Abstract
Mutations in DJ-1, a human gene with homologues in organisms from all kingdoms of life, have been shown to be associated with autosomal recessive, early onset Parkinson's disease (PARK7). We report here the three-dimensional structure of the DJ-1 protein, determined at a resolution of 1.1 Å by x-ray crystallography. The chain fold of DJ-1 resembles those of a bacterial protein, PfpI, that has been annotated as a cysteine protease, and of a domain of a bacterial catalase whose role in the activity of that enzyme is uncertain. In contrast to PfpI, a hexameric protein whose oligomeric structure is essential for its putative proteolytic activity, DJ-1 is a dimer with completely different intersubunit contacts. The proposed catalytic triad of PfpI is absent from the corresponding region of the structure of DJ-1, and biochemical assays fail to detect any protease activity for purified DJ-1. A highly conserved cysteine residue, which is catalytically essential in homologues of DJ-1, shows an extreme sensitivity to radiation damage and may be subject to other forms of oxidative modification as well. The structure suggests that the loss of function caused by the Parkinson's-associated mutation L166P in DJ-1 is due to destabilization of the dimer interface. Taken together, the crystal structure of human DJ-1 plus other observations suggest the possible involvement of this protein in the cellular oxidative stress response and a general etiology of neurodegenerative diseases.
Parkinson's disease (PD) is a chronic, progressive neurological disorder characterized by the gradual degeneration of dopanergic neurons in the substantia nigra, the portion of the midbrain basal ganglia that controls the smoothness of movement (1). The disease is named after Dr. James Parkinson, who first described “the shaking palsy” in 1817 (2). Symptoms include trembling in the limbs, jaw, and face, muscle rigidity, slowness of movement (bradykinesia), and impaired balance and coordination. Several types, both sporadic and familial, have been characterized with different apparent causes and presentation. Some toxins [e.g., 1-methyl 4-phenyl-1,2,3,6,-tetrahydropyridine (MPTP) and some neuroleptic drugs] can induce PD symptoms in humans (3) and animals (4).
Sporadic and congenital PD is characterized biochemically and cellularly by the loss of a characteristic black pigment in the substantia nigra (5) because of dopanergic neuronal death by apoptosis, low levels of dopamine in the striatum, increased levels of brain iron and decreased ferritin, and the formation of Lewy bodies (6), which are filmentous aggregates containing the proteins α-synuclein and ubiquitin (these are also present in some Alzheimer's disease patients; ref. 7). Sporadic PD is not genetic, but it affects ≈1% of those over age 60, the mean age of onset (8). In the U.S., 1.5 million people are affected, and the total cost from PD in the U.S. alone is $5.6 billion per year (9).
The causes of sporadic PD are unknown but several genes have been identified whose mutation leads to rare familial PD, and 15–20% of patients with sporadic PD have a close relative with at least one symptom (10). A recent study of Icelanders (11) mapped a susceptibility locus for late-onset idiopathic PD (PARK10) to chromosome 1 (around location 1p32; logarithm of odds score: 4.9). The PARK1–9 genes are genetic disorders characterized by inheritable, early onset PD (10); most are inherited in an autosomal dominant fashion. The best characterized is PARK1, associated with mutations in the gene for α-synuclein (12), the presynaptic protein that accumulates in Lewy bodies (13). PARK5, another autosomal dominant early onset form of PD, appears to be caused by mutations in a gene for a ubiquitin C-terminal hydrolase (14). Interestingly, PARK2, inherited as autosomal recessive, is associated with mutations in another gene involved in ubiquitin-dependent protein degradation, PARKIN, encoding an E3 ubiquitin-protein ligase (15). These genes were the only familial PD genes identified until the recent report from Bonifati et al. (16) that mutations correlated with the PARK7 form of autosomal recessive early onset parkinsonism in two different families map to location 1p36.2-3 on the human genome. This locus is for the DJ-1 gene, which encodes a protein of 189 aa of unknown function. One family had loss of the first three exons in the gene; the other a point mutation that results in a change of Leu-166 to Pro. Bonifati et al. (16) conclude that the DJ-1 protein (the protein mutated in autosomal recessive early onset PD) is lacking in the first family and is functionally inactivated in the other. We report here the crystal structure of human DJ-1 at 1.1-Å resolution. The structure resembles in many respects a model of the monomer of the protein built by Bonifati et al. (16), but the protein is a dimer and the intersubunit interactions are unexpected and important for understanding both the likely functions of the protein and the effects of some PD-associated mutations.
DJ-1 was originally identified as a gene that transformed mouse NIH 3T3 cells when cotransfected with H-ras or c-myc (17). The same group later identified PIASxα (protein inhibitor of activated STAT) as a DJ-1-binding protein, and found that DJ-1 restored androgen receptor (AR) transcription activity that was repressed by PIASxα (18). Another DJ-1-binding protein, DJBP, was cloned by the yeast two-hybrid system. DJBP was also found to bind in vitro and in vivo to the DNA-binding domain of AR in a testosterone-dependent manner and to be colocalized with DJ-1 or AR in the nucleus. Coimmunoprecipitation showed that the formation of a ternary complex between DJ-1, DJBP, and the AR occurs in cells in which DJ-1 bound to the AR through DJBP, that DJBP repressed a testosterone-dependent AR transactivation activity in monkey Cos1 cells by recruiting a histone deacetylase complex, and that DJ-1 partially restored activity, suggesting that AR is positively regulated by DJ-1, which antagonizes the function of negative regulators, including DJBP (18). DJ-1 was independently identified as RS, a 20-kDa micro-tubule-associated protein complexed with an RNA-binding protein (RBP) in FTO-2B rat hepatoma cells; the RNA-binding activity of RBP was stimulated on the dissociation of RS/DJ-1 (19). Recombinant DJ-1 expressed in Escherichia coli binds RBP and inhibits its RNA-binding activity. DJ-1 was also identified as a sperm protein that appears to play an important role in fertilization (20, 21). The addition of anti-mouse DJ-1 serum to sperm solution before in vitro fertilization resulted in a decrease in the efficiency of fertilization to about one-third of that when preimmune serum was added (21). DJ-1 is ubiquitously expressed in various human tissues, its expression is induced by growth stimuli, and it translocates from cytoplasm to nucleus in the S phase of the cell cycle (17). Finally, uncharacterized oxidized forms of DJ-1 were detected on two-dimensional gels in response of human endothelial cells to sublethal levels of hydrogen peroxide and paraquat, compounds that produce reactive oxygen species (ROS) in cells (22). The same result was obtained with endogenous production of ROS in murine macrophages under endotoxin-induced inflammatory conditions (23). Taken together, these observations suggest that DJ-1 may have multiple cellular functions, depending on where it is expressed (24).
A search of the sequence databases for homologues of DJ-1 finds >250 likely relatives in all kingdoms of life. The amino acid sequence of DJ-1 is ≈35% identical on average with members of the ThiJ family of bacterial enzymes that are believed to be involved in 4-methyl-5(β-hydroxyethyl)-thiazole monophosphate biosynthesis (25). ThiJ itself catalyzes the phosphorylation of hydroxymethylpyrimidine (HMP) to HMP monophosphate (25); such thiamine derivatives are thought to protect cells against singlet oxygen and other types of ROS (26, 27). However DJ-1 is also ≈30% identical on average with members of the PfpI family of putative intracellular cysteine proteases (28), and many of the same residues are conserved between both the ThiJ and PfpI families. The three-dimensional structure of PfpI was determined as part of a structural genomics initiative (29); it shows a dimer of trimeric subunits with the active sites located at subunit interfaces. The PfpI family in turn appears to be distantly related to the Type I glutamine amidotransferase superfamily, whose members also use a catalytic Cys/His/Glu-Asp triad similar to that used by the serine and cysteine proteases (30, 31). Finally, some DJ-1 homologues in lower organisms have been annotated as containing a helix–turn–helix motif and thus possibly being involved in transcriptional regulation (32). Recently EcHsp31, the E. coli yedU (hchA) gene product, a representative member of a family of chaperones that alleviates protein misfolding by interacting with early unfolding intermediates, was shown to have a domain with a fold resembling that of PfpI (33). DJ-1 also shows sequence similarity to a yeast protein of unknown function, YDR533c, whose transcription we found to be up-regulated when yeast cells enter the quiescent (G0) state because of carbon starvation or the presence of misfolded proteins (34). YDR533c expression is also up-regulated by various forms of stress, including oxidative stress and heat shock. Thus, despite sequence similarity to a number of proteins whose biochemical or cellular functions have been annotated, including some of known three-dimensional structure, the biochemical function(s) of DJ-1 remain unknown. In the hope that structural information might shed light on the role of DJ-1 in the cell, we undertook the determination of its crystal structure.
Methods
Protein Expression and Purification. E. coli strain BL21(DE3) was transformed with a pET-15b vector (Novagen) harboring the full-length human DJ-1 sequence between the NcoI and BamHI sites, thus eliminating the encoding sequence for His-Tag in the original vector. Cells were grown to an OD600 of 0.8 in LB medium broth supplemented with 100 μg/ml ampicillin at 37°C. Protein overexpression was induced with the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) followed by incubation for 3 h. Cells were harvested by centrifugation and stored at –80°C.
Frozen cells were thawed on ice and resuspended in 50 mM Tris·HCl, pH 7.5/2 mM EDTA/1 mM DTT. The suspended cells were kept on ice and lysed by the addition of lysozyme to a final concentration of 10 mg/ml for 30 min at 4°C, followed by sonication. The bacterially expressed DJ-1 was purified from the cleared lysate essentially as described for the human protein (19). Briefly, ammonium sulfate was added to 70% saturation and the lysate was incubated at 4°C for 30 min, and was then cleared by centrifugation. The supernatant was loaded onto a phenyl Sepharose (Bio-Rad) column that had been equilibrated with 70% saturated ammonium sulfate, 10 mM potassium phosphate (pH 6.8) and 1 mM DTT. Protein was eluted with a 70–0% saturated ammonium sulfate gradient over 10-column volumes. The fractions containing DJ-1 were identified with SDS/PAGE and peak fractions were pooled and dialysed overnight against 10 mM potassium phosphate (pH 6.8), 1 mM DTT. The dialysed sample was loaded onto a hydroxy-apatite column (Calbiochem) that had been equilibrated with 10 mM potassium phosphate, 1 mM DTT and eluted with a 10-column volume gradient of 10–500 mM potassium phosphate (pH 6.8). Fractions containing DJ-1 were identified with SDS/PAGE, pooled, dialysed overnight against 10 mM potassium phosphate (pH 6.8), 1 mM DTT and concentrated to 23 mg/ml as determined by using bicinchoninic acid (BCA) assay (Pierce). Purified DJ-1 ran as a single band on overloaded SDS/PAGE stained with Coomassie blue R-250.
Crystal Growth, Data Collection, Structure Solution, and Refinement Statistics. Crystals of DJ-1 in space group P3121 with one monomer in the asymmetric unit were obtained by using the hanging drop vapor diffusion method by mixing 2 μl of DJ-1 at 23 mg/ml and 2 μl of reservoir solution (30% PEG 400/100 mM Tris·HCl, pH 8.5/200 mM sodium citrate). Crystals measuring ≈0.2 mm × 0.35 mm × 0.2 mm appeared after 3 days of incubation at room temperature and were soaked in 32% PEG 400/100 mM Tris·HCl, pH 8.5/200 mM sodium citrate before shock-cooling by immersion into liquid nitrogen.
Native diffraction data were collected in both high- and low-resolution passes from a single crystal maintained at 100 K and were integrated and scaled by using denzo and scalepack, respectively (35). Experimental phase information was obtained from a single ethylmercury phosphate (EMP) derivative prepared by soaking a crystal in a 2-mM solution of EMP in 32% PEG 400/100 mM Tris·HCl, pH 8.5/200 mM sodium citrate for 12 h, followed by a short backsoak. Because the EMP-soaked crystal was not isomorphous with the native crystal, a separate pseudonative data set was collected from a crystal that had been soaked in stabilizing solution (see above), but was not derivatized. The two Hg sites were located and single isomorphous replacement/anomalous scattering (SIRAS) phases extending to 1.5-Å resolution were calculated by using solve (36, 37). The resulting experimental phases were significantly improved by density modification in resolve (38), producing electron density maps of outstanding clarity. The protein and solvent models were manually built into these density-modified electron density maps by using o (39). The resulting model was subjected to stereochemically restrained positional and anisotropic displacement parameter (ADP) refinement in shelx (40) by using all measured native intensity data extending to 1.1-Å resolution. Estimated SDs (ESDs) were obtained by inversion of the unrestrained least-squares matrix, and the reported averages in Table 1 include all atoms except those protein atoms in alternate conformations.
Table 1. Data collection, phasing, and refinement statistics.
| Native | Hg | |
|---|---|---|
| Data collection | ||
| Beamline | APS 14 BM-C | APS 14 ID-B |
| Space group | P3121 | P3121 |
| Unit cell dimensions, Å | a = b = 75.31, | a = b = 75.06, |
| c = 75.42 | c = 75.70 | |
| Resolution, Å | 33-1.1 | 26-1.5 |
| Unique reflections | 100,971 | 39,963 |
| Completeness, % | 99.3 (95.8) | 99.6 (100) |
| Multiplicity | 6.1 (3.3) | 5.4 (5.3) |
| I/σ(I) | 36.4 (2.6) | 19.2 (5.6) |
| Rmerge, % | 5.4 (54.2) | 7.4 (34.3) |
| Phasing | ||
| Sites | 2 | |
| FOMSOLVE | 0.42 (0.25) | |
| FOMRESOLVE | 0.59 (0.28) | |
| Refinement statistics | ||
| Total residues | 186 | |
| Discretely disordered residues | 18 | |
| Water molecules | 252 | |
| Rwork/Rfree, % | 13.9/16.1 | |
| Mean protein B factor, Å2 | 22.8 | |
| Mean water B factor, Å2 | 39.7 | |
| Mean protein/water anisotropy | 0.62/0.49 | |
| ESD protein atoms, Å | 0.042 | |
| ESD water atoms, Å | 0.046 |
Numbers in parentheses indicate the value in the highest-resolution shell: native data, 1.14-1.10 Å, ethylmercury phosphate (EMP) data, 1.55-1.50 Å. FOM, figure of merit; ESD, estimated SD.
The model quality was assessed by using procheck (41); a Ramachandran plot shows only one residue outside the allowed regions, Cys-106. The corresponding residue is an outlier in every other crystal structure of proteins in the PfpI/ThiJ/DJ-1 superfamily. See Table 1 for data and model statistics.
Protease Assay. Protease activity was assayed in the same manner as that used for PfpI (28, 29). A 0.1% azocasein solution in 50 mM sodium phosphate buffer (pH 7.3) was incubated with ≈0.25 mg/ml protein [DJ-1, YDR533p, YOR391p, or papain (a cysteine protease control)] at 37°C for 30 min. The reaction was quenched with 500 μl of 15% trichloroacetic acid and placed on ice for 15 min. The solution was then centrifuged to remove the precipitate and the OD440 of the supernatant was measured. This assay was performed with various concentrations of protein, ranging from 0.2 to 2 mg/ml and a pH ranging from 6.0 to 8.5. In all experiments, protease activity was detected only in the papain control.
Gel Filtration Chromatography. Purified DJ-1 (0.75 ml at 10 mg/ml) was applied to a 125-ml S-100 (Amersham Pharmacia Biosciences, Piscataway, NJ) gel filtration column at a flow rate of 0.25 ml/min in 10 mM Hepes, pH 7.5, 100 mM KCl at 4°C. Previous calibration of the column was performed under identical running conditions using a calibration mixture of the following: blue dextran (void), chicken ovalbumin (43 kDa), Lactococcus lactis galactose mutarotase (38 kDa), bovine chymotrypsinogen A (25 kDa), and bovine RNase A (14 kDa).
Results and Discussion
Structure Description. The polypeptide chain fold of DJ-1 consists of a six-stranded parallel β-sheet (1, 2, 5, 6, 7, and 11) sandwiched by eight α-helices and with a β-hairpin (3 and 4) on one end and a three-stranded (8, 9, and 10) antiparallel β-sheet on the opposite end. There is a significant kink dividing helices C and D and a cis-Pro at position 66. The overall fold is essentially identical to that of the homology model proposed by Bonifati et al. (16) and to the structure (29) of the putative bacterial cysteine protease Pfp-I (Cα rms deviation of 1.6 Å), on which their homology model was based. One major difference is the presence of an additional α-helix (H) at the C terminus that contributes to the dimer interface (see below). The structure is also similar to the fold of a domain of the bacterial catalase HPII (42). This domain is not required for catalase activity and its role in that protein is unknown (43).
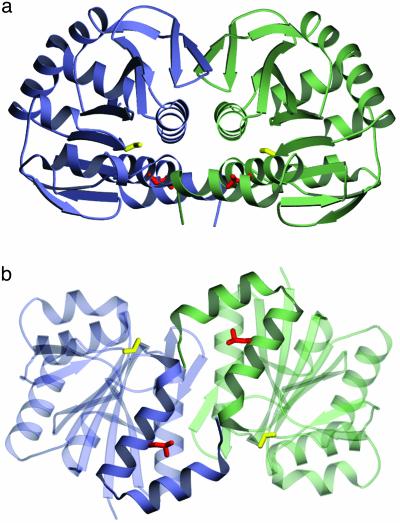
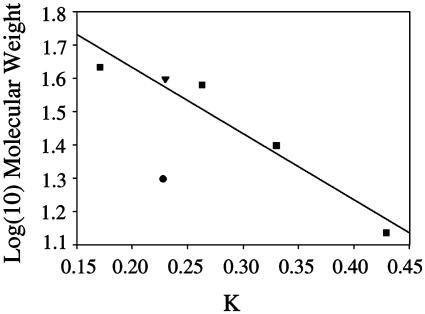
Dimeric Structure of DJ-1. The crystal structure of human DJ-1 indicates that the protein exists as a dimer (Fig. 1). This dimer, in which the two subunits are related by a crystallographic symmetry operation (2-fold rotation), likely represents the oligomerization state of the protein in solution because it buries 2,615 Å2 of surface area and the intermolecular interface excludes water. In addition, purified human DJ-1 migrates as a single species with an apparent molecular mass of 40 kDa in gel filtration chromatography (Fig. 2), almost exactly equal to the expected molecular mass of the dimeric species. Because the function of DJ-1 is unknown, it is difficult to speculate as to the functional importance of its oligomeric form; however, the extensive network of packing interactions between monomers observed in this structure suggests that DJ-1 may be an obligate dimer.
Fig. 1.
(a and b) Two views of the DJ-1 crystallographic dimer. Monomer A is purple and monomer B is green. The view in b is rotated by 90° with respect to the view in a. In both views, Cys-106 is yellow and Leu-166, which is mutated to proline in PARK7 familial PD, is red. In b, the unusual coaxial arrangement of the two C-terminal α-helices (G and H) at the dimer interface is highlighted. The figure was made with povscript+ (50).
Fig. 2.
Gel filtration chromatography indicates that DJ-1 is a dimer in solution. The observed elution of DJ-1 is plotted against the expected molecular mass of the monomer (•) and the dimer (▾). The dimeric molecular weight agrees well with the best-fit line for the calibration standards (▪), whereas the monomer does not.
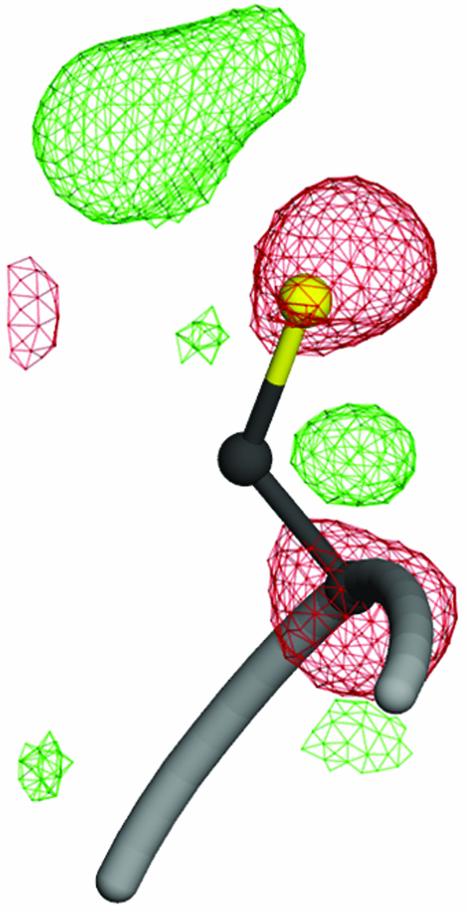
Modification of Cysteine 106. The crystal structure also suggests an explanation for the uncharacterized acidic isoform of DJ-1 that is formed in vivo in response to oxidative stress (22, 23). Cys-106, a highly conserved residue located at the “nucleophile elbow” region of DJ-1, was found to be very sensitive to radiation damage during synchrotron data collection. Fig. 3 shows that this residue is surrounded by both positive and negative difference electron density, indicating that it has been structurally modified by exposure to x-ray radiation. In particular, the presence of strong negative difference electron density on both Sγ and Cα indicates that this residue has undergone radiolysis with partial loss of the thiol group and possible cleavage of the polypeptide backbone. Cys-106 is in a strained backbone conformation (φ = 77°, ϕ = –101°) in this and in every other protein of known structure in the PfpI/ThiJ/DJ-1 superfamily, likely contributing to the unusual reactivity of this residue. The extreme radiation sensitivity of this residue suggests that it may also be a favored target of modification by ROS generated by oxidative stress, yielding the acidic isoform of DJ-1, possibly by the formation of a sulfinic acid at Cys-106. It further suggests that this cysteine may be a site of regulation of DJ-1 activity by oxidation, making the protein a potential sensor for the presence of certain types of ROS in the cell.
Fig. 3.
A view of radiation damage around Cys-106 at the nucleophile elbow region. Fourier difference (Fo – Fc) electron density is shown contoured at +3.0 σ (green) and –3.0 σ (red). The pronounced difference electron density around this residue indicates that it is particularly sensitive to radiation-induced structural changes and possibly prone to other oxidative modification. The figure was made with POVSCRIPT+ (50).
Atomic Displacement Parameter Analysis. The very high resolution of the data obtained from crystals of DJ-1 permits the refinement of ADPs to describe atomic mobility in the protein. ADPs contain information about both the magnitude and preferred direction of displacement for every atom in the structure, and therefore contain more information than the isotropic displacement parameters that are typically refined against lower resolution data. Overall, the atomic displacements in DJ-1 are notably more isotropic than are typically found in proteins that diffract to comparable resolution, with a mean protein atom anisotropy of 0.62 (σ = 0.16). The average anisotropy of proteins that diffract to better than 1.4-Å resolution is 0.45 (σ = 0.15), where anisotropy is defined as the ratio of the smallest to the largest eigenvalues of the ADP tensor.
The most highly anisotropic displacements in DJ-1 are in the three most mobile regions of the protein, including residues 37–45, 50–70, and 85–100. Mobility in the remainder of the protein is restricted and is largely isotropic in character, suggesting the absence of any pronounced segmental flexibility in DJ-1. Furthermore, Rosenfield difference analysis of the ADPs indicates that the increased mobility of residues 37–45, 50–70, and 85–100 is not consistent with overall rigid-body motion of the protein in the crystal lattice (data not shown). The mobile regions of DJ-1 comprise a discrete surface of the protein that contains helices B and D. These two helices define a narrow crevice on the periphery of the protein that connects with the cleft containing Cys-106. It is tempting to speculate that this local region of flexibility may be involved in macromolecular binding or recognition, either with an unidentified substrate or with one or more of the proteins that have been shown to bind to DJ-1.
Functional Consequences of the Structure. All homologues of DJ-1, including PfpI, ThiJ, the yeast protein YDR533c, and the amidotransferases, have a conserved Cys residue (Cys-106 in human DJ-1). This residue has been proposed to be the attacking nucleophile in the proteolytic reaction catalyzed by PfpI, the glutaminase reaction catalyzed by the amidotransferases, and the kinase reaction catalyzed by ThiJ (29). EcHsp31 also contains a cysteine at the same position in the structure but its role in that chaperone is unclear (33).
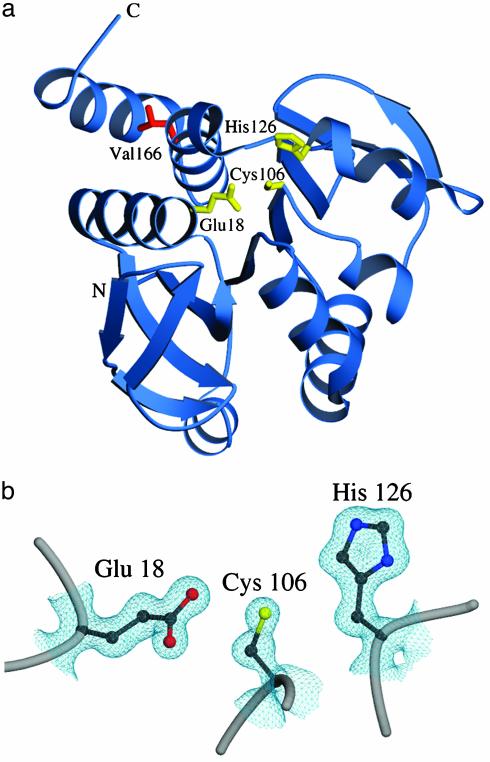
The crystal structure indicates that DJ-1 is not an intracellular cysteine protease. PfpI in its crystal is a dimer of trimers with three putative active sites existing at dimer interfaces within the hexamer. Each site contains a cysteine, histidine, and glutamate residue arranged as in the canonical “catalytic triads” of the serine and cysteine proteases, with the glutamate being contributed by the adjacent subunit (29). These same residue types (Glu-18, His-126, and Cys-106) are found in a deep crevice in the surface of DJ-1, but all three residues come from the same subunit in DJ-1 and their arrangement is permuted relative to that required for the proton transfers that must occur in the cysteine or serine protease mechanism (Fig. 4). Protease assays using a variety of synthetic and natural substrates fail to detect any proteolytic activity for purified human DJ-1, or for its yeast counterpart YDR533c (data not shown). We conclude that either the proteolytic activity measured for PfpI is an accident of its unusual oligomeric structure or that the superfamily of proteins to which both PfpI and DJ-1 belong consists of a number of families with different biochemical and cellular functions. The latter explanation seems likely, in view of the existence of more distant relatives (ThiJ and the glutamine amidotransferases) with distinct functions. Hence, the DJ-1 superfamily can be subdivided into bacterial enzymes catalyzing intracellular proteolysis (PfpI family), ubiquitous amidotransferases catalyzing transfer of the amide nitrogen of glutamine to a variety of substrates (GATase family), bacterial enzymes catalyzing the phosphorylation of B1 and B6 vitamers (ThiJ family), and so on.
Fig. 4.
Residues near the nucleophile elbow region of DJ-1 do not comprise a catalytic triad. (a) A ribbon diagram of the DJ-1 monomer is shown, with residues Glu-18, Cys-106, and His-126 in yellow. Leu-166, which is mutated to proline in PARK7 familial PD, is red. (b) A closer view of the nucleophile elbow region with 2Fo – Fc electron density contoured at 1.0 σ. Although the identity of the residues is correct for a catalytic triad, their conformation prohibits proton shuttling by the canonical mechanism of a serine/cysteine protease or a glutamine amidotransferase. The figure was made with povscript+ (50).
Attempts to detect kinase activity for purified DJ-1 with HMP and other thiamine or pyridoxine derivatives as substrates were unsuccessful. Amidotransferase activity has not yet been tested, but the absence in the structure of DJ-1 of the catalytic triad found in the active site of all GATase family members would appear to rule out such activity as well. It must be emphasized that although the absence of a catalytic triad in DJ-1 seems to rule out protease or amidotransferase activity, it does not exclude the possibility of other types of enzymatic activity for this protein. In this regard, thematics detects several residues with perturbed ionization profiles in DJ-1, including a cluster of highly conserved acidic residues (Glu-15, Glu-16, and Glu-18) near Cys-106. Residues with perturbed ionization profiles in thematics have been shown to correlate with catalytically active residues in a number of test systems (44) and this result indicates that Glu-15, Glu-16, and Glu-18 are of special interest.
DJ-1 and its paralogues in higher eukaryotes appear, from our structural data, to represent a distinct family with as yet uncharacterized biochemical function. Based on the established interactions of DJ-1 with PIASxα (45), DJBP (18), RBP (19), and its indirect association with the AR, transcriptional regulation is one likely function. Very speculatively, it is also interesting to note that DJ-1 and both the thiol peroxidases and the peptide methionine sulfoxide reductases share some similarities, including the proximity of acidic residues to a reactive Cys (46), involvement in oxidative stress, and possible roles in cellular regulation (47). Such an activity for DJ-1 might also explain why defects in this protein result in dopaminergic neuron death in PD, due to the accumulation of damaged proteins in the cell.
Structural Consequences of the L166P Mutation. In comparing the DJ-1 structure with those PfpI and the model of DJ-1 derived from PfpI by Bonifati et al. (16), a major difference is the presence of an additional α-helix at the C terminus that, in combination with helix G, mediates dimerization of human DJ-1. The unusual planar arrangement formed by these four helices in the dimer is shown in Fig. 1b. Leu-166, which is mutated to Pro in PARK7 familial PD, is located in the middle of helix G and significant structural deformation in this helix is expected to result from such a substitution at this position. Based on our crystal structure, perturbations in helix G will interfere with the extensive packing interactions at the dimer interface, thereby disrupting the DJ-1 dimer. Recent biochemical studies have shown that the L166P mutation abolishes DJ-1 self-interaction and the resulting monomeric protein is targeted for ubiquitin-mediated degradation, supporting this hypothesis (48).
Conclusion
Cookson (24) has proposed that the underlying dysfunction in PD is a failure of ubiquitin-mediated protein degradation and that evidence implicating both oxidative stress and proteasome inhibition in Parkinson's, Huntington's, and Alzheimer's diseases imply a commonality between different neurodegenerative disorders, a view shared by Sherman and Goldberg (49). In this model, misfolded proteins trigger all of these diseases, with different proteins for each one. The mechanisms that follow and the effects on different neurons result from specific protein partners in various types of neurons that change either the rate of accumulation of misfolded proteins or the cellular responses to them. The crystal structure of DJ-1 and expression data from its homologues in yeast are broadly consistent with this proposal. The structure suggests that DJ-1 has some important biochemical function, possibly involving regulation of transcription in response to oxidative stress that is compromised in PD-associated mutations, due to loss of the integrity of the dimer. This putative function would be more important in quiescent (G0) cells such as neurons, which cannot dilute out protein damage by rapid turnover and resynthesis or by proliferation. Loss of DJ-1 function would then be predicted to increase the level of damaged proteins, possibly including synuclein, in such cells, rendering them susceptible to apoptotic death.
Acknowledgments
We thank Tim Fenn and Dr. Todd Holyoak for assistance with data collection and useful suggestions; Dr. James Clifton for assisting with the gel filtration experiment; and Dr. Mark R. Cookson and his coworkers for advice, encouragement, and sharing of data before publication. This work was supported by a generous grant from the Ellison Medical Research Foundation (to G.A.P.). Use of the Advanced Photon Source at Argonne National Laboratories for synchrotron x-ray data collection was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of the Bio-CARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources Grant RR07707.
Abbreviations: PD, Parkinson's disease; DJ-1, the protein mutated in autosomal recessive early onset PD; DJBP, DJ-1-binding protein; AR, androgen receptor; RBP, RNA-binding protein; ROS, reactive oxygen species; ADP, anisotropic displacement parameter.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1P5F).
See commentary on page 9111.
Note. As this manuscript was being prepared for submission, Tao and Tong (51) described a crystal structure of human DJ-1 to 1.8 Å.
References
- 1.Olanow, C. W. & Tatton, W. G. (1999) Annu. Rev. Neurosci. 22, 123–144. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson, J. (1817) An Essay on the Shaking Palsy (Sherwood, Neely, and Jones, London).
- 3.Langston, J. W., Ballard, P., Tetrud, J. W. & Irwin, I. (1983) Science 219, 979–980. [DOI] [PubMed] [Google Scholar]
- 4.Gershanik, O. S. (2002) in Parkinson's Disease and Movement Disorders, eds. Jankovic, J. J. & Tolosa, E. (Lippincott, Williams & Wilkins, Baltimore), pp. 331–357.
- 5.Tretiakoff, C. (1919) Thèse de Médicine (University of Paris, Paris).
- 6.Lewy, F. (1912) in Handbuch der Neurologie, eds. Lewandowski, M. & Abelsdorff, G. (Springer, Berlin), Vol. 3, pp. 920–933. [Google Scholar]
- 7.Kotzbauer, P. T., Trojanowski, J. Q. & Lee, V. M. (2001) J. Mol. Neurosci. 17, 225–232. [DOI] [PubMed] [Google Scholar]
- 8.Schrag, A. (2002) in Parkinson's Disease and Movement Disorders, eds. Jankovic, J. J. & Tolosa, E. (Lippincott, Williams & Wilkins, Baltimore), pp. 73–89.
- 9.Lang, A. E. & Lozano, A. M. (1998) N. Engl. J. Med. 339, 1044–1053. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, T. M. & Dawson, V. L. (2003) J. Clin. Invest. 111, 145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hicks, A. A., Petursson, H., Jonsson, T., Stefansson, H., Johannsdottir, H. S., Sainz, J., Frigge, M. L., Kong, A., Gulcher, J. R., Stefansson, K. & Sveinbjornsdottir, S. (2002) Ann. Neurol. 52, 549–555. [DOI] [PubMed] [Google Scholar]
- 12.Polymeropoulos, M. H., Lavedan, C., Leroy, E., Ide, S. E., Dehejia, A., Dutra, A., Pike, B., Root, H., Rubenstein, J., Boyer, R., et al. (1997) Science 276, 2045–2047. [DOI] [PubMed] [Google Scholar]
- 13.Spillantini, M. G., Schmidt, M. L., Lee, V. M., Trojanowski, J. Q., Jakes, R. & Goedert, M. (1997) Nature 388, 839–840. [DOI] [PubMed] [Google Scholar]
- 14.Leroy, E., Boyer, R., Auburger, G., Leube, B., Ulm, G., Mezey, E., Harta, G., Brownstein, M. J., Jonnalagada, S., Chernova, T., et al. (1998) Nature 395, 451–452. [DOI] [PubMed] [Google Scholar]
- 15.Shimura, H., Hattori, N., Kubo, S., Mizuno, Y., Asakawa, S., Minoshima, S., Shimizu, N., Iwai, K., Chiba, T., Tanaka, K. & Suzuki, T. (2000) Nat. Genet. 25, 302–305. [DOI] [PubMed] [Google Scholar]
- 16.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256–259. [DOI] [PubMed] [Google Scholar]
- 17.Nagakubo, D., Taira, T., Kitaura, H., Ikeda, M., Tamai, K., Iguchi-Ariga, S. M. & Ariga, H. (1997) Biochem. Biophys. Res. Commun. 231, 509–513. [DOI] [PubMed] [Google Scholar]
- 18.Niki, T., Takahashi-Niki, K., Taira, T., Iguchi-Ariga, S. M. & Ariga, H. (2003) Mol. Cancer Res. 1, 247–261. [PubMed] [Google Scholar]
- 19.Hod, Y., Pentyala, S. N., Whyard, T. C. & El-Maghrabi, M. R. (1999) J. Cell Biochem. 72, 435–444. [PubMed] [Google Scholar]
- 20.Welch, J. E., Barbee, R. R., Roberts, N. L., Suarez, J. D. & Klinefelter, G. R. (1998) 19, 385–393. [PubMed]
- 21.Wagenfeld, A., Gromoll, J. & Cooper, T. G. (1998) Biochem. Biophys. Res. Commun. 251, 545–549. [DOI] [PubMed] [Google Scholar]
- 22.Mitsumoto, A., Nakagawa, Y., Takeuchi, A., Okawa, K., Iwamatsu, A. & Takanezawa, Y. (2001) Free Radical Res. 35, 301–310. [DOI] [PubMed] [Google Scholar]
- 23.Mitsumoto, A. & Nakagawa, Y. (2001) Free Radical Res. 35, 885–893. [DOI] [PubMed] [Google Scholar]
- 24.Cookson, M. R. (2003) Neuron 37, 7–10. [DOI] [PubMed] [Google Scholar]
- 25.Mizote, T., Tsuda, M., Nakazawa, T. & Nakayama, H. (1996) Microbiology 142, 2969–2974. [DOI] [PubMed] [Google Scholar]
- 26.Osmani, A. H., May, G. S. & Osmani, S. A. (1999) J. Biol. Chem. 274, 23565–23569. [DOI] [PubMed] [Google Scholar]
- 27.Bilski, P., Li, M. Y., Ehrenshaft, M., Daub, M. E. & Chignell, C. F. (2000) Photochem. Photobiol. 71, 129–134. [DOI] [PubMed] [Google Scholar]
- 28.Halio, S. B., Blumentals, I. I., Short, S. A., Merrill, B. M. & Kelly, R. M. (1996) J. Bacteriol. 178, 2605–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Du, X., Choi, I. G., Kim, R., Wang, W., Jancarik, J., Yokota, H. & Kim, S. H. (2000) Proc. Natl. Acad. Sci. USA 97, 14079–14084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raushel, F. M., Thoden, J. B. & Holden, H. M. (1999) Biochemistry 38, 7891–7899. [DOI] [PubMed] [Google Scholar]
- 31.Zalkin, H. & Smith, J. L. (1998) Adv. Enzymol. Relat. Areas Mol. Biol. 72, 87–144. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H., Shiba, T., Sakaki, Y., Hattori, M. & Omura, S. (2003) Nat. Biotechnol. 21, 526–531. [DOI] [PubMed] [Google Scholar]
- 33.Quigley, P. M., Korotkov, K., Baneyx, F. & Hol, W. G. (2003) Proc. Natl. Acad. Sci. USA 100, 3137–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trotter, E. W., Kao, C. M., Berenfeld, L., Botstein, D., Petsko, G. A. & Gray, J. V. (2002) J. Biol. Chem. 277, 44817–44825. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger, T. C., Kim, S. H. & Eisenberg, D. (1987) Acta Crystallogr. A 43, 1–5. [Google Scholar]
- 38.Terwilliger, T. C. (2001) Acta Crystallogr. D 57, 1763–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 40.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319–343. [PubMed] [Google Scholar]
- 41.Laskowski, R. A., Macarthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 42.Horvath, M. M. & Grishin, N. V. (2001) Proteins 42, 230–236. [PubMed] [Google Scholar]
- 43.Loewen, P. (1996) Gene 179, 39–44. [DOI] [PubMed] [Google Scholar]
- 44.Ondrechen, M. J., Clifton, J. G. & Ringe, D. (2001) Proc. Natl. Acad. Sci. USA 98, 12473–12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi, K., Taira, T., Niki, T., Seino, C., Iguchi-Ariga, S. M. & Ariga, H. (2001) J. Biol. Chem. 276, 37556–37563. [DOI] [PubMed] [Google Scholar]
- 46.Lowther, W. T., Brot, N., Weissbach, H. & Matthews, B. W. (2000) Biochemistry 39, 13307–13312. [DOI] [PubMed] [Google Scholar]
- 47.Weissbach, H., Etienne, F., Hoshi, T., Heinemann, S. H., Lowther, W. T., Matthews, B., St. John, G., Nathan, C. & Brot, N. (2002) Arch. Biochem. Biophys. 397, 172–178. [DOI] [PubMed] [Google Scholar]
- 48.Miller, D. W., Ahmad, R., Hague, S., Baptista, M. J., Canet-Aviles, R., McLendon, C., Carter, D. M., Zhu, P.-P., Stadler, J., Klinefelter, G. R., et al. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 49.Sherman, M. Y. & Goldberg, A. L. (2001) Neuron 29, 15–32. [DOI] [PubMed] [Google Scholar]
- 50.Fenn, T. D., Ringe, D. & Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944–947. [Google Scholar]
- 51.Tao, X. & Tong, L. (2003) J. Biol. Chem., in press. [DOI] [PubMed]