Abstract
Coliphage N4 virion RNA polymerase (vRNAP), the most distantly related member of the T7-like family of RNA polymerases, is responsible for transcription of the early genes of the linear double-stranded DNA phage genome. Escherichia coli single-stranded DNA-binding protein (EcoSSB) is required for N4 early transcription in vivo, as well as for in vitro transcription on super-coiled DNA templates containing vRNAP promoters. In contrast to other DNA-dependent RNA polymerases, vRNAP initiates transcription on single-stranded, promoter-containing templates with in vivo specificity; however, the RNA product is not displaced, thus limiting template usage to one round. We show that EcoSSB activates vRNAP transcription at limiting single-stranded template concentrations through template recycling. EcoSSB binds to the template and to the nascent transcript and prevents the formation of a transcriptionally inert RNA:DNA hybrid. Using C-terminally truncated EcoSSB mutant proteins, human mitochondrial SSB (Hsmt SSB), phage P1 SSB, and F episome-encoded SSB, as well as a Hsmt-EcoSSB chimera, we have mapped a determinant of template recycling to the C-terminal amino acids of EcoSSB. T7 RNAP contains an amino-terminal domain responsible for binding the RNA product as it exits from the enzyme. No sequence similarity to this domain exists in vRNAP. Hereby, we propose a unique role for EcoSSB: It functionally substitutes in N4 vRNAP for the N-terminal domain of T7 RNAP responsible for RNA binding.
Bacteriophage N4 virion RNA polymerase (vRNAP), responsible for transcription of the phage early genes, is injected into the host cell with the phage genome upon infection (1). vRNAP is inactive on linear double-stranded templates but transcribes denatured genomic N4 DNA or promoter-containing single-stranded DNA (ssDNA) with in vivo specificity (2, 3). vRNAP promoters contain a 5- to 7-bp stem, 3-nt loop hairpin, and conserved sequences (3, 4).
In vivo, N4 early transcription requires Escherichia coli DNA gyrase and E. coli ssDNA-binding protein (EcoSSB), which provide an “active” promoter structure (5). Other SSBs (T7 gp 2.5, T4 gp 32, and N4 SSB) fail to stimulate vRNAP transcription in vitro because they destabilize the promoter hairpin (6). vRNAP transcripts generated from ssDNA templates are retained as RNA·DNA hybrids, suggesting that vRNAP cannot displace the RNA product (2). We show that EcoSSB activates vRNAP transcription on ssDNA templates through displacement of the RNA product and, therefore, through template recycling.
The 177-aa EcoSSB polypeptide consists of a 115-aa N-terminal domain responsible for DNA binding and an ≈50-aa Pro- and Gly-rich sequence followed by a 10-aa acidic tail, highly conserved in eubacterial SSBs (7, 8). Human mitochondrial (Hsmt) SSB is similar in sequence and structure to the N-terminal domain of EcoSSB but lacks the conserved C-terminal sequence (9–11). We show that Hsmt SSB did not activate vRNAP transcription although it did not disrupt the promoter hairpin. We determined that the 10 C-terminal amino acids of EcoSSB are essential for activation through the use of C-terminally truncated EcoSSB proteins, Hsmt SSB-EcoSSB chimeras (12), and bacteriophage P1 (13) and F episome-encoded (14) SSBs. The implications of these findings, which suggest a novel role for a ssDNA-binding protein, are discussed.
Materials and Methods
Proteins. vRNAP from N4 virions and N-terminally His-6-tagged mini-vRNAP were purified as described (15). EcoSSB was purchased from Amersham Pharmacia or purified as described below. Hsmt SSB, the truncated EcoSSB proteins, and the Hsmt SSB-EcoSSB chimeric protein were provided by Ute Curth and Claus Urbanke (Hannover Medical School, Hannover, Germany). EcoSSB, P1 SSB, and F SSB were purified as described (16) with modifications. E. coli K-12 RDP268 cells, a AB1157 derivative where the ssb gene has been replaced by a DNA fragment carrying the aphA gene encoding kanamycin and neomycin resistance (17), bearing pRPZ146 expressing EcoSSB, pKAC50 expressing F SSB (16), or pHAL253 expressing P1 SSB (13) were used as the starting material. Cells (1 liter) grown in LB and 50 μg/ml kanamycin, were collected by centrifugation and sonicated in 300 mM NaCl buffer S (20 mM Tris·HCl, pH 8.0/0.2 mM EDTA/2 mM DTT) on ice. The cleared supernatant was loaded onto a 5-ml denatured DNA-agarose column (Pharmacia Biotech); the column was washed with 15 ml of 500 mM NaCl buffer S, and the SSB protein was eluted by 2 M NaCl buffer S. Fractions containing SSB protein were pooled, diluted 10-fold with buffer S, and applied onto a 1-ml HiTrap Q column (Pharmacia Biotech). Concentrated protein was eluted with 500 mM NaCl buffer S, mixed with an equal volume of 100% glycerol, and stored at –20°C. All three proteins were at least 99% pure and nuclease-free.
Runoff Transcription Assay. Reaction mixtures (5 μl) contained 1 mM each ATP, CTP, and GTP, 0.1 mM UTP, 1 μCi of [α-32P]UTP (3,000 Ci/mmol, Amersham Pharmacia), 10 nM vRNAP, and 1 nM DNA template (unless otherwise stated) in buffer A (10 mM Tris·HCl, pH 7.8/10 mM MgCl2/50 mM NaCl/1 mM DTT). Template 12-P2–52 (3′-GAAATTTCTCTTCTTCGAGGCGAAGAAAACCTACTTCATTACGTAGGTTAGGACATACTTTAAGAATTGGAAACAGACCCT-5′, the hairpin inverted repeat and position + 1 are underlined) is a synthetic deoxyoligonucleotide containing 12-nt upstream of the promoter hairpin, the N4 early promoter P2, and 52 nt of transcribed region. Where indicated, no labeled NTP was added and 5′ 32P-labeled DNA template was used. EcoSSB, Hsmt SSB, and SSBs deletions or chimeras were present at concentrations specified in the figure legends. Reactions were incubated at 37°C for 5 min unless otherwise stated, terminated by addition of 7 μl of stop solution (95% formamide/20 mM EDTA/0.05% bromophenol blue/0.05% xylene cyanol), and analyzed by electrophoresis on 8 M urea/8% polyacrylamide gels and phosphorimaging. Derivatives of template 12-P2–52 used to determine the effect of template and/or product length on EcoSSB activation contained no sequences upstream of the P2 hairpin and 24 (0-P2–24), 34 (0-P2–34), 40 (0-P2–40), or 52 (0-P2–52) nt of transcribed sequence. Template 34-P2–24 contains 22 additional nucleotides of upstream sequence (3′-GGCTATCAAGTTATTGATAGAT). All sequences were derived from the N4 genome.
Analysis of the State of the Template and RNA Product by Native Gel Electrophoresis. Reactions were carried out as described above. Some of the samples were treated with 0.2 units/μl RNase H or 1 unit/μl Nuclease S1 (GIBCO/BRL) before analysis. After addition of glycerol to 10%, 5-μl aliquots were analyzed by electrophoresis at 4°C on 6% polyacryamide (80:1) gels (50 mM Tris base/380 mM glycine/2 mM EDTA, pH 8.5/2.5% glycerol), and autoradiography.
DNase I Protection Assays. Reaction mixtures (5 μl) contained 50 nM 5′ 32P-labeled DNA in buffer A. EcoSSB or Hsmt SSB (2 μM final concentration) were added, and the mixtures were incubated on ice for 10 min and treated for 15 min at 37°C with DNase I (0.01 units/μl final concentration, GIBCO/BRL). After addition of 7 μl of Stop solution, samples were analyzed by electrophoresis on 8 M urea/8% polyacrylamide gels and autoradiography.
RNA–Protein Crosslinking. Stalled mini-vRNAP elongation complexes (SECs) were prepared in transcription reactions without CTP by using a set of DNA templates containing the P2 promoter and a 23-nt transcribed region that ends in four G residues. Eight variants of this sequence in which an A residue replaced one of the T residues in the transcribed region were used. The sequence of the full-length template is: 3′-CTTCGAGGCGAAGAAAACCTTCTTCTTTTCCTTCCTGGGG-5′ (hairpin inverted repeat and position + 1 are underlined, and the T residue replaced with an A residue at positions +3, +6, +9, +10, +11, +12, +15, or +19, in the variant templates are in bold). Then 1 μM N-terminally His-6-tagged mini-vRNAP, 1 μM DNA oligonucleotide, 2 μM EcoSSB, 0.5 mM 5-IodoUTP (Sigma), 1 mM GTP, 0.1 mM ATP, 2 μCi of [α-32P]ATP (3,000 Ci/mmol, Amersham Pharmacia), and buffer A were mixed in a total volume of 5 μl in a 15-μl well of a polystyrene MicroWell minitray with lid (Nunc) on ice. The minitray was placed on a 37°C metal heat block for 5 min and then on a Spectroline TM-312A UV Transilluminator (Spectronics, Westbury, NY) lid down with an ice pack on top. After irradiation for 40 min at 312 nm (19), the samples were mixed with 5 μl of 2× loading buffer (100 mM Tris·HCl, pH 6.8/200 mM DTT/4% SDS/0.2% bromophenol blue/20% glycerol), heated at 96°C for 2 min, and analyzed by SDS/PAGE and phosphorimaging.
Results
EcoSSB Activates vRNAP Transcription at Limiting ssDNA Template Concentrations. To determine the effect of EcoSSB on vRNAP transcription of ssDNA templates, runoff transcription reactions were carried out on a synthetic oligonucleotide template containing vRNAP early promoter P2 and a 52-nt transcribed region. In the absence of EcoSSB and at the lowest template concentration, RNAP transcription yields three major products: the 52-nt runoff product and two additional products differing at the 3′ end due to nontemplated nucleotide addition (Fig. 1A, lane 1). Longer products are also synthesized (Fig. 1 A, lanes 3 and 5). In the presence of EcoSSB, activation occurs at low template concentrations (compare lanes 1 and 2 to 5 and 6), and the synthesis of longer transcripts is abolished (compare lanes 3 and 5 to 4 and 6).
Fig. 1.
Effect of EcoSSB on N4 vRNAP runoff transcription of the ssDNA template 12-P2–52. (A) Where indicated, EcoSSB and vRNAP were present at 1 μM and 10 nM, respectively. (B) A 100-μl complete transcription mixture containing 1 nM template but no EcoSSB was divided into four aliquots (I, II, III, and IV) before incubation at 37°C. I, no additions; II, 50 nM template was added after 20 min incubation; III, 1 μM EcoSSB was added before incubation; and IV, 1 μM EcoSSB was added after 20 min of incubation. At the indicated times, 5-μl aliquots were analyzed.
We have previously shown that the product of vRNAP transcription on ssDNA templates is retained in an RNA·DNA hybrid (2). Therefore, we considered that EcoSSB activates transcription at limiting template concentrations through recycling of the template. To explore this possibility, we determined the time course of RNA synthesis in three reactions at low template concentration. In the absence of EcoSSB (Fig. 1B, I), no further RNA synthesis was observed beyond 20 min of incubation. This effect was due to template limitation because RNA synthesis resumed on further addition of template (Fig. 1B, II). In the presence of EcoSSB, the rate of RNA synthesis was constant during the course of the reaction, indicating that template limitation was overridden by the presence of EcoSSB (Fig. 1B, III). The possibility that template degradation might occur in the absence of EcoSSB was ruled out because EcoSSB activated transcription after RNA synthesis had reached a plateau (Fig. 1B, IV). The slow rate of RNA synthesis after EcoSSB addition reflects the rate of melting of the RNA·DNA hybrid, as determined by the change in sensitivity of the isolated 32P-RNA·DNA hybrid to RNase H and nuclease S1 upon EcoSSB addition (not shown). These results indicate that EcoSSB prevents formation of an RNA·DNA hybrid during transcription.
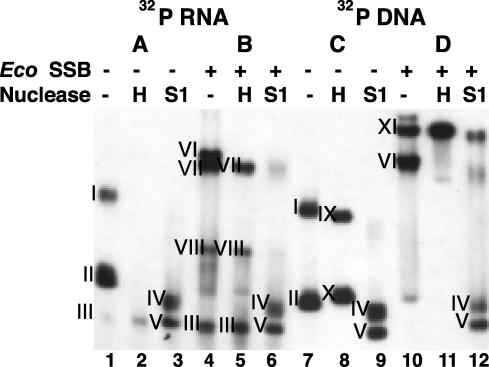
EcoSSB Activates Transcription by Binding to the Template and the RNA Product. To determine whether EcoSSB prevents the formation of an RNA·DNA hybrid, we analyzed the physical state of the template and the transcript synthesized in the absence or presence of SSB by native gel electrophoresis (Fig. 2). Transcription reactions were carried out with unlabeled template and labeled NTPs (A and B), or with labeled template and unlabeled NTPs (C and D) to analyze the state of the product and template, respectively. Before incubation, 1 μM EcoSSB was added to one-half of each reaction mixture (B and D). At the end of the transcription reaction, each reaction mixture was divided into three aliquots: a control sample, a sample with RNase H added to degrade RNA in RNA·DNA hybrids, and a third sample with nuclease S1 added to degrade single-stranded nucleic acids.
Fig. 2.
Effect of EcoSSB on the state of the DNA template and the RNA product. (A and B) Transcription reactions contained 5 nM unlabeled 12-P2–52 DNA template and [α-32P]UTP. (C and D) Transcription reactions contained 5 nM 5′ 32P-labeled template and unlabeled NTPs. Where indicated, EcoSSB was present at 1 μM. After a 15-min incubation, each mixture was divided into three aliquots and treated for 15 min at 37°C with no additions, RNase H, or Nuclease S1, as indicated, and analyzed by native PAGE.
In the absence of EcoSSB, the majority of the RNA product (Fig. 2 A, lane 1, bands I and II) and the DNA template (Fig. 2C, lane 7, bands I and II) were present in RNA:DNA hybrids as revealed by the disappearance of the labeled RNA (Fig. 2 A, lane 2) and the change in mobility of the labeled template (Fig. 2C, lane 8) after RNase H treatment. Band III, which is barely detected under these conditions, corresponds to free RNA; band X corresponds to template DNA; band IX is not detected after heating and quick cooling of the template before electrophoresis, indicating that it is the product of intermolecular template interaction. Treatment with nuclease S1 yielded the same complexes (bands IV and V) whether the template or transcript was labeled, indicating the presence of RNA·DNA hybrids.
In the presence of EcoSSB, a significant fraction of the RNA was RNase H resistant (compare Fig. 2 A and B, lanes 2 and 5, band VII and below), indicating the absence of a DNA·RNA hybrid. However, a species sensitive to RNase H (Fig. 2B, lane 4, band VI), corresponding to a RNA·DNA·EcoSSB complex intermediate, was present. Under these conditions, the labeled template formed two different complexes with EcoSSB (Fig. 2D, lane 10, bands XI and VI). Complex VI (Fig. 2D, lane 10), contains RNA (B, lane 4) and was sensitive to RNase H treatment, indicating that it is the RNA·DNA:EcoSSB complex intermediate. The lower mobility complex (Fig. 2D, lane 10, band XI), which increased in amount after RNase H treatment and was partially degraded after nuclease S1 treatment, corresponds to a DNA·EcoSSB complex (Fig. 2D, lanes 11 and 12). In all cases, treatment with nuclease S1 resulted in the same S1-resistant complexes (bands IV and V).
We conclude that the RNA product of vRNAP transcription on ssDNA templates remains in an RNA·DNA hybrid, which limits template usage to a single round. In the presence of EcoSSB, both the transcript and the template are present in complexes with EcoSSB.
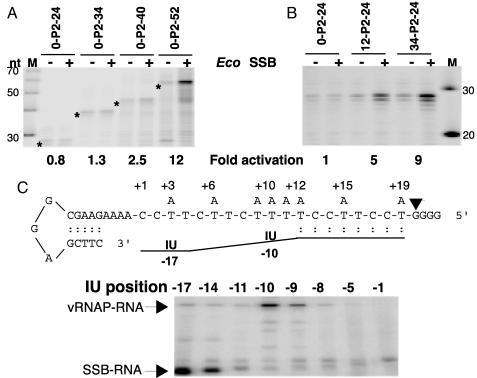
EcoSSB Activation Depends on Template Length, and EcoSSB Interacts with the Nascent Product. EcoSSB exists in solution and binds to ssDNA as a homotetramer (20). Three different stable binding modes, which depend on the type of salt and its concentration and are referred to as (SSB)35, (SSB)56, and (SSB)65, have been defined (21, 22). In the (SSB)35-binding mode only two of the subunits interact with ssDNA, whereas in the (SSB)56- and (SSB)65-binding modes, all four subunits contact ssDNA (21, 23). Similar binding modes have been observed with poly(U) and poly(A) (ref. 24, and W. Bujalowski, L. B. Overman, and T. M. Lohman, unpublished results) although the affinity of EcoSSB for ribopolynucleotides is much lower than for deoxypolynucleotides (25). Based on the nucleic acid binding properties of EcoSSB, we expected the effect of EcoSSB to be dependent on the length of the template or of the transcript. The results of an experiment with templates containing promoter P2 and 24, 34, 40, or 52 transcribed nucleotides are shown in Fig. 3A. EcoSSB had no effect when the transcripts were 24 or 34 nt long, whereas 2.5- and 12-fold activation was detected when the transcripts were 40 and 52 nt long, respectively. These results do not differentiate between the role of the length of the transcript or the template. Therefore, we tested activation on templates where the total length of the DNA was increased by addition of sequences upstream of the promoter. Although no activation was detected on the 0-P2–24 template, activation was observed with templates 12-P2–24 and 34-P2–24 (Fig. 3B), indicating that the length of the DNA template available for EcoSSB binding is a determinant of EcoSSB assisted-template recycling.
Fig. 3.
(A and B) Effect of template length on EcoSBB activation. Runoff transcription reactions contained 1 nM template, 1 μM EcoSSB, and 1 μM mini-vRNAP and were incubated for 10 min at 37°C. (C) Interactions of the nascent transcript with mini-vRNAP and EcoSSB. (Upper) DNA template and nascent RNA arrangement in SEC. DNA templates bearing a single A in the transcribed region at the indicated positions numbered relative to the transcription start site (+1) were used in mini-vRNAP transcription reactions with 5-I-UTP, GTP, and [α-32P]ATP. Arrowhead, position of the active center in the SEC. RNA is represented by a line. IU at positions –10 and –17 of the RNA in SEC is shown. (Lower) Crosslinking of the nascent RNA containing IU at different positions to mini-vRNAP and EcoSSB in SEC. Positions containing IU in nascent RNA of the SEC are numbered relative to the 3′ end (–1). Arrows indicate RNA crosslinked to mini-vRNAP and to EcoSSB. IU, 5-iodo-uridine.
To determine contacts between the nascent transcript and EcoSSB, SECs were synthesized on templates containing a single A at specific positions, no G residues up to position +19, followed by four G residues (Fig. 3C). Transcription reactions were carried out in the presence of 5-Iodo-UTP, [α-32P]ATP, and GTP. SECs were irradiated with UV light, and the proteins were analyzed by SDS/PAGE. Maximal crosslinking to mini-vRNAP occurred when the photoactive ribonucleotide was present 10-nt upstream of the 3′ end of the transcript (Fig. 3C) and was not dependent on the presence of EcoSSB (not shown). Crosslinking to EcoSSB was evident when the analog was present 14 nt and was maximal when present at 17 nt from the 3′ end (Fig. 3C). These results strongly suggest that the transcript binds to EcoSSB as it exits from the enzyme.
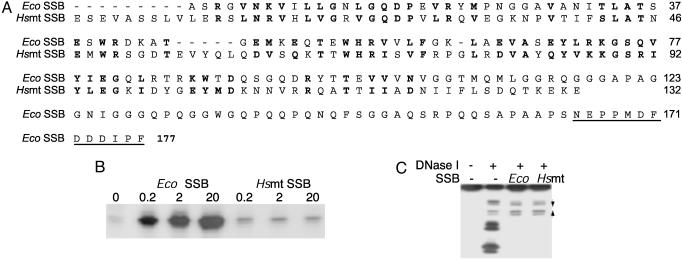
The C-Terminal Region of EcoSSB Is Required for Activation of Transcription. We have previously shown that SSBs such as T4 gp32 (26), fd gpV (27), T7 gp 2.5 (28), and N4 SSB (29) cannot stimulate, but actually inhibit, vRNAP transcription on ssDNA templates because they destabilize the vRNAP promoter hairpin (6). The ssDNA-binding determinants of EcoSSB reside in the 115 N-terminal amino acids (Fig. 4A) (8). The 132-aa Hsmt SSB (30) also binds to ssDNA as a homotetramer (9) and shares significant sequence homology to the DNA-binding domain of EcoSSB (Fig. 4A). Hsmt SSB and EcoSSB have similar DNA-binding properties (9); the structures of the DNA-binding domain of EcoSSB and of Hsmt SSB have been solved (11, 31, 32) and show striking similarities (10). EcoSSB and Hsmt SSB cannot form heterotetramers (12) because of differences in amino acids present in the dimer interface (10). We tested the effect of Hsmt SSB on vRNAP transcription of ssDNA templates. In contrast to EcoSSB, Hsmt SSB did not activate transcription. However, Hsmt SSB did not inhibit transcription even at the highest concentration tested (Fig. 4B), suggesting that that Hsmt SSB does not destabilize the promoter hairpin.
Fig. 4.
Comparison of EcoSSB and Hsmt SSB. (A) Amino acid sequences of EcoSSB and Hsmt SSBs (7, 30). Conserved amino acids are shown in bold. The EcoSSB acidic C terminus is underlined. (B) Effect of Eco and Hsmt SSBs on vRNAP transcription. (C) DNase I footprinting of template DNA with EcoSSB and Hsmt SSB. Converging arrows indicate the promoter hairpin inverted repeat.
To determine the effect of Hsmt SSB binding on promoter structure, we compared the patterns of EcoSSB and Hsmt SSB protection of a 5′ end-labeled promoter-containing DNA from DNase I cleavage (Fig. 4C). Both proteins yielded the same patterns of DNase I cleavage at the hairpin region and of protection on the rest of the fragment. We conclude that the inability of Hsmt SSB to activate vRNAP transcription is not due to destabilization of the promoter hairpin and cannot be easily explained by the mode of Hsmt SSB DNA binding.
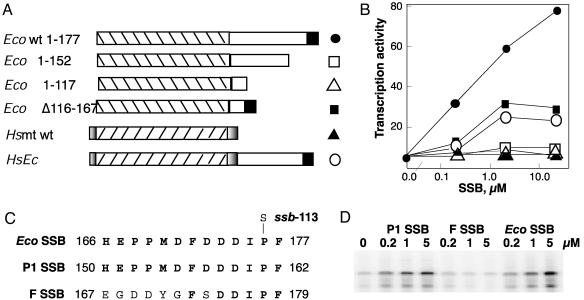
To define the EcoSSB sequence determinants of vRNAP transcript displacement, we used a series of mutant EcoSSBs, with deletions in the C-terminal third of the protein (12). Two of the mutant proteins, EcoI-152 and EcoI-117 (Fig. 5A) that are truncated by 25 and 50 aa, show higher affinity for ssDNA (12). EcoSSB Δ116–167 lacks the Pro- and Gly-rich region but contains the C-terminal 10 aa (Fig. 5A). In addition, we used a chimeric protein (Hsmt-EcoSSB), in which amino acids 113–176 of EcoSSB have been fused to the C terminus of Hsmt SSB (Fig. 5A) (12).
Fig. 5.
Mutant and chimeric SSB proteins and their effect on vRNAP transcription. (A) Schematic representation of mutant and chimeric proteins (12). DNA-binding domains (striped); EcoSSB C-terminal 10-aa region (filled); linker sequence between Eco SSB DNA-binding domain and the C-terminal 10-aa region (open); and Hsmt SSB unique N-terminal and C-terminal sequences (shaded). (B) The effect of mutant and chimeric SSB proteins on vRNAP transcription. Symbols are presented to the right of schematic representation in A. (C) Sequence of the C-terminal 13 aa of the EcoSSB (7), P1 SSB (13), and F SSB (33). (D) Effect of P1 SSB and F SSB on vRNAP transcription. Reaction mixtures containing no SSB or the indicated concentrations of different SSB proteins, 3 nM 12-P2–52 DNA template and 1 μM mini-vRNAP were incubated for 5 min at 37°C, and the products were analyzed on 8 M urea/8% polyacrylamide gels.
We determined the effects of these SSBs on vRNAP transcription of ssDNA templates at three protein concentrations (Fig. 5B). Under the conditions tested, WT EcoSSB showed 20-fold activation of transcription at the highest concentration. Eco Δ116–167 activated to 50% of WT EcoSSB levels. The two C-terminally truncated EcoSSBs, EcoI-152 and EcoI-117, did not activate transcription, suggesting that determinants for transcript displacement are localized to the C terminus of EcoSSB. In support of this idea, the chimeric protein containing the C-terminal one-third of EcoSSB fused to the C terminus of Hsmt SSB was proficient in transcription activation (Fig. 5B). The inability of EcoSSB Δ116–167 and Hsmt SSB-EcoSSB to activate to the same extent as WT EcoSSB is most likely due to their decreased affinity for ssDNA (12).
The DNA-binding domains of P1 SSB and F SSB share 91% and 85% homology, respectively, with the EcoSSB DNA-binding domain (13, 33) and P1 and F SSBs can complement a strain lacking EcoSSB (13, 18). However, although P1 SSB and EcoSSB have identical sequences at their 13 C-terminal amino acids (13), the sequence of F SSB differs in 7 of 13 positions (33) (Fig. 5C). We tested the effect of purified P1 and F SSBs on transcription of vRNAP. P1 SSB was as active as EcoSSB in stimulating vRNAP transcription of ssDNA templates; in contrast, F SSB was inactive (Fig. 5D). These results reinforce our conclusion that the C terminus of EcoSSB is essential for displacement of the RNA product from vRNAP-elongating complexes.
Discussion
N4 vRNAP is unable to displace the RNA product when transcribing ssDNA templates (2). Here, we showed that EcoSSB activates vRNAP transcription of ssDNA templates by binding to the DNA template and the RNA product, preventing formation of the RNA·DNA hybrid, and allowing template recycling. Moreover, activation is dependent on the length of the template and the C-terminal amino acids of EcoSSB.
EcoSSB exists in solution and binds to ssDNA as a homotetramer (20, 21). Although formation of the EcoSSB tetramer is not required for EcoSSB binding to ssDNA, the affinity of the monomer is ≈10-fold lower than that of the promoter (34). The ssb-1 mutation (H55Y) destabilizes tetramer formation (20). We have previously shown that cells carrying the ssb-1 mutation cannot support N4 early transcription, indicating that tetramer formation is required for EcoSSB activation of vRNAP transcription (5). Our results raise three questions: (i) How is EcoSSB recruited to the elongating transcription complex? (ii) Is EcoSSB-assisted template recycling due to binding to the DNA template, to the RNA product or both? (iii) What binding mode is EcoSSB using in the process? Based on the results presented in Fig. 3 A and B, we suggest that EcoSSB is recruited to the transcription complex through interaction with the template in the (SSB)35-binding mode because activation is observed when 36 or more nucleotides are available for EcoSSB binding.
We have presented several lines of evidence indicating that the C-terminal amino acids of EcoSSB are required for template recycling. Interactions between the EcoSSB C terminus and other proteins have been suggested in several cases (reviewed in ref. 35). Direct EcoSSB–protein interactions have been shown with the plasmid homolog of E. coli UmuC, MucB (36), with the χψ subunits of DNA polymerase III holoenzyme (37), and with exonuclease I (38). In the last two cases, the interaction was greatly reduced (39) or abolished (38) by a substitution of Ser for Pro at the penultimate residue (40) (Fig. 5C); although the mutant protein, SSB-113, has slightly increased affinity for ssDNA (41). In contrast, N4 early transcription is not affected by the ssb-113 mutation (5), indicating that, although the EcoSSB C terminus is required, a Pro at position 176 is not essential for EcoSSB-assisted template recycling. Further mutagenesis of the C-terminal 10 aa of EcoSSB will allow us to identify specific residues of EcoSSB required for template recycling.
N4 vRNAP is a 3,500-aa-long polypeptide that lacks extensive sequence similarity to either of the two known families of DNA-dependent RNA polymerases (15). However, vRNAP contains four short motifs (TxxGR, A, B, and C) characteristic of the T7-like RNAP family, which includes phage-, mitochondrial-, some chloroplast nuclear-, and linear plasmid-encoded enzymes (15, 42). We have defined a stable and transcriptionally active 1,106-aa long domain (mini-vRNAP) located at the center of the vRNAP polypeptide, which possesses the same initiation, elongation, termination, and product displacement properties as full-length vRNAP (15). Mutational, biochemical, and phylogenetic analyses indicate that N4 mini-vRNAP is a highly evolutionarily diverged member of the T7 RNAP family (15).
T7 RNAP uses ssDNA templates containing a double-stranded promoter (–17 to +1) (43), and undergoes multiple rounds of transcription, indicating product displacement (43). The structure of T7 RNAP is often compared to a cupped right hand with subdomains designated thumb, palm, and fingers (44). T7 RNAP possesses an N-terminal domain (amino acids 1–325), located in front of the palm/thumb junction, which walls off the front of the catalytic cleft of the enzyme (44). This domain displays RNA-binding ability (45), and also plays a role in transition from synthesis of abortive products to processive elongation (46). Photochemical crosslinking of RNA to T7 RNAP in transcription complexes maps a major contact site to the 5′ end of the transcript to the amino acid 144–168 region and a minor contact site to amino acids 1–93 (45). Moreover, as the RNA is displaced from the RNA·DNA hybrid, it is directed toward a positively charged surface in T7 RNAP, surrounding Lys-302 and Lys-303 (47). The Glu-148 to Ala (E148A) substitution in T7 RNAP leads to premature abortive transcription and defective RNA binding (48). No sequence similarities are detected between the T7 RNAP and N4 mini-vRNAP up to T7 RNAP amino acid 410 (15). Moreover, analysis of the crystal structure of mini-vRNAP reveals the existence of an N-terminal domain with no structural similarities to the T7 RNAP N-terminal domain (K. Murakami, unpublished results). Finally, neither vRNAP nor mini-vRNAP binds to RNA (E.K.D., unpublished results). The results presented in this paper indicate that EcoSSB binds to nascent RNA as it exits from the enzyme. Therefore, we propose that EcoSSB functionally replaces the N-terminal domain in T7 RNAP responsible for RNA binding.
Previously, we have proposed that supercoiling of the template facilitates extrusion of the promoter hairpin required for vRNAP binding and provides a site of invasion for EcoSSB binding to the DNA at the promoter (49). Tethering of EcoSSB through template binding would allow interaction with vRNAP through C-terminal amino acids of EcoSSB. However, thus far, we have been unable to detect vRNAP–EcoSSB interactions by using purified proteins. Therefore we suggest that, upon promoter clearance, vRNAP undergoes an extensive structural rearrangement similar to that observed in T7 RNAP during the transition from initiation to elongation (47, 50, 51). In vRNAP, this event would occur when the transcript is 12-nt long (E.K.D., unpublished results). This structural reorganization within vRNAP will either display or create the site of interaction of the C-terminal region of EcoSSB, resulting in recruitment of EcoSSB to the vRNAP elongation complex and EcoSSB binding to the RNA as it exits from the enzyme. Because EcoSSB is a tetramer, it is possible that it interacts simultaneously with the template and the RNA product. Experiments are in progress to identify the site of EcoSSB interaction on vRNAP in the template/vRNAP/RNA complex.
Acknowledgments
We thank Drs. U. Curth and C. Urbanke for generously providing the EcoSSB mutant proteins and the Hsmt SSB-EcoSSB chimera; R. Porter (Pennsylvania State University, University Park) and H. Lehnherr and J. Bendtsen (Ernst-Moritz-Arndt-University, Greifswald, Germany) for strains; and W. Epstein and T. Lohman for valuable comments on the manuscript. This work was supported by National Institutes of Health Grant AI 12575 (to L.B.R.-D.).
Abbreviations: RNAP, RNA polymerase; vRNAP, virion RNAP; ssDNA, single-stranded DNA; SSB, ssDNA-binding protein; EcoSSB, Escherichia coli SSB; SEC, stalled elongation complex; Hsmt SSB, human mitochondrial SSB.
References
- 1.Falco, S. C., VanderLaan, K. & Rothman-Denes, L. B. (1977) Proc. Natl. Acad. Sci. USA 74, 520–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falco, S. C., Zivin, R. & Rothman-Denes, L. B. (1978) Proc. Natl. Acad. Sci. USA 75, 3220–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haynes, L. L. & Rothman-Denes, L. B. (1985) Cell 41, 597–605. [DOI] [PubMed] [Google Scholar]
- 4.Glucksmann, M. A., Markiewicz, P., Malone, C. & Rothman-Denes, L. B. (1992) Cell 70, 491–500. [DOI] [PubMed] [Google Scholar]
- 5.Markiewicz, P., Malone, C., Chase, J. W. & Rothman-Denes, L. B. (1992) Genes Dev. 6, 2010–2019. [DOI] [PubMed] [Google Scholar]
- 6.Glucksmann-Kuis, M. A., Dai, X., Markiewicz, P. & Rothman-Denes, L. B. (1996) Cell 84, 147–154. [DOI] [PubMed] [Google Scholar]
- 7.Sancar, A., Williams, K. R., Chase, J. W. & Rupp, W. D. (1981) Proc. Natl. Acad. Sci. USA 78, 4272–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams, K. R., Spicer, E. K., LoPresti, M., Guggenheimer, R. A. & Chase, J. W. (1983) J. Biol. Chem. 258, 3346–3355. [PubMed] [Google Scholar]
- 9.Curth, U., Urbanke, C., Greipel, J., Gerberding, H., Tiranti, V. & Zeviani, M. (1994) Eur. J. Biochem. 221, 435–443. [DOI] [PubMed] [Google Scholar]
- 10.Webster, G., Genschel, J., Curth, U., Urbanke, C., Kang, C. & Hilgenfeld, R. (1997) FEBS Lett. 411, 313–316. [DOI] [PubMed] [Google Scholar]
- 11.Yang, C., Curth, U., Urbanke, C. & Kang, C. (1997) Nat. Struct. Biol. 4, 153–157. [DOI] [PubMed] [Google Scholar]
- 12.Curth, U., Genschel, J., Urbanke, C. & Greipel, J. (1996) Nucleic Acids Res. 24, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehnherr, H., Bendtsen, J. D., Preuss, F. & Ilvyna, T. V. (1999) J. Bacteriol. 181, 6463–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolodkin, A. L., Capage, M. A., Golub, E. I. & Low, K. B. (1983) Proc. Natl. Acad. Sci. USA 80, 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazmierczak, K. M., Davydova, E. K., Mustaev, A. A. & Rothman-Denes, L. B. (2002) EMBO J. 21, 5815–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohman, T. M., Green, J. M. & Beyer, R. S. (1986) Biochemistry 25, 21–25. [DOI] [PubMed] [Google Scholar]
- 17.Porter, R. D., Black, S., Pannuri, S. & Carlson, A. (1990) Bio/Technology 8, 47–51. [DOI] [PubMed] [Google Scholar]
- 18.Porter, R. D. & Black, S. (1991) J. Bacteriol. 173, 2720–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willis, M. C., Hicke, B. J., Uhlenbeck, O. C., Cech, T. R. & Koch, T. H. (1993) Science 262, 1255–1257. [DOI] [PubMed] [Google Scholar]
- 20.Williams, K. R., Murphy, J. B. & Chase, J. W. (1984) J. Biol. Chem. 259, 11804–11811. [PubMed] [Google Scholar]
- 21.Lohman, T. M. & Overman, L. B. (1985) J. Biol. Chem. 260, 3594–3603. [PubMed] [Google Scholar]
- 22.Bujalowski, W. & Lohman, T. M. (1986) Biochemistry 25, 7799–7802. [DOI] [PubMed] [Google Scholar]
- 23.Bujalowski, W. & Lohman, T. M. (1989) J. Mol. Biol. 207, 249–268. [DOI] [PubMed] [Google Scholar]
- 24.Lohman, T. M. & Bujalowski, W. (1994) Biochemistry 33, 6167–6176. [DOI] [PubMed] [Google Scholar]
- 25.Overman, L. B., Bujalowski, W. & Lohman, T. M. (1988) Biochemistry 27, 456–471. [DOI] [PubMed] [Google Scholar]
- 26.Kowalczykowski, S. C., Lonberg, N., Newport, J. W. & von Hippel, P. H. (1981) J. Mol. Biol. 145, 75–104. [DOI] [PubMed] [Google Scholar]
- 27.Alberts, B. M., Frey, L. & Delius, H. (1972) J. Mol. Biol. 68, 139–152. [DOI] [PubMed] [Google Scholar]
- 28.Kim, Y. T., Tabor, S., Bortner, C., Griffith, J. D. & Richardson, C. C. (1992) J. Biol. Chem. 267, 15022–15031. [PubMed] [Google Scholar]
- 29.Lindberg, G. J., Kowalczykowski, S. C., Rist, J. K., Sugino, A. & Rothman-Denes, L. B. (1989) J. Biol. Chem. 264, 12700–12708. [PubMed] [Google Scholar]
- 30.Tiranti, V., Rocchi, M., DiDonato, S. & Zeviani, M. (1993) Gene 126, 219–225. [DOI] [PubMed] [Google Scholar]
- 31.Raghunathan, S., Ricard, C. S., Lohman, T. M. & Waksman, G. (1997) Proc. Natl. Acad. Sci. USA 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raghunathan, S., Kozlov, A. G., Lohman, T. M. & Waksman, G. (2000) Nat. Struct. Biol. 7, 648–652. [DOI] [PubMed] [Google Scholar]
- 33.Chase, J. W., Merril, B. M. & Williams, K. R. (1983) Proc. Natl. Acad. Sci. USA 80, 5480–5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bujalowski, W. & Lohman, T. M. (1991) J. Mol. Biol. 217, 63–74. [DOI] [PubMed] [Google Scholar]
- 35.Lohman, T. M. & Ferrari, M. E. (1994) Annu. Rev. Biochem. 63, 527–570. [DOI] [PubMed] [Google Scholar]
- 36.Sarov-Blat, L. S. & Livneh, Z. (1998) J. Biol. Chem. 273, 5520–5527. [DOI] [PubMed] [Google Scholar]
- 37.Glover, B. P. & McHenry, C. S. (1998) J. Biol. Chem. 273, 23476–23484. [DOI] [PubMed] [Google Scholar]
- 38.Genschel, J., Curth, U. & Urbanke, C. (2000) Biol. Chem. 381, 183–192. [DOI] [PubMed] [Google Scholar]
- 39.Kelman, Z., Yuzhakov, A., Andjelkovic, J. & O'Donnell, M. (1998) EMBO J. 17, 2436–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Glassberg, J., Meyer, R. & Kornberg, A. (1979) J. Bacteriol. 140, 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chase, J. W., L'Italien, J. J., Murphy, J. B., Spicer, E. K. & Williams, K. R. (1984) J. Biol. Chem. 259, 805–814. [PubMed] [Google Scholar]
- 42.Cermakian, N., Ikeda, T. M., Miramontes, P., Lang, B. F., Gray, M. W. & Cedergren, R. (1997) J. Mol. Evol. 45, 671–681. [DOI] [PubMed] [Google Scholar]
- 43.Milligan, J. F., Groebe, D. R., Witherell, G. W. & Uhlenbeck, O. C. (1987) Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeruzalmi, D. & Steitz, T. A. (1998) EMBO J. 17, 4101–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sastry, S. & Ross, B. M. (1998) Proc. Natl. Acad. Sci. USA 95, 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muller, D. K., Martin, C. T. & Coleman, J. E. (1988) Biochemistry 27, 5763–5771. [DOI] [PubMed] [Google Scholar]
- 47.Ma, K., Temiakov, D., Jiang, M., Anikin, M. & McAllister, W. T. (2002) J. Biol. Chem. 277, 43206–43215. [DOI] [PubMed] [Google Scholar]
- 48.He, B., Rong, M., Durbin, R. K. & McAllister, W. T. (1997) J. Mol. Biol. 265, 275–288. [DOI] [PubMed] [Google Scholar]
- 49.Dai, X. & Rothman-Denes, L. B. (1998) Genes Dev. 12, 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tahirov, T. H., Temiakov, D., Anikin, M., Patlan, V., McAllister, W. T. & Vassylyev, D. G. (2002) Nature 420, 43–50. [DOI] [PubMed] [Google Scholar]
- 51.Yin, Y. W. & Steitz, T. A. (2002) Science 298, 1387–1395. [DOI] [PubMed] [Google Scholar]