Abstract
We have recently identified an RNA aptamer for the transcription factor NF-κB p50 homodimer [(p50)2], which exhibits little sequence resemblance to the consensus DNA target for (p50)2, but binds (p50)2 with an affinity similar to that of the optimal DNA target. We describe here the 2.45-Å resolution x-ray crystal structure of the p50 RHR/RNA aptamer complex. The structure reveals that two RNA molecules bind independent of each other to the p50 N-terminal Ig-like domains. The RNA secondary structure is comprised of a stem and a stem–loop separated by an internal loop folded into a kinked helix because of the cross-strand stacking of three internal loop guanines. These guanines, placed at the edge of the 3′ helix, together with the major groove of the irregular 3′ helix, form the binding surface for p50. Each p50 monomer uses the same surface to recognize the distorted RNA major groove as observed in the κB DNA/p50 RHR complex, resulting in strikingly similar interfaces. The structure reveals how the aptamer specifically selects p50 and discriminates against p65. We also discuss the physiological implications of RNA binding by (p50)2.
The Rel/NF-κB family of inducible transcription factors mediates cellular responses to a large array of stimuli by regulating the expression of hundreds of genes with distinct functions. These cellular activities include immune and inflammatory responses, programmed cell death, growth, proliferation, and development (1, 2). The mammalian Rel/NF-κB family is comprised of five members [p50, p52, p65 (RelA), and c-Rel and RelB] that can form combinatorial dimers. These proteins share an ≈300-residue-long homologous region referred to as the Rel homology region (RHR). This N-terminal RHR contains all of the critical elements for dimerization, DNA binding, and nuclear import (Fig. 1a). Three-dimensional x-ray crystal structures revealed that the NF-κB RHR is organized into two folded Ig-like domains (3, 4). The N-terminal domain (NTD) is responsible for sequence-specific DNA binding, whereas the C-terminal domain (CTD) is primarily responsible for subunit association. Residues emanating from loops in the dimerization domain (DD) also make nonspecific DNA contacts.
Fig. 1.
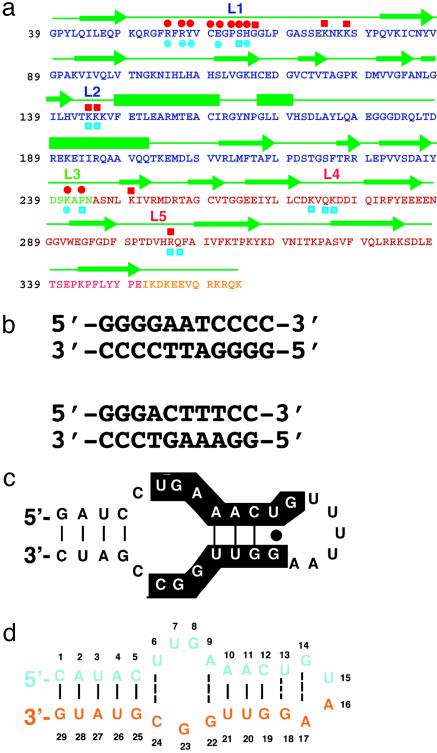
Sequence features of p50 RHR and its DNA and RNA targets. (a) Sequence of the RHR of murine p50. Single-letter amino acid codes for the NTD, DD, linker, and flexible region at the C terminus are denoted by blue, green, red, and yellow, respectively. Green arrows denote residues in β-strands, and bars are α-helices. The nucleic acid-binding loops L1, L2, and L3 are shown. Residues of p50 that contact RNA bases through hydrogen bonds (red circles) and backbone phosphates (red squares) are shown above the protein sequence. The DNA base-specific (blue circles) and backbone (blue squares) contacts are denoted below the protein sequence. (b) κB-DNA sequences that preferentially bind the p50 homodimer (Upper) and p50/p65 heterodimer (Lower) are shown. The 11-mer κB-DNA sequence (Upper) is located in the promoter of the H2 gene and binds p50 RHR homodimer. The 10-mer κB-DNA sequence (Lower) is located in the promoter of the Ig-κ light-chain gene and binds p50/p65. (c) The 31-nt RNA aptamer originally identified by in vitro-selection for binding to (p50)2. Nucleotides conserved among selected aptamer variants are highlighted. (d) The optimized 29-nt RNA aptamer used in the cocrystallization experiment.
NF-κB dimers bind transcriptional regulatory DNA elements that are collectively referred to as κB-DNA sites (Fig. 1b). DNA-bound x-ray structures of several of the NF-κB dimers have been solved (3–8). From these structural studies it is possible to discern the rules that govern target site selection by the NF-κB dimers. (p50)2 prefers an 11-bp target of sequence 5′-GGGRN A/T NYCCC (Fig. 1b Upper). p50 monomers bind to the 5′ half site comprised of the five base pairs 5′-GGGRN. p65, on the other hand, does not require the first G:C base pair. The p65 half site therefore consists of four base pairs, 5′-TTCC, resulting in a 10-bp target site for p50/p65 (Fig. 1b Lower). In NF-κB, DNA-contacting residues are contributed by protein loops connecting the secondary structures. The most important of these loops is referred to as loop L1, which contributes five base-contacting amino acids in p50 and four base-contacting residues in p65 (6, 9).
Using SELEX, we have recently identified an RNA aptamer specific for the NF-κB p50 homodimer from a pool of ≈1014 randomized RNA sequences (ref. 10 and Fig. 1c). The aptamer binds both (p50)2 and p50/p65 with high affinities that are indistinguishable from the affinities of the same dimers toward their optimal κB-DNA sequences. We also showed that the RNA aptamer can efficiently block DNA binding in vitro (10), suggesting that both RNA and DNA bind NF-κB in a similar manner. In addition, the 31-nt aptamer selectively binds (p50)2 in a yeast three-hybrid system (11). The 31-nt aptamer sequence clearly differs from the κB-DNA sequences (Fig. 1 b and c). Furthermore, the RNA sequence does not bear any symmetry-related half sites, unlike the κB-DNA sequences. Therefore, it is unclear how the RNA specifically binds the two symmetrical, or nearly symmetrical, subunits of an NF-κB dimer. To address the above questions we have crystallized a complex containing the p50 homodimer and an optimized 29-nt RNA aptamer. This aptamer (Fig. 1d) is an improved variant of the original 31-nt aptamer (Fig. 1c), selected in yeast for improved interactions with (p50)2 in a yeast three-hybrid selection system (12). This optimized RNA aptamer has been shown to block (p50)2 binding to DNA in vivo in a yeast one-hybrid system (12). We describe here the 2.45-Å resolution crystal structure of the NF-κB p50 RHR homodimer bound to this high-affinity RNA aptamer.
Methods
Protein and RNA Purifications. The truncated murine NF-κB p50 cDNA (residues 39–364) was purified by using the procedure described(3). Theα-p50RNAaptamer(5′-CAUACU2GA3CUGUA2G2U2G2CGUAUG) was synthesized by Dharmacon Research (Lafayette, CO), deprotected according to the protocol suggested by the supplier, and purified by denaturing PAGE and elution. RNA concentration was determined by UV absorbance at 260 nm, by using the concentration conversion factor recommended by the supplier.
Crystallization and Structure Solution. The complex between p50 homodimer and the RNA aptamer was formed by mixing protein and RNA in a 1:1.1 molar ratio for crystallization. The structure was solved by combining molecular replacement and single isomorphous replacement (SIR) methods. A detailed description of crystallization, structure solution, and crystallographic analyses is in Supporting Text and Table 2, which are published as supporting information on the PNAS web site, www.pnas.org. The coordinates have been deposited into the Protein Data Bank (PDB ID code 1OOA).
Binding Affinity Measurement. Equilibrium dissociation constants for various RNA aptamer/NF-κB complexes were estimated by nitrocellulose filter binding experiments. Purified proteins (see legend to Table 1) were quantitated by Bradford assay and diluted into binding buffer (10 mM Hepes, pH 7.5/100 mM NaCl/1 mM DTT) containing 50 μg/ml BSA. For a detailed description, see Supporting Text and Table 2.
Table 1. RNA aptamer affinities for different forms of murine NF-κB.
| Construct | (p50)2* | p50/p65† | (p65)2‡ | p50dd/p65§ | p50/p65dd¶ | p50dd/p65dd∥ |
|---|---|---|---|---|---|---|
| Kd ± SD** | 5.4 ± 2.2 | 22 ± 7 | 3,500 ± 2,300 | 187 ± 115 | 10 ± 4 | NB†† |
Equilibrium dissociation constant estimates (nM) were obtained by nitrocellulose filter binding as described in Methods.
Homodimer of p50 RHR (amino acids 39-363).
Heterodimer of p50 RHR (amino acids 37-363) and p65 RHR (amino acids 19-291).
Homodimer of p65 RHR (amino acids 1-325).
Heterodimer of p50 dimerization domain (amino acids 245-350) and p65 RHR (amino acids 19-304).
Heterodimer of p65 dimerization domain (amino acids 191-304) and p50 RHR (amino acids 39-350).
Heterodimer of p50 dimerization domain (amino acids 245-350) and p65 dimerization domain (amino acids 191-304).
Mean ± SD based on at least two determinations.
NB, no binding detected.
Results and Discussion
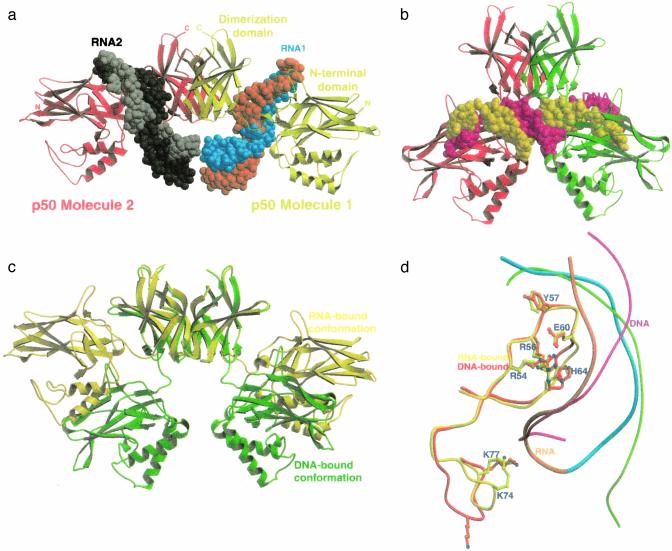
Overall Structure. The overall structure of the complex and its comparison with DNA-bound (p50)2 is shown in Fig. 2. In the RNA/p50 RHR complex, one RNA molecule binds to each of the p50 monomers of the homodimer. This binding occurs almost exclusively through the NTD and the short linker connecting the NTD to the DD of the p50 RHR (Fig. 2a). The RNA uses its internal loop and the 3′ stem–loop structure to bind the protein. These RNA secondary structures are folded into an irregular helix with a long and wide major groove that perfectly complements the protein surface both in size and shape. The complex displays extensive base-specific contacts but few nonspecific contacts with the phosphodiester backbone. Base-specific contacts are mediated by the protein side chains emanating mostly from loop L1 of p50. The RNA aptamer has no internal motifs that are twofold symmetry-related, making it impossible for each monomer of the symmetrical protein homodimer to bind simultaneously to a single RNA molecule. The p50 NTDs are splayed away from each other and from the twofold symmetry axis, resulting in a relatively open conformation for the complex (Fig. 2 a and c). This finding is in contrast to the DNA/p50 RHR complex in which one DNA molecule binds per p50 homodimer. Because the two p50 NTDs in the homodimer bind to two roughly symmetrical, closely spaced half sites in the same DNA molecule, the NTDs closely approach the symmetry axis, providing a relatively closed conformation for the complex (Fig. 2 b and c). Although the RNA molecule does not bear a strong resemblance to the consensus κB-DNA sequence, the chemistry of the core interface is remarkably conserved (Fig. 2d).
Fig. 2.
Overall structure of the p50 RHR bound to the RNA aptamer and its comparison with the DNA-bound complex. (a) Ribbon presentation of the entire RNA/p50 RHR complex. Two p50 subunits are shown in red and yellow. The RNA molecules are shown in space-filling models. The RNA strands are cyan and orange in molecule 1 and black and gray in molecule 2. (b) Ribbon presentation of the DNA/p50 RHR complex. The orientations of the p50 RHR dimerization domains in a and b are identical. (c) Superposition of the p50 RHRs aligned based on the dimerization domain. A 40° rotation and 10-Å translation are required to superpose the NTDs. (d) Superposition of loop L1 of the p50 RHR in the RNA- and DNA-bound complexes. Shown is the near-perfect alignment of the side chains of all five base-contacting residues located in loop L1. The better overall complementation to the protein surface of the RNA, in blue and orange, compared with DNA, in green and magenta, is noticeable here.
Stability of the Complex. The isolated κB-DNA sequence binds the p50 RHR/p65 RHR heterodimer with ≈1–5 nM affinity (13, 14). We observed that in the DNA/NF-κB complex, both protein subunits must contact the same DNA molecule to form a complex of appreciable stability. In the RNA/p50 RHR complex, each RNA molecule bound to a p50 monomer in the complex does not directly contact the other RNA or the opposing protein subunit. This result suggests that the two RNA molecules bind independent of each other. To test this hypothesis, we have determined the binding affinities of the RNA aptamer for the p50 RHR homodimer, the p50 RHR/p65 RHR heterodimer, the p50 RHR/p65 DD, and the p50 DD/p65 RHR. The binding affinities are listed in Table 1. We observed that RNA binds to the p50 RHR homodimer with an ≈4- to 5-fold higher affinity than to the p50 RHR/p65 RHR. The p65 RHR homodimer binds RNA extremely weakly, indicating that the RNA aptamer is p50 RHR specific, and discriminates against p65. These results suggest that in the RNA/p50 RHR/p65 RHR complex, only the p50 subunit binds to RNA. This idea is further supported by the fact that the removal of the p65 NTD from the heterodimer does not reduce its affinity for RNA. As expected, RNA does not bind either p50 DD/p65 RHR or p50 DD/p65 DD. Therefore, most of the binding affinity results from direct RNA-protein contacts between the RNA aptamer and the N-terminal domain of a p50 monomer.
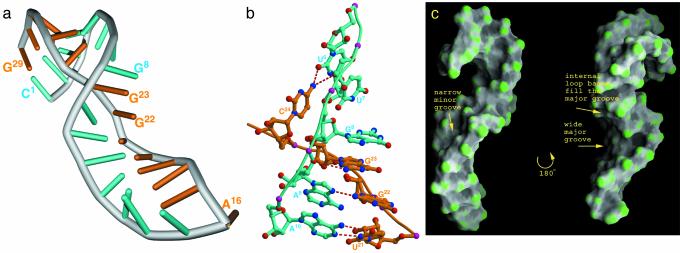
RNA Structure. The optimized RNA aptamer has a guanine-rich, asymmetric internal loop with four nucleotides in one strand (U6U7G8A9) and three nucleotides (G22G23C24) in the other (Fig. 1d). In its folded structure, this internal loop forms an irregular double-stranded motif highlighted by the cross-strand stacking of three guanines, G8, G23, and G22 (Fig. 3 a and b). The projection of bases from both stands into one side of the duplex forces the two strands to closely approach, reducing the strand-to-strand distance by >8 Å, compared with a regular A-helix minor groove width (Fig. 3c). The two short duplexes flanking the internal loop assume A-helical geometry, although the 3′ helix is more irregular than a typical A-helix. The flanking helices are oriented in an ≈40° angle, resulting in an overall bent shape for the RNA. U6 and C24 of the internal loop form a noncanonical base pair. Both U7 and A9 remain mainly within the helical axis, and are involved in stacking interactions with the neighboring bases U6 and A10, respectively (Fig. 3b). Deviation of the 3′ helix from a normal A-helix results from distortion of the duplex structure at one end and the G14:A17 sheared base pair at the other end of the helix. The G14:A17 base pair of the GUAA tetraloop sequence is the final base pair of the 3′ helix. This sheared base pair and stacking of U15 underneath the G14:A17 base pair is a conserved feature of all GNRA-type tetraloops (15, 16). A16 is flipped out of the loop and is not engaged in any interactions.
Fig. 3.
Structure of the RNA aptamer. (a) Overall structure of the α-p50 RNA aptamer. Residues 1–15 representing one strand are cyan, and residues 16–29 representing the other strand are orange. (b) A close-up view of the folded structure of the internal loop. Three cross-strand-stacked guanines are indicated. The non-Watson–Crick hydrogen bond pairing between U6 and C24 and between A9 and G22 of the internal loop are indicated. (c) Surface presentation (23) of the RNA in two orientations related by 180° rotation around the long axis. The image on the left is in the same orientation as a and shows the narrow minor-groove section near the internal loop. The image on the right shows the protein-binding groove of the RNA.
The RNA/Protein Complex Interface. Hydrogen-bonded interactions at the protein–RNA interface are shown schematically in Fig. 4. Superposition of p50 NTDs from the RNA- and DNA-bound complexes reveals an rms deviation of only 1.2 Å. The similarities extend even to the base-contacting side chains (Fig. 2d). Therefore, the p50 NTD in its free state presents a rigid surface that can bind nucleic acids without undergoing a major conformational change. The in vitro selection for RNA aptamers against (p50)2 identified the optimal RNA that folds into a distorted A-helix, which neatly complements the surface of the p50 NTD in shape, dimensions, and chemistry (Fig. 2d). This feature of a folded RNA major groove accommodating the protein molecule is observed in many RNA/protein complexes (17). To accommodate the entire protein-binding surface, the major groove of the RNA helix extends into the duplex formed by the internal loop residues. The cross-strand stacked guanines G8, G22, and G23 form the expanded major groove. The bases at the major groove facing the stack along one side are G18, G19, U20, U21, G22, G23, and G8. The bases that stack on the other side of the major groove are A11, C12, U13, and G14. Remarkably, these 11 bases were absolutely conserved in all RNAs that survived multiple rounds of the initial in vitro selection (ref. 10 and Fig. 1c).
Fig. 4.
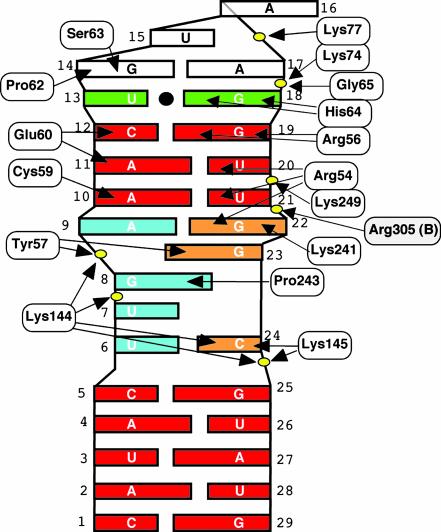
Summary of contacts between the RNA aptamer and a p50 monomer. Red, canonical RNA Watson–Crick base pairs; cyan, 5′ half of predicted internal loop; green, wobble pair; white, GNRA-type tetraloop; and orange, 3′ half of predicted internal loop. Phosphates in the RNA backbone contacted by p50 residues are represented as yellow ovals.
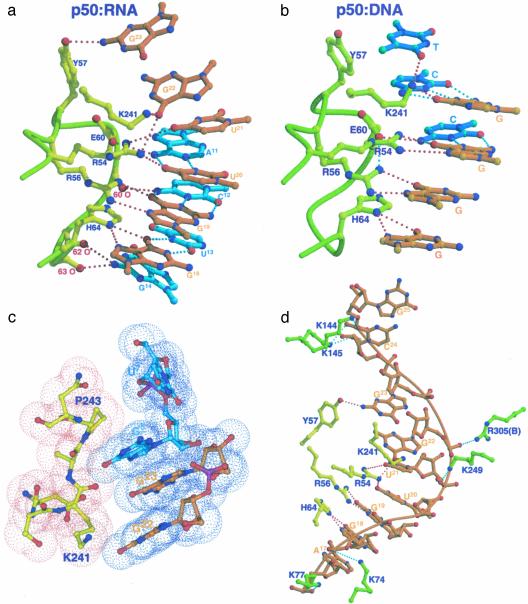
Base-specific contacts. The most striking feature of the complex is that the chemistry of the core RNA/p50 RHR complex interface is essentially identical to that of the κB-DNA/p50 RHR interface (Fig. 5a). The five DNA bases that contact amino acids R54, R56, E60, H64, and K241 are projected identically in this complex and interact with the RNA in a similar fashion (Fig. 5b). The distorted, irregular, and long RNA major groove presents the sequence 5′-G18G19U20U21G22 for direct contact by p50 residues H64, R56, R54, and K241. Contacts between H64 and G18 and between R56 and G19 observed here are conserved both in sequence and chemistry in all nucleic acid/NF-κB complexes. In the RNA complex, however, R54 is involved in more elaborate contacts with RNA than is observed in DNA complexes. The guanidinium group of R54 makes simultaneous contacts with carbonyl oxygens of three bases, U20, U21, and G22. As indicated earlier, G22 projects out of the helical axis into the major groove, tilted toward U20 and U21. This protrusion brings G22 close to the protein surface, such that it is possible for R54 to directly contact all three bases simultaneously (Fig. 5a). K241, the only residue contributed by the domain linker segment, directly contacts the carbonyl oxygen of G22. A11 (in a pair with U21) donates a hydrogen bond to the carboxylate oxygen of E60. This hydrogen-bonding contact is reminiscent of the interaction between E60 and 4N of the C of a G:C bp in DNA/NF-κB complexes.
Fig. 5.
The detailed chemistry of the RNA/p50 RHR complex interface. (a) Direct base-specific hydrogen-bonding contacts between p50 and the RNA aptamer at the core of the protein–RNA interface. (b) Direct base-specific hydrogen-bonding contacts between p50 and p50-specific DNA half site observed in the x-ray structure of p50 homodimer bound to MHC-κB DNA. (c) Stacking interactions between residues in the linker (P243 and K241) and RNA bases (G8 and G22) shown by van der Waals surface model. (d) Contacts between p50 and RNA along one side of the RNA duplex. Nonspecific RNA backbone contacts by the protein are highlighted in three places of the RNA aptamer. Protein residues in yellow denote specific contacts.
These interactions at the core of the complex are capped by direct van der Waals contacts between P243 of the linker and the flipped-out G8 at one end (Fig. 5c), and the backbone carbonyls of residues at positions 62, 63, and G14 at the other end (Fig. 5a). The hydroxyl of Y57 contacts the NH2 of G23 in this complex. In all DNA/p50 RHR complexes, Y57 makes van der Waals contacts with the methyl groups of two thymines located near the center of κB-DNA sites (Fig. 5b).
Backbone contacts. Contacts between the RNA phosphodiester backbone and four lysine side chains define the boundary of the RNA–protein interface (Fig. 5d). K144 and K145 both contact the phosphate of G25, the last RNA residue at the 5′ side. These contacts are reminiscent of minor groove contacts by the same residues in DNA complexes. The extreme 3′ end of the RNA is contacted by K74 and K77 from the back of loop L1. K249 makes a salt bridge with the phosphate at position 21. This residue is the only one from the dimerization domain that mediates any contact with the RNA.
NF-κB subunit-specific interactions. The clearest difference in base-specific contacts between this complex and all DNA/NF-κB complexes is the set of RNA contacts with the main chain of the p50 RHR-encompassing residues from E60 to H64. Superposition of the NTDs from RNA- and DNA-bound complexes reveals that the backbone moves closer by 2–4.5 Å to the major groove-reaching residues A11, C12, U13, and G14 (Fig. 2d). These residues pair with residues in the other strand that make extensive protein contacts. The carbonyl of E60 contacts the NH2 of C12, and the carbonyls of P62 and S63 contact the NH2 of G14 (Figs. 4 and 5a). These residues simply cannot contact the DNA bases because of the relatively rigid helical conformation and groove geometry of DNA duplexes. Sequence differences between p50 and p65 in this segment (E60-H64) are the likely reason why p65 binds so poorly to this aptamer. Glycine, proline, and histidine at positions 61, 62, and 64 are replaced by valine, arginine, and alanine, respectively, in p65. In the absence of the glycine-proline sequence, it is not possible for p65 to assume such a structure. We believe that these interactions between RNA and backbone carbonyls are critical for the stability of the complex. P62 makes van der Waals contacts with G14 (Fig. 5d). In addition, H64 of p50 is substituted by alanine in p65. Thus, the direct interactions between H64 and G14 cannot take place in p65.
Implications for Biological Regulation and Inhibitor Design. Although no naturally occurring RNA that binds p50 or any other NF-κB dimer has yet been identified, there is emerging evidence that many other transcription factors play dual roles in regulating gene expression by binding both specific DNA and RNA sequences (11). The classic example is Xenopus laevis TFIIIA. The transcription of 5S rRNA by RNA polymerase III requires the essential factor TFIIIA. However, the same factor binds the resulting 5S transcript, a component of ribosomes (18–21). Several other transcription factors such as TRA-1, bicoid, and p53 have been shown or implicated to bind to specific RNA sequences (22). More intriguing still is the finding that the transcription factor STAT-1 appears to be capable of binding a natural untranslated RNA (19, 20). Complex formation between STAT-1 and this RNA has been proposed to repress the expression of the major histocompatibility complex (MHC) gene in human trophoblasts. Interestingly, three-dimensional structures of STAT-1 and NF-κB, as well as their DNA recognition, appear to be evolutionarily related. The putative STAT-1-binding RNA contains multiple motifs similar to specific STAT-1-binding DNA sequences, suggesting that the protein employs a similar mechanism in binding both DNA and RNA. This binding might be analogous to our current observation of similar p50 interactions with physiological κB sites and an in vitro-selected RNA aptamer. We believe that it is not unlikely that many untranslated RNAs or other RNAs within the cell might interact with NF-κB, thus adding another level of complexity to gene regulation.
Our RNA/(p50)2 cocrystal structure reveals the existence of a previously unknown “open” NF-κB conformation incompatible with DNA binding. The spatial relationship between NTDs and DDs in this NF-κB form creates clefts and binding surfaces that might be bound and stabilized by small molecules. Such binding events could stabilize this open conformation, inhibiting the ability of NF-κB to bind DNA and activate gene expression. This NF-κB conformation can therefore serve as a target for virtual docking by using a database of small molecules. Lead compounds predicted to have high differential affinity for the open NF-κB conformation might then be rapidly tested in vitro and in vivo for inhibition of DNA binding by the p50 homodimer and p50/p65 forms of NF-κB.
In conclusion, we present the structure of an in vitro-selected RNA aptamer bound to the NF-κB p50 homodimer. This represents an example in which both RNA- and DNA-bound structures of a transcription factor are known. Together these two structures reveal a strikingly conserved protein–nucleic acid interface, although the primary nucleotide sequences of κB-DNA and the RNA aptamer are clearly different. The ability of an RNA molecule with a different sequence than κB-DNA to fold and bind p50 with striking resemblance suggests that specific cellular RNAs may serve as target for NF-κB to add another level of physiological regulation. At the same time, information derived from these structures also provides clues for the identification of small-molecule, subunit-specific NF-κB inhibitors.
Supplementary Material
Acknowledgments
We thank Tom Huxford, Rashmi Talwar, Anu Moorthy, and Chris Phelps for reading the manuscript, and Lori Lebruska for contributions to the early stages of this work. This work is supported by the Mayo Foundation (to L.J.M.) and grants from the National Cancer Institute (to G.G.) and American Cancer Society (to L.J.M.). L.A.C. was a Howard Hughes Medical Institute Predoctoral Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RHR, Rel homology region; NTD, N-terminal domain; DD, dimerization domain.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1OOA).
References
- 1.Karin, M. & Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18, 621–663. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225–260. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh, G., van Duyne, G., Ghosh, S. & Sigler, P. B. (1995) Nature 373, 303–310. [DOI] [PubMed] [Google Scholar]
- 4.Muller, C. W., Rey, F. A., Sodeoka, M., Verdine, G. L. & Harrison, S. C. (1995) Nature 373, 311–317. [DOI] [PubMed] [Google Scholar]
- 5.Cramer, P., Larson, C. J., Verdine, G. L. & Muller, C. W. (1997) EMBO J. 16, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. Q., Ghosh S. & Ghosh, G. (1998) Nat. Struct. Biol. 5, 67–73. [DOI] [PubMed] [Google Scholar]
- 7.Chen, F. E., Huang, D.-B., Chen, Y.-Q. & Ghosh, G. (1998) Nature 391, 410–413. [DOI] [PubMed] [Google Scholar]
- 8.Huang, D.-B., Chen, Y.-Q., Ruetsche, M., Phelps, C. B. & Ghosh, G. (2001) Structure (London) 9, 669–678. [DOI] [PubMed] [Google Scholar]
- 9.Chen, F. E. & Ghosh, G. (1999) Oncogene 18, 6845–6852. [DOI] [PubMed] [Google Scholar]
- 10.Lebruska, L. L. & Maher, L. J., III (1999) Biochemistry 38, 3168–3174. [DOI] [PubMed] [Google Scholar]
- 11.Cassiday, L. A. & Maher, L. J., III (2001) Biochemistry 40, 2433–2438. [DOI] [PubMed] [Google Scholar]
- 12.Cassiday, L. A. & Maher, L. J., III (2003) Proc. Natl. Acad. Sci. USA 100, 3930–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart, D. J., Speight, R. E., Cooper, M. A., Sutherland, J. D. & Blackburn, J. M. (1999) Nucleic Acids Res. 27, 1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phelps, C. B., Sengchanthalangsy, L. L., Malek, S. & Ghosh, G. (2000) J. Biol. Chem. 275, 24392–24399. [DOI] [PubMed] [Google Scholar]
- 15.Heus, H. A. & Pardi, A. (1991) Science 253, 191–194. [DOI] [PubMed] [Google Scholar]
- 16.Hermann, T. & Patel, D. J. (1999) J. Mol. Biol. 294, 829–849. [DOI] [PubMed] [Google Scholar]
- 17.Draper, D. E. (1999) J. Mol. Biol. 293, 255–270. [DOI] [PubMed] [Google Scholar]
- 18.Pelham, H. R. & Brown, D. D. (1980) Proc. Natl. Acad. Sci. USA 77, 4170–4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyman, J. A. (1999) Biol. Reprod. 60, 23–31. [DOI] [PubMed] [Google Scholar]
- 20.Peyman, J. A. (2001) Am. J. Reprod. Immunol. 45, 382–392. [DOI] [PubMed] [Google Scholar]
- 21.Pittman, R. H., Andrews, M. T. & Setzer, D. R. (1999) J. Biol. Chem. 274, 33198–33201. [DOI] [PubMed] [Google Scholar]
- 22.Cassiday, L. A. & Maher, L. J., III (2002) Nucleic Acids Res. 30, 4118–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholls, A., Bharadwaj, R. & Honig, B. (1993) Biophys. J. 64, A116–A125. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.