a more closed conformation and binds a nucleotide diphosphate ligand at the interdomain cleft (Fig. 3b), even though no such ligand was added during crystallization. A slightly less well resolved DVV molecule also occupies the site in the N-terminal domain of molecule B. The open and closed enzyme conformations differ almost exclusively by a rigid body hinge motion of ≈10°. The pivot points for the rigid body rotation are localized at residue 204 of the interdomain linker peptide and residue 373 of the long C-terminal tail.
Superimposing the N-terminal domains of molecules A and B yields a rms deviation (rmsd) of only 0.52 Å. The C-terminal domains superimpose with a rmsd of 1.47 Å; however, this is due almost entirely to a large shift in a single loop comprised of residues 259–269 (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). When this loop is disregarded, the rmsd between the C-terminal domains drops to 0.52 Å. The structures of the individual domains thus remain nearly identical in the presence and absence of ligands (discussed below in more detail).
Acceptor-Binding Site. The glycopeptide DVV binds on a highly exposed surface of the N-terminal domain (Fig. 4); the cleft closure seen in molecule B results in no additional protein–substrate interactions. However, in the closed form, the ligand is less well ordered, suggesting that the binding site is partially occupied. This difference in the binding site occupancy can be explained by a difference in crystal packing. In molecule A, the DVV is stabilized by close crystal contacts. In the closed form (molecule B), the site is freely accessible to a large solvent channel running the length of the crystal lattice, explaining why the occupancy of the site is particularly sensitive to changes in DVV concentration during crystal handling.
Fig. 4.
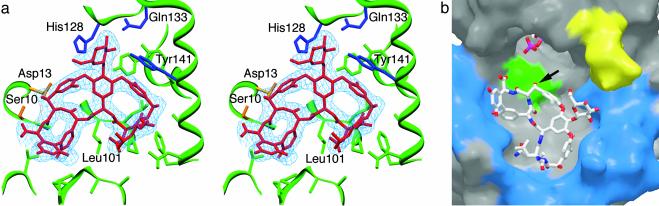
(a) Stereoview showing difference electron density (3σ contour level) for DVV bound to molecule A. Highlighted are the putative catalytic residues Ser-10 and Asp-13 (in gold) and the side chains binding glucose moiety (in blue). (b) Molecular surface view of the GtfA ternary complex (molecule B). DVV binds against surface of N-terminal domain with the attacking hydroxyl (arrow) pointed toward the β-phosphate of TDP. The relative positions of the potential catalytic residues (green), hypervariable regions in the N-terminal domain (blue), and poorly ordered loop in the C-terminal domain (yellow) are shown. The image in a was prepared by using setor (29). The image in b was prepared by using spock (30) and raster3d (28).
GtfA binds the cup-shaped aglycone core similarly for both DVV and vancomycin, with the ligands' convex surface packing against a shallow hydrophobic pocket on the N-terminal domain (Fig. 4b). The floor of the pocket is formed by the connecting peptide turn between the fourth β-strand (Nβ4) and helical (Nα4) elements of the Rossmann motif, which includes the hydrophobic side chains of Leu-101, Leu-102, and Pro-103. Flanking this turn, the more elaborated connecting loops following strands Nβ3 (residues 57–72) and Nβ5 (125–159) create a broad binding surface lined by hydrophobic residues (Val-57, Pro-68, Pro-69, Gly-70, Ala-71, Gly-144, Ala-145, Leu-148, Phe-149, and Tyr-141). Among these, the Tyr-141 phenyl ring forms a parallel stacking interaction with the substrate chlorotyrosine moiety at residue 2.
A few hydrogen bonds also contribute to substrate binding. Notably, the reactive hydroxyl group at residue 6 of the substrate is in good hydrogen bonding distance to the GtfA side chains of both Ser-10 and Asp-13. A potential hydrogen bond is also observed between the Asn-3 side chain of DVV and the carbonyl oxygen of GtfA Gly-70. Finally, the DVV main chain carbonyl groups of residues 3 and 6 hydrogen bond to the GtfA main chain amide groups of Leu-102 and Ser-10, respectively. These latter interactions slightly distort the largely rigid heptapeptide core, when compared with the crystal structure of the free vancomycin dimer (20).
The binding modes of DVV and vancomycin differ markedly in the region of the carbohydrate attachment (Fig. 3b). The single glucosyl moiety of DVV binds in a subsite formed by residues 128–141, near the edge of the interdomain cleft (Fig. 4a). The hexose O2 hydroxyl group is buried in the enzyme, hydrogen bonding to the side chains of Tyr-141 and Gln-133, the latter of which interacts with the O3 hydroxyl group as well. The O6 hydroxyl is directed toward the solvent, but is stabilized by a hydrogen bond with the adjacent His-128 side chain. In the vancomycin complex, the larger disaccharide cannot be accommodated in same manner. The glucosyl moiety is rotated by 180°, which thus projects the O2 hydroxyl and attached vancosamine sugar into the solvent, but buries the O6 hydroxyl within the enzyme subsite. In this orientation, the ligand makes different hydrogen bonds with enzyme, which induces minor shifts in the surrounding protein structure. Because the vancosamine sugar of vancomycin is entirely exposed in the surrounding solvent, no stabilizing interactions with the protein are made.
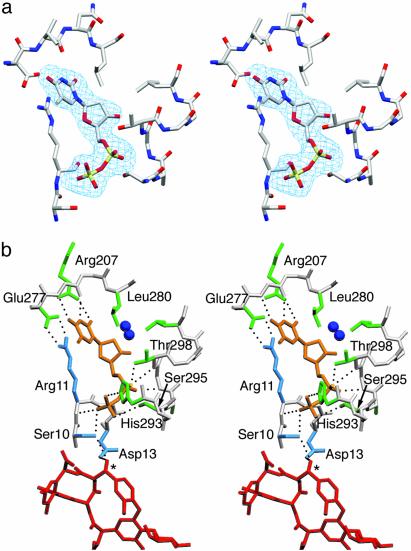
TDP-Binding Site. In molecule B, clear electron density indicates that an unidentified nucleotide diphosphate ligand with a pyrimidine base is bound at the interdomain cleft (Fig. 5a). Because this compound was not added to the crystallization buffer, we conclude that this species originated in the expression system and remained tightly bound to the enzyme throughout protein purification. The homologous BGT and MurG enzymes bind UDP-Glc and UDP-GlcNAc, respectively, in an analogous position (8, 10). Thus, the natural TDP-epi-vancosamine substrate for GtfA would be also expected to bind to this site.
Fig. 5.
(a) Difference electron density (3σ level) for TDP in the interdomain cleft. (b) Binding interactions of TDP (gold) with residues of the N-terminal (cyan) and C-terminal (green) domains including the 292HHXXAGT298 loop. Ribose moiety interacts only via buried water molecules (blue spheres). Interdomain aromatic capping interaction between Arg-11 and Glu-277 and position of attacking hydroxyl (asterisk) of the bound DVV (red) are also shown. The images were prepared by using setor (29).
Although the resolution of the x-ray data is insufficient for a conclusive identification of the base, the electron density is consistent with a thymine moiety. TDP was used to model the ligand because no defined electron density is observed for an attached sugar. However, it remains possible that the species is in fact a mixture of TDP/TDP-sugar species, or that a sugar moiety displays multiple conformations or disorder. Supporting this latter possibility, multiple conformations have been previously observed for the carbohydrate portion of UDP-GlcNAc in the MurG complex (8).
Binding interactions for the TDP ligand are shown in Fig. 5b. The closed conformation of GtfA allows the pyrophosphate moiety to interact directly with the N termini (positive dipole) of α-helices from both domains. The α phosphate is positioned near the Cα4 helix, making hydrogen bonds with two residues in the 292HHXXAGT298 loop, a highly conserved motif in the antibiotic Gtfs (6). Here, Gly-297 and Thr-298 bind the ligand via the main chain amide groups and the Thr side chain hydroxyl. Interestingly, a sulfate anion observed in the GtfB crystal structure (6) binds in nearly the identical position. The β phosphate interacts with the N-terminal domain, making hydrogen bonds with the Arg-11 and Gly-12 amide groups of the Nα1 helix, as well as the Ser-10 side chain hydroxyl oxygen. Additional hydrogen bonds are provided by the Ser-230 amide and side chain hydroxyl group, as well as by the side chains of His-293 and Ser-295. All residues in GtfA that interact with the pyrophosphate are conserved among antibiotic Gtf enzymes, with the exception of Ser-295. The 292HHXXAGT298 loop is a variant of the Gly-rich Rossmann nucleotide-binding motif; residue 295 is generally a conserved glycine in the Gtf subfamily and in MurG, (6).
The thymine base of TDP apparently hydrogen bonds to both main chain amide and carbonyl groups of Va 278, as well as the Arg-207 side chain. Equivalent main chain interactions were reported in the binary complexes of both BGT and MurG. The thymine ring is sandwiched between the Leu-280 side chain and an ion pair formed by the Glu-277 and Arg-11 side chains, which closes over the ring in a parallel stacking arrangement (Fig. 5). The Glu-277-Arg-11 interaction, involving highly conserved residues, is analogous to the aromatic “capping” by Phe observed in the MurG complex with UDP-GlcNAc (8). The TDP ribose moiety has no direct interactions with the protein: the single hydroxyl group hydrogen bonds to two buried water molecules.
Although a donor sugar is not observed in the GtfA crystal structure, its likely position can be approximated by comparison to the MurG complex with UDP-GlcNAc. The sugar moiety would be positioned in the interdomain cleft near the reactive hydroxyl of the bound aglycone, where the side chains of Ser-231 and Ser-295 could provide potential hydrogen bonding interactions with the hexose. In the crystal structure, Ser-295 hydrogen bonds the β phosphate of TDP and is directed away from the presumed sugar position. However, this side chain is often substituted by a Gly in closely related homologs, and thus may not be essential for phosphate binding. Ser-295 may instead rotate to interact with the donor sugar moiety when present. Supporting its possible role in sugar binding, Ser-231 and Ser-295 are conserved in GtfA and GtfD, both of which transfer either vancosamine or epi-vancosamine, but are not conserved in the GtfB and GtfE homologs, which are specific for glucose.
In the MurG complex with UDP-GlcNAc, two consecutive Gln side chains located on the loop between the Cβ5 strand and Cα5 helix make direct binding interactions with sugar moiety (8). Among the wider family of antibiotic Gtfs and in all enzymes from the vancomycin class biosynthetic pathways, these residues are highly conserved as an Asp-Gln sequence. In the crystal structure of the GtfB homolog, this loop adopts a similar conformation to that in the MurG complex, with both Asp and Gln side chains in good position to interact with the donor sugar (6). In the GtfA crystal structure, this loop, corresponding to residues 314–324, is poorly ordered and extends away from the putative sugar subsite (Fig. 3). However, the presence of the donor sugar could induce a similar closure of this flexible loop and allow binding interactions that may help to properly orient the substrate in the true catalytic ternary complex. Interestingly, the GtfA isoforms are unique among closely related Gtfs in having a 4-aa (NVVE) insertion between the conserved Asp-317 and Gln-322 residues. This insertion may be necessary to accommodate the adjacent carbohydrate constituent of substrate DVV, and to possibly assist in its binding. In this case, the inserted Glu-321 may now assume the binding role otherwise provided by the conserved Asp.
Conformational Changes on TDP Binding. Clearly, binding of the donor substrate triggers the closure of the GtfA cleft, thereby allowing the pyrophosphate to interact with both N- and C-terminal domains simultaneously. Both BGT and MurG undergo a similar change from open to closed conformations on ligand binding as well (8, 9). However in these enzymes, the cleft does not close as tightly, and the bound nucleotide ligands interact only with the C-terminal domain. In GtfA, closure of the cleft introduces direct interdomain interactions across the cleft, which may serve to further stabilize the catalytically active conformation. Among these interactions is the ion pair formed between the side chains of Arg-11 and Glu-277, which covers the bound TDP thymine base (Fig. 5b). This ion pair is part of an intricate hydrogen bonding network, including an additional crossdomain interaction between Glu-45 and Arg-207, as well as bonding with the side chains of Glu-15, Arg-42, and Asp-205, residues that are all conserved among the closely related glycopeptide Gtfs.
Although the structure of the C-terminal domain remains largely unchanged between the open and closed conformations, a large shift in position by up to 6 Å is observed for residues 259–269. This flexible loop is located between the Cβ2 and Cβ3 strands of the domain's Rossmann fold (Fig. 6). In the open conformation, some stabilizing intraloop hydrogen bonds are observed, including a β-hairpin type interaction between the Gly-259 carbonyl and Asp-262 amide groups, but the loop is not stabilized with respect to rest of protein structure. Moreover, the Trp-260 side chain hydrogen bonds to the Glu-277 side chain, but otherwise is highly exposed in the surrounding solvent. In the closed conformation, the loops swings dramatically, which, along with a rotation of the Trp-260 side chain, serves to bury the indole ring between loops 57–60 and 229–231 of the N- and C-terminal domains, respectively. In doing so, all hydrogen bonds within the loop are lost, and the Glu-277 side chain is consequently free to make the salt bridge with Arg-11. The shift of loop 259–269 also requires a concerted shift in the adjacent loop 229–231 by 2 Å, bringing the Ser-230 side chain in a suitable position to hydrogen bond with the bound NDP, as well as the Arg-11 amide group.
Implications for Catalysis and Substrate Recognition. Little is understood about the catalytic mechanism of the antibiotic Gtfs or any other member of the GT-B enzyme superfamily. For MurG (21) and the related Gtf OleD, from the biosynthetic pathway of oleandomycin (22, 23), catalysis is reported to proceed via a compulsory ordered Bi-Bi mechanism. The nucleotide-sugar donor appears to binds first to MurG, whereas the macrolide acceptor binds first for OleD. However, the current GtfA crystal structure reveals that for this enzyme binding of the two substrates can occur independently to create a glycopeptide acceptor binary complex or a nucleotide donor complex where the occupancy of the acceptor site can be varied.
The essential catalytic residues have not been definitively identified in GtfA or the other closely related Gtfs. According to the mechanism proposed for other GT-B enzymes, a general base initiates catalysis by abstracting a proton from the reactive aglycone hydroxyl group; a direct nucleophilic attack by the reactive hydroxyl then occurs at the donor sugar C1 carbon center (24). Inspection of the GtfA complexes with DVV and vancomycin reveal only two protein residues in an appropriate position to serve as the catalytic base: Ser-10 and Asp-13. Both residues hydrogen bond to the reactive hydroxyl of the bound glycopeptide ligand, and the Ser-10–Arg-11–Asp-13 motif is highly conserved among vancomycin group Gtfs.
For the GtfB enzyme, mutation of Asp-13 did not significantly lower the rate of catalysis (6). However, the His residue corresponding to Asp-13 in GtfA was found to be important for catalysis in MurG (8). The Ser-10 side chain is also a possible candidate for this catalytic role. Although not a typical base, the simultaneous interaction of Ser-10 with the TDP pyrophosphate may impart the greater basic character needed for the hydroxyl to serve this function. Further mutational analysis of both residues, in this or a closely related system, will be important to define their roles in catalysis.
Despite a low overall sequence homology, the GT-B glycosyltransferases share a small number of conserved amino acids, including a common His-X7-Glu motif (25) that overlaps with the 292HHXXAGT298 loop. The His residue in the His-X7-Glu motif has been proposed to be important in stabilizing the leaving group during catalysis (26). In the GtfA complex with TDP, the conserved His-293 side chain makes a hydrogen bond with the nucleotide β phosphate (Fig. 5b), consistent with this proposal. The Glu residue in this motif is proposed to play a role in nucleotide recognition by hydrogen bonding the ribose hydroxyls of the UDP moiety (26). In GtfA and the other Gtfs of the vancomycin class pathways, this residue is not conserved, but is rather substituted by Val or Leu. These substitutions, which offer no hydrogen bonding potential, are likely determinants for the preference for deoxy nucleotides by these enzymes.
Finally, the GtfA complex offers the first direct observation of acceptor cosubstrate binding to the N-terminal domain for a GT-B glycosyltransferase, and provides some initial insights into the structural basis of aglycone recognition and specificity. These structures readily explain the preference of GtfA for the DVV acceptor substrate. Although both DVV and the diglycosylated vancomycin bind successfully to the enzyme, the presence of the second sugar and altered binding mode are likely to obstruct proper closure of the flexible loop, which we predict to occur in the catalytically relevant complex.
For the antibiotic Gtfs as a family, one particularly intriguing question is how these homologous enzymes have evolved to expand their repertoire of glycosylation reactions, not only via the acceptance of different sugar donor and acceptor substrates, but by developing alternative regiospecificities. For example, in chloroeremomycin biosynthesis, GtfC transfers the final aminodeoxy sugar onto the C2-OH of the glucosyl moiety of DVV (Fig. 1), whereas GtfA transfers the same sugar onto β-OH of residue 6 of DVV. Hence, the two structurally homologous enzymes glycosylate spatially distinct sites on the same acceptor with the same sugar.
These enzymes must have distinctly different binding modes for the same ligand. The molecular basis for this behavior lies in the structure of the N-terminal domain. Protein loops 57–75 and 125–152, which form much of the aglycone-binding site in GtfA, are hypervariable regions among this family of enzymes, varying greatly in both size and amino acid sequence (6). How these structural differences may reprogram binding to allow glycosylation at residue 4 of the acceptor peptide is not clear. We previously modeled an aglycone substrate into the unliganded GtfB crystal structure (6), positioned against the same N-terminal domain surface in a manner generally similar to that observed for the GtfA complex. Although the modeled aglycone in the GtfB structure had a quite complementary fit to the observed enzyme conformation, both variable loops, now demonstrated to be critical in defining the shape and chemical character of the binding site, were disordered and unobserved in the GtfB crystal structure. Thus, the true binding mode for the aglycone in GtfB may indeed be more consistent with that seen for GtfA. Further crystallographic analysis of acceptor complexes for other members of this Gtf family will be important in better understanding the determinants underlying substrate recognition and regiospecificity.
Supplementary Material
Acknowledgments
We thank Dr. Stephen Ginell and Dr. Michael Becker at APS SBC and NSLS beamlines, respectively, for their assistance. We thank Daniel Kahne and colleagues (Princeton University, Princeton) for the sample of DVV. Portions of this research were carried out at the National Synchrotron Light Source, Brookhaven National Laboratory (supported by U.S. Department of Energy Contract DE-AC02-98CH10886), DuPont–Northwestern–Dow Collaborative Access Team (DND-CAT) Synchrotron Research Center at the Advanced Photon Source (APS), and the Argonne National Laboratory Structural Biology Center (supported by U.S. Department of Energy Contract W-31-109-ENG-38) at the APS. DND-CAT is supported by E.I. DuPont de Nemours, Dow Chemical Company, National Science Foundation Grant DMR-9304725, and the State of Illinois (IBHE HECA NWU 96). This work was supported by National Institutes of Health Grant 49338 (to C.T.W.) and the Michigan Life Sciences Corridor (R.M.G.).
Abbreviations: Gtf, glycosyltransferase; rmsd, rms deviation; DVV, desvancosaminyl vancomycin; Se-Met, selenomethionine; bGtfA, balhimycin GtfA.
Data Deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB codes 1PN3 and 1PNV).
References
- 1.Solenberg, P. J., Matsushima, P., Stack, D. R., Wilkie, S. C., Thompson, R. C. & Baltz, R. H. (1997) Chem. Biol. 4, 195–202. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez, M., Snyder, N., Zweifel, M., Wilkie, S. C., Stack, D. R., Cooper, R. D. G., Nicas, T., Mullen, D., Butler, T. & Thompson, R. C. (1998) J. Antibiot. 51, 560–569. [DOI] [PubMed] [Google Scholar]
- 3.Ge, M., Chen, Z., Onishi, H. R., Kohler, J., Silver, L. L., Kerns, R., Fukuzawa, S., Thompson, C. & Kahne, D. (1999) Science 284, 507–511. [DOI] [PubMed] [Google Scholar]
- 4.Losey, H. C., Peczuh, M. W., Chen, Z., Eggert, U. S., Dong, S. D., Pelczer, I., Kahne, D. & Walsh, C. T. (2001) Biochemistry 40, 4745–4755. [DOI] [PubMed] [Google Scholar]
- 5.Losey, H. C., Jiang, J., Biggins, J. B., Oberthur, M., Ye, X.-Y., Dong, S. D., Kahne, D., Thorson, J. S. & Walsh, C. T. (2002) Chem. Biol. 9, 1305–1314. [DOI] [PubMed] [Google Scholar]
- 6.Mulichak, A. M., Losey, H. C., Walsh, C. T. & Garavito, R. M. (2001) Structure (London) 9, 547–557. [DOI] [PubMed] [Google Scholar]
- 7.Ha, S., Walker, D., Shi, Y. & Walker, S. (2000) Protein Sci. 9, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu, Y., Chen, L., Ha, S., Gross, B., Falcone, B., Walker, D., Mokhtarzadeh, M. & Walker, S. (2003) Proc. Natl. Acad. Sci. USA 100, 845–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrielink, A., Ruger, W., Driessen, H. P. C. & Freemont, P. S. (1994) EMBO J. 13, 3413–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morera, S., Imberty, A., Aschke-Sonnenborn, U., Ruger, W. & Freemont, P. S. (1999) J. Mol. Biol. 292, 717–730. [DOI] [PubMed] [Google Scholar]
- 11.Gulick, A. M., Schmidt, D. M., Gerlt, J. A. & Rayment, I. (2001) Biochemistry 40, 15716–15724. [DOI] [PubMed] [Google Scholar]
- 12.Pelzer, S., Sussmuth, R., Heckmann, D., Recktenwald, J., Huber, P., Jung, G. & Wohlleben, W. (1999) Antimicrob. Agents Chemother. 43, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., Thomas, M. G., Hubbard, B. K., Losey, H. C., Walsh, C. T. & Burkart, M. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11942–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 15.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowtan, K. (1994) Protein Crystallogr. 31, 34–38. [Google Scholar]
- 17.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 18.Sack, J. S. (1995) CHAIN: Crystallographic Modeling Program Version 7.0 (Baylor College of Medicine, Waco, TX).
- 19.Brunger, A. T. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 20.Schafer, M., Schneider, T. R. & Sheldrick, G. M. (1996) Structure (London) 4, 1509–1515. [DOI] [PubMed] [Google Scholar]
- 21.Chen, L., Men, H., Ha, S., Ye, X.-Y., Brunner, L., Hu, Y. & Walker, S. (2002) Biochemistry 41, 6824–6833. [DOI] [PubMed] [Google Scholar]
- 22.Quiros, L. M. & Salas, J. A. (1995) J. Biol. Chem. 270, 18234–18239. [DOI] [PubMed] [Google Scholar]
- 23.Quiros, L. M., Carbajo, R. J., Brana, A. F. & Salas, J. A. (2000) J. Biol. Chem. 275, 11713–11720. [DOI] [PubMed] [Google Scholar]
- 24.Unligil, U. M. & Rini, J. M. (2000) Curr. Opin. Struct. Biol. 10, 510–517. [DOI] [PubMed] [Google Scholar]
- 25.Kapitonov, D. & Yu, R. K. (1999) Glycobiology 9, 961–978. [DOI] [PubMed] [Google Scholar]
- 26.Ha, S., Gross, B. & Walker, S. (2001) Curr. Drug Targets Infect. Disord. 1, 201–213. [DOI] [PubMed] [Google Scholar]
- 27.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 28.Merritt, E. A. & Murphy, M. E. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 29.Evans, S. V. (1993) J. Mol. Graphics 11, 134–138. [DOI] [PubMed] [Google Scholar]
- 30.Christopher, J. A. (1997) SPOCK: The Structural Properties Observation and Calculation Kit (Texas A&M Univ., College Station).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.