Abstract
GRIM-19 (gene associated with retinoid-IFN-induced mortality 19), isolated as a cell death activator in a genetic screen used to define mechanisms involved in IFN-β- and retinoic acid-induced cell death, codes for a ≈16-kDa protein that induces apoptosis in a number of cell lines. Antisense ablation of GRIM-19 caused resistance to cell death induced by IFN plus retinoic acid and conferred a growth advantage to cells. To understand the molecular bases for its cell death regulatory activity, we used a yeast two-hybrid screen and identified that the transcription factor STAT3 (signal transducer and activator of transcription 3) binds to GRIM-19. GRIM-19 inhibits transcription driven by activation of STAT3, but not STAT1. It neither inhibits the ligand-induced activation of STAT3 nor blocks its ability to bind to DNA. Mutational analysis indicates that the transactivation domain of STAT3, especially residue S727, is required for GRIM-19 binding. Because GRIM-19 does not bind significantly to other STATs, our studies identify a specific inhibitor of STAT3. Because constitutively active STAT3 up-regulates antiapoptotic genes to promote tumor survival, its inhibition by GRIM-19 also demonstrates an antioncogenic effect exerted by biological therapeutics.
Keywords: cytokines, cell growth, apoptosis, immune response
Interferons inhibit cell growth by stimulating the synthesis of growth inhibitors or activating apoptosis (1, 2). For example, the transcription factors STAT1 (signal transducer and activator of transcription 1), IRF1, and IRF8/ICSBP act as tumor suppressors by regulating the expression of several genes (3–6). In addition, activation of the expression of transcription factors belonging to the murine 200-gene family inhibits cell growth (7, 8). Given the complexity of IFN action and their pleiotropic effects on various cell types, several other as yet undiscovered inhibitors of cell growth may be used by these cytokines. Although IFNs themselves strongly inhibit cell growth, many tumor cells are insensitive to IFN-induced growth arrest (9). In many tumor cells moderately sensitive to IFNs, pretreatment with all-trans retinoic acid (RA), a vitamin A metabolite, enhances sensitivity to IFN-induced growth inhibition (10, 11). However, the mechanisms that regulate IFN/RA-induced growth inhibition are poorly defined.
Previously, we have shown that the IFN/RA combination induces cell death in a number of tumor cell lines, unlike the individual agents (11, 12). To understand the molecular bases for the synergistic antitumor action of IFN and RA, we used a genetic technique to identify cell death-associated genes, based on the ability of antisense RNAs to ablate the expression of critical cell death associated proteins after transfection of a total cDNA library cloned in an antisense episomal expression vector (12, 13). Cells expressing death-specific antisense genes survive treatment with IFN/RA and continue to grow. The gene of interest is then rescued and sequenced. Based on their original function, these genes were named gene(s) associated with retinoid-IFN-induced mortality (GRIM). Among several genes identified in this manner, we found GRIM-19, which coded for a 16-kDa protein present both in the nucleus and cytoplasm (14). Overexpression of GRIM-19 caused apoptosis, and cells that expressed moderate levels of this protein grew more slowly than cells transfected with the empty vector. To define its mechanism of action, we screened a yeast two-hybrid library with GRIM-19 and identified STAT3 as one of its binding partners. Here we show that GRIM-19 interacts specifically with the transcription factor STAT3 and inhibits STAT3-dependent gene expression. The transactivation domain (TAD) of STAT3 appears to be a direct target of GRIM-19. Because STAT3 is activated constitutively by oncogenes and autocrine growth factors (15), GRIM-19 may prove to be a novel antioncogenic protein.
Materials and Methods
Plasmids, Cell Lines, and Antibodies. WT and mutant STAT3 proteins were expressed with N-terminally flag-epitope tags, by using the pCMV2-flag vector. Deletion and substitution mutations were engineered with the aid of PCR. Expression-verified mutants were used for further experiments. STAT3-DB (16) and mAbs against GRIM-19 (17) have been described. Antibodies against native, tyrosine (STAT3-YP) and serine (STAT3-SP) phosphorylated forms of STAT3 (Cell Signaling, Beverly, MA), STAT1, STAT2, STAT5a, and c-Jun (Santa Cruz Biotechnology), and hemagglutinin tag, myc tag, and actin (Sigma) were used. Soluble IL-6 receptor, IL-6 (R & D Systems), human IFN-β (Ares-Serono, Geneva), RA (Sigma), and murine IFN-γ (Pestka Biomedical Labs, Piscataway, NJ) were used as necessary. MCF-7, WT, STAT3–/– (18), and STAT1–/– (19) cells were grown in DMEM with 5% charcoal-stripped FBS. An immortalized human mammary epithelial cell line hTERT-HME (CLONTECH) was grown as suggested by the supplier. The pLEGFP-Stat3-WT or pLEGFP-Stat3-Y705F vectors were generated by inserting WT Stat3 or Y705F-Stat3 cDNA into the HindIII site of a retroviral vector pLEGFP-N1 (CLONTECH). To obtain infectious retrovirus, each construct was transfected into BOSC23 (CLONTECH) packaging cells, and after 48 h supernatants were used to infect hTERT-HME1 cells on 3 consecutive days. Stably transduced hTERT-HME1 cell pools were shown to express STAT3 proteins of the expected size by Western analysis.
Gene Expression Analyses. Western analysis, immunoprecipitation (IP), electrophoretic mobility-shift assay (EMSA), luciferase assays, and β-galactosidase reporter assays were performed as described (12, 20). For Northern analysis, total RNA was probed with 32P-labeled DNAs corresponding to cdc2 or cyclin B1 (21–23). The cDNA probes were generated from 4 μg of hTERT-HME1 total RNA, using Moloney murine leukemia virus reverse transcriptase (Life Technologies, Rockville, MD). RT-PCR amplification was carried out for one cycle at 95°C for 5 min, followed by 32 cycles at 95°C for 45 s, 62°C for 45 s, and 72°C for 45 s. PCR primers used were as follows: 5′-ATCGGGGAACCTCTGATTTT-3′ (sense) and 5′-TCACACACAGGCACCTTCTC-3′ (antisense) for cyclin B1 (24), and 5′-GTCAGTCTTCAGGATGTGCT-3′ (sense) and 5′-GGCCACACTTCATTATTGGG-3′ (antisense) for cdc2 (25). A 32P-labeled double-stranded oligonucleotide with the sequence 5′-GATCCTTCTGGGAATTCCTAGATC-3′ was used for STAT3 EMSA. The S3-Luc construct bears three copies of the Stat3 binding site (similar to the one found in human C-reactive protein gene promoter) cloned upstream of a luciferase reporter driven by the thymidine kinase (TK) promoter. Palindromic IFN-response element (pIRE)-Luc contains three copies of the pIRE of the IRF1 promoter upstream of the TK promoter. Luciferase reporters driven by the cyclin B1 and cdc2 promoters have been described (26, 27). The β-actin–β-galactosidase reporter was used as an internal control for normalizing variations in transfection efficiency.
Yeast Two-Hybrid Screen. The commercially available Matchmaker 3 system (CLONTECH) was used for isolating GRIM-19-interacting proteins. Briefly, the full-length GRIM-19 ORF was expressed as a myc-tagged fusion protein with the DNA binding domain (DBD) of GAL4 from the plasmid pGBKT7 in Saccharomyces cerevisiae strain AH109. A pretransformed (yeast strain Y187) human bone marrow cDNA library, expressed as hemagglutinin epitope-tagged fusion proteins with the TAD of GAL4 in pAct2, was used as a source of GRIM-19 binding partners. The two strains were mated, and positive clones were isolated after selection on minimal medium lacking leucine, tryptophan, and histidine. The specificity of interaction was validated by performing cotransformation of the individual plasmids identified in the first round with GRIM-19. Positive clones were sequenced to identify the gene.
Results
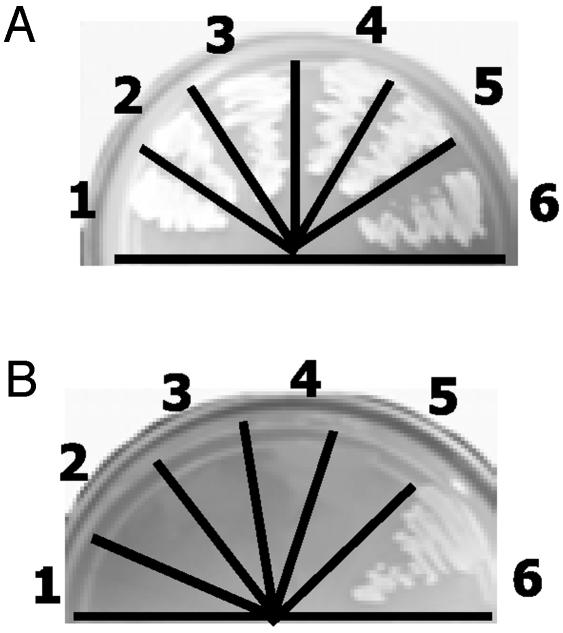
Identification of STAT3 as a GRIM-19 Binding Protein. To identify targets of GRIM-19, we used a yeast two-hybrid screen and identified 16 individual positive colonies from a library that expressed ≈1 × 106 cDNAs in the first round. Among these, four independent clones were identified later as STAT3. All of the rescued STAT3 plasmids lacked the N-terminal 40 aa, eliminating an essential role for this domain in the interaction with GRIM-19. We have prioritized an analysis of the interactions between GRIM-19 and STAT3 because Stat3 is activated constitutively by a number of oncogenes and growth factors in tumor cells (15, 28). The other clones remain to be characterized. Whereas the single and double yeast transformants grew on media containing leucine, tryptophan, and histidine (Fig. 1A), as expected, only double transformants expressing STAT3 and GRIM-19 survived in minimal medium lacking these nutrients (Fig. 1B). We also confirmed the interaction between STAT3 and GRIM-19 in yeast extracts (data not shown).
Fig. 1.
Identification of STAT3 as a GRIM-19 binding protein. Yeast cells transformed with the indicated expression vectors were grown on media containing (A) or lacking (B) tryptophan, leucine, and histidine for 1 week. Plasmids transformed in each case were as follows: 1, pAct2; 2, pGBKT7; 3, STAT3; 4, GRIM-19; 5, GRIM-19+pACT2; 6, STAT3+GRIM-19. pGBKT7 and pAct2 express only the GAL4-DBD and GAL4-TAD, respectively.
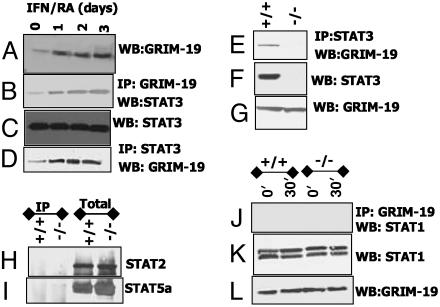
Interaction Between STAT3 and GRIM-19 in Mammalian Cells. To demonstrate the physiological relevance of the yeast two-hybrid data in mammalian cells, human breast carcinoma MCF-7 cells were stimulated with IFN/RA (known inducers of GRIM-19), and cell lysates were immunoprecipitated with GRIM-19-specific antibodies. The products were probed with STAT3-specific antibodies in a Western analysis. As shown in Fig. 2, STAT3 binds to GRIM-19 in the unstimulated state. After IFN/RA treatment, more STAT3 was coimmunoprecipitated with GRIM-19 (Fig. 2B). This increase appears, in part, to be caused by a rise in cellular GRIM-19 content (Fig. 2A). There was no change in STAT3 levels under these conditions. Conversely, GRIM-19 was coimmunoprecipitated by STAT3 antibodies (Fig. 2D). A similar profile of interaction between GRIM-19 and STAT3 was observed in HeLa, T47D, and BT20 cells (data not shown). We examined the specificity of this interaction further by performing an IP with STAT3 antibodies using lysates from WT and STAT3–/– mouse embryonic fibroblasts. STAT3 antibodies failed to coimmunoprecipitate GRIM-19 from the mutant but not WT cells (Fig. 2E). No difference in the steady-state expression of GRIM-19 was noted between these two cell types (Fig. 2G). STAT3 was also coimmunoprecipitated with GRIM-19 from STAT1–/– cells (data not shown). Thus, the GRIM-19–STAT3 interactions are specific and occur under physiological conditions.
Fig. 2.
Endogenous GRIM-19 binds to STAT3 specifically. (A–D) MCF-7 cells stimulated with IFN-β (500 units/ml) and RA (1 μM) were lysed, and IP or Western (WB) analysis was performed with specific antibodies. (E–G) Specific IP of STAT3 with GRIM-19. The STAT3 genotypes of cells are indicated. (H and I) GRIM-19 does not bind to other STATs. Lanes 1 and 2, lysates were immunoprecipitated with GRIM-19 antibodies. Lanes 3 and 4, lysates without IP. Western transfers were probed with the indicated antibodies. (J–L) STAT1 does not bind to GRIM-19. Cells were stimulated with murine IFN-γ (200 units/ml) for 30 min; lysates were processed and analyzed as indicated.
GRIM-19 Does Not Bind to Other STATs. IP with GRIM-19 antibodies, followed by Western analysis of the products with STAT-specific antibodies, did not reveal an appreciable interaction between GRIM-19 and other STATs (Fig. 2 H–L). We performed these experiments in STAT3–/– cells to avoid a potential interaction of STAT3 with other STATs, which could permit the indirect IP of these STATs with GRIM-19 through STAT3. In both WT and STAT3–/– cells, no strong interaction between GRIM-19 and STAT1, STAT2, or STAT5a was detected, despite their abundant expression. Similar results were obtained in other cell types (data not shown). The STAT1 antibody detects both the α and β isoforms, which can be seen in the Western assay (Fig. 3I). IFN-γ treatment, which activates STAT1α and STAT1β (29), did not enhance these interactions (Fig. 3J). However, upon prolonged exposure, we detected weak STAT1α and STAT1β bands (data not shown). A comparable level of GRIM-19 was present in both cell types (Fig. 3K), indicating that this difference could not be attributed to lack of GRIM-19.
Fig. 3.
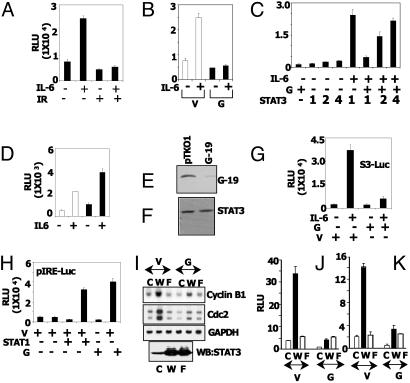
GRIM-19 inhibits STAT3-dependent gene expression. (A) MCF-7 cells were transfected with S3-Luc and treated with the indicated agents for 16 h. Luciferase activity in the lysates was measured. Each bar represents mean relative light units (RLU) ± SE of triplicate transfectants in the same experiment. IL-6 (10 ng/ml), IFN-β (500 units/ml), and RA (1 μM) were used in this experiment. IR, IFN-β/RA combination. (B) Transfection was performed as in A, in the presence of the empty vector pCXN2 or the GRIM-19 (G-19) expression plasmid. (C) Increasing amounts of STAT3 plasmid overcome GRIM-19-induced inhibition. Luciferase activity was determined as in A. Numbers across the STAT3 row indicate the fold excess of STAT3 used, relative to GRIM-19, in the cotransfection assays. GRIM-19 and the reporter were used at 100 ng per transfection in the STAT3–/– cells. Cells were treated with soluble IL-6 receptor (100 ng/ml) and IL-6 (10 ng/ml). (D) Antisense GRIM-19 promotes STAT3-dependent gene expression. Cells stably expressing empty pTKO1 vector (empty bars) and antisense GRIM-19 (filled bars) were used for transfection analysis as in A. (E and F) The same cell lysates were monitored for GRIM-19 and STAT3 expression by Western analysis. (G and H) The specific inhibitory effect of GRIM-19 on STAT3. STAT1–/– cells were transfected with S3-Luc or pIRE Luc along with GRIM-19, and cells were stimulated with the indicated agents. IFN-γ was used at 200 units/ml in H. STAT1 expression was reconstituted along with reporter where indicated. (I) Expression of STAT3-regulated genes in hTERT-HME cells is inhibited by GRIM-19. Stable cell lines expressing WT and Y705F mutant forms of STAT3 were transfected with mammalian expression vector pCXN2 (V) or the same vector expressing GRIM-19 (G). RNA was analyzed by the Northern method. (J and K) The same cells were transfected with luciferase reporters driven by the cyclin B1 and cdc2 promoters, and the influence of GRIM-19 on luciferase expression was measured. c, Control vector; W, WT mutant of STAT3; F, Y705F mutant of STAT3.
GRIM-19 Is an Inhibitor of STAT3-Dependent Gene Expression. We next examined the biological significance of GRIM-19/STAT3 interactions (Fig. 3). A luciferase reporter (S3-Luc) driven by a minimal STAT3-responsive element was used in transient transfection studies in MCF-7 cells. First, we examined the effect of IFN/RA on the IL-6-induced expression of S3-Luc. Luciferase expression was strongly induced upon IL-6 treatment, which was inhibited by IFN/RA (Fig. 3A). IFN/RA itself did not induce the reporter. In a separate experiment, cells were transfected with GRIM-19 and the effects on IL-6 induced expression of S3-Luc were studied. Although the empty expression vector had no effect on IL-6-induced expression of S3-Luc, GRIM-19 ablated it (Fig. 3B). Basal expression of the reporter was repressed to a slight, but significant, extent. We have tested whether overexpression of STAT3 could overcome GRIM-19-dependent repression. In this experiment, STAT3 and GRIM-19 were coexpressed with the reporter in STAT3–/– cells (18) to avoid any contribution of endogenous STAT3. As expected, GRIM-19 inhibited the IL-6-induced STAT3-dependent expression of the reporter and increasing doses of STAT3 overcame the inhibition (Fig. 3C). In a complementary experiment, we compared the effect of IL-6 in cells transfected with empty vector pTKO1 or expressing antisense GRIM-19 (Fig. 3D). IL-6 induced the reporter significantly more in cells expressing antisense GRIM-19 than in control pTKO1 cells. GRIM-19 expression was suppressed strongly in cells expressing antisense GRIM-19 (Fig. 3E). There was no difference in STAT3 expression in these cells (Fig. 3F).
Because IL-6 also induces STAT1 in some cell types (30), it is unclear from these experiments whether these inhibitory effects are directed entirely toward STAT3-dependent transcription. To test this point, S3-Luc was transfected into STAT1–/– mouse embryonic fibroblasts, and the effect of GRIM-19 on the luciferase gene was measured. IL-6 induced the luciferase reporter normally, and this induction was inhibited by GRIM-19 (Fig. 3G). These results rule out the possibility that STAT1 is involved in this repression. We next tested the effect of GRIM-19 on STAT1-inducible expression through the pIRE. In this experiment, STAT1 was coexpressed with the pIRE-Luc reporter, and the cells were treated with IFN-γ. In parallel, we studied the effect of coexpressed GRIM-19 on luciferase expression (Fig. 3H). As expected, STAT1 coexpression permitted a robust IFN-γ-dependent induction of the reporter, which was not repressed by GRIM-19. On the contrary, a stimulatory effect of GRIM-19 was seen. These data show an exclusive negative regulatory effect of GRIM-19 on STAT3-dependent gene expression.
To further demonstrate the negative effects of GRIM-19, we measured the expression of endogenous STAT3-regulated genes in human hTERT-HME cells stably expressing a control vector (pLEGFPN1), the WT, or the Y705F mutant of STAT3. Two separate experiments were performed. In the first, the empty vector pCXN2 or the same vector expressing GRIM-19 (14) was introduced into the cells, using the Fugene 6 reagent. Total RNA was prepared, and Northern analysis was performed for the expression of the STAT3-regulated genes cyclin B1 and cdc2 (Fig. 3I). Expression of these genes was strongly up-regulated in WT cells compared with control vector transfected cells. The Y705F mutant failed to induce these two genes. The cdc2 probe detected two mRNAs (1.8 and 1.6 kb), and both were induced by STAT3. In contrast to the pCXN2 control, GRIM-19 expression substantially (≈90% of the control) inhibited the STAT3-induced expression of cyclin B1 and cdc2 mRNAs. GRIM-19 had no effect on GAPDH mRNA expression. Importantly, even basal expression was inhibited by GRIM-19. The Y705F mutant of STAT3 failed to induce these mRNAs, and GRIM-19 had no effect. The difference between WT and Y705F cells does not appear to be caused by a variation in STAT3 protein expression (Fig. 3I Bottom). The results shown in Fig. 3I were confirmed in a second experiment using luciferase reporters driven by the native cyclin B1 (Fig. 3J) or cdc2 (Fig. 3K) promoters. Basal and WT STAT3-dependent induction of these reporters was strongly ablated by GRIM-19. As expected, the Y705F mutant did not induce the reporters strongly and GRIM-19 had no effect. A similar inhibition of the expression of endogenous STAT3-dependent genes BcL2 and BcL-XL was observed in MCF-7 cells stably transfected with GRIM-19 (data not shown).
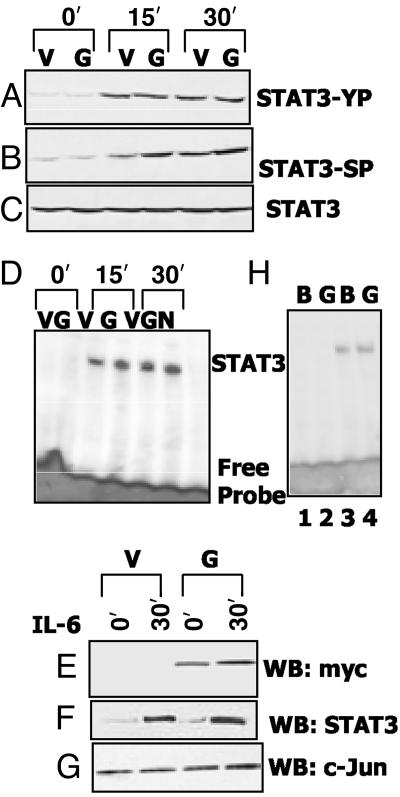
GRIM-19 Does Not Inhibit the Activation or DNA Binding of STAT3. In light of the negative regulatory effects of GRIM-19 on STAT3-inducible gene expression, we next examined whether GRIM-19 blocked the activation and DNA binding of STAT3 in response to IL-6. MCF-7 cells stably expressing GRIM-19 were exposed to IL-6 for various times, and a Western analysis of cell extracts was performed with phospho-specific STAT3 antibodies. Both serine and tyrosine phosphorylation of STAT3 increased after IL-6 treatment. However, in response to IL-6, no significant difference in the tyrosine or serine phosphorylation of STAT3 was observed between the control and GRIM-19-expressing cells (Fig. 4 A and B). As expected, there was no difference in the steady-state levels of STAT3 in these cells (Fig. 4C). Thus, GRIM-19 does not prevent STAT3 activation. We next examined nuclear extracts for the DNA binding of STAT3 by using EMSA (Fig. 4C). Again, there was no significant difference in the ability of STAT3 to bind to the STAT3-responsive element between cells expressing GRIM-19 or controls. The identity of the EMSA band was established by supershifting this complex with STAT3 antibodies and by the failure of a mutant STAT3-responsive element to compete with the WT sequence (data not shown). We also confirmed the stable overexpression of GRIM-19 in the nuclear extracts (Fig. 4D). As expected, more STAT3 migrated to the nucleus in IL-6-treated cells, compared with untreated cells. However, there was no significant difference in the amount of STAT3 in the nuclear extracts, whether the cells overexpressed GRIM-19 or not (Fig. 4F). Antibodies against c-Jun were used to probe these nuclear extracts, as an internal control (Fig. 4G). c-Jun was constitutively present in the nuclei of both cell types. To test whether GRIM-19 interferes with DNA binding further, IL-6-treated nuclear extracts were incubated with recombinant GRIM-19 before EMSAs (Fig. 4H). No inhibition of DNA binding was observed, compared with the control (Fig. 4H, compare lanes 3 and 4). GRIM-19 itself did not form any complex with the probe (Fig. 4H, compare lanes 1 and 2). These results show that the negative effects of GRIM-19 are not caused by inhibition of the nuclear translocation of STAT3 or interference with its binding to DNA.
Fig. 4.
Effect of GRIM-19 on STAT3 activation and DNA binding. (A–C) MCF-7 cells stably transfected with empty vector (V) or vector expressing myc-tagged GRIM-19 (G) were stimulated with IL-6 (10 ng/ml). Cell lysates were analyzed by the Western method (WB) with antibodies specific for native or phospho-STAT3. (D) Nuclear extracts (5 μg) from IL-6-stimulated cells were used for EMSA with a STAT3 binding element. N, no extract. (E–G) Total nuclear protein (70 μg) was used in Western analyses. (H) The effect of recombinant GRIM-19 on DNA binding of STAT3. Reactions were incubated with buffer (B) or 4 μg of recombinant GRIM-19 (G). Lanes 1 and 2, no nuclear extract; lanes 3 and 4, nuclear extract from IL-6-treated cells.
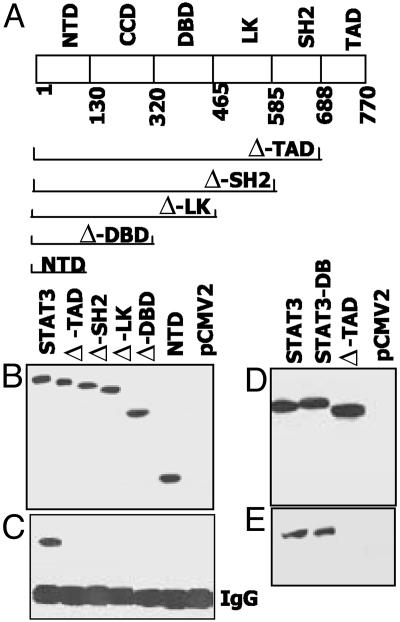
GRIM-19 Binding to STAT3 Requires the TAD and the Ser-727 Residue. Because GRIM-19 inhibits STAT3-inducible gene expression without significantly altering its activation, we next determined which region of STAT3 is targeted by GRIM-19. Several STAT3 mutants, each lacking specific functional modules (Fig. 5A), were generated. These mutants were transfected into STAT3–/– cells to avoid any interference from endogenous STAT3, and the cell extracts were immunoprecipitated with GRIM-19-specific antibodies. The IP products were analyzed by the Western method and probed with antibody to the flag epitope tag. As shown in Fig. 5B, all mutants were expressed equivalently. WT STAT3 bound to GRIM-19 normally. However, any deletion extending beyond the TAD prevented the co-IP of STAT3 with GRIM-19 (Fig. 5C). Lastly, STAT3DB, a mutant that fails to bind to DNA (16), but not the TAD mutant, bound to GRIM-19 similarly to WT STAT3 (Fig. 5E). These observations indicate that the STAT3 TAD is critical for GRIM-19 binding.
Fig. 5.
The TAD of STAT3 is required for GRIM-19 binding. (A) Diagram of the modular structure of STAT3. Amino acid positions at which stop codons are introduced into the mutants and the coordinates of various domains are indicated. CCD, coiled-coil domain; LK, linker domain; NTD, N-terminal domain; SH2, Src homology 2 domain. (B and D) The N-terminally flag-tagged mutants were transfected into STAT3–/– cells, and 60μg of lysate from each sample was used for Western analysis with anti-flag antibodies. STATDB has been described (16). (C and E) Total lysate (250 μg) was used for IP with GRIM-19 specific antibodies, followed by Western analysis with anti-flag antibodies.
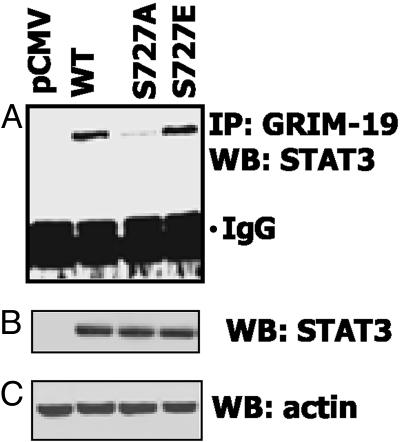
The STAT3 TAD bears a serine at residue 727, which is critical for optimal transactivation (31, 32). Because GRIM-19 requires the STAT3 TAD for binding, we studied the impact of S727 mutation on this process. Substitution mutants bearing an alanine or a phosphomimetic glutamic acid residue at residue 727 were cotransfected with GRIM-19 into STAT3–/– cells (Fig. 6A). The S727A mutant lost almost 95% of GRIM-19 binding capacity compared with the WT protein. The S727E mutant bound to GRIM-19 normally, indicating the importance of a negative charge at this site for an efficient interaction. The differences in binding were not caused by differential expression of STAT3, because all mutants were expressed similarly (Fig. 6B).
Fig. 6.
The S727 residue is important for GRIM-19 binding to STAT3. (A) STAT3–/– cells were transfected with flag-tagged WT or mutant STAT3 constructs and an IP with GRIM-19-specific antibodies was performed as in Fig. 5. The same lysates were analyzed for the expression of STAT3 (B) and actin (C). WB, Western analysis.
Discussion
Transcription factor STAT3 (31, 32) is central to several diverse biological activities in mammalian cells including cytokine-induced responses, differentiation, embryonic growth, and cell survival (33). In normal cells, its activity is tightly regulated by feedback inhibitory mechanisms that prevent excessive signaling. In contrast, constitutive activation of STAT3 promotes the growth of cells transformed by viruses, oncogenes, and autocrine growth factors (15, 34). Two distinct types of proteins, the suppressor of cytokine signaling (SOCS) proteins (35) and protein inhibitor of activated STAT3 (PIAS3) (36), inhibit activated STAT3. The SOCS proteins inhibit the Janus tyrosine kinases (JAKs) and cause postinduction inhibition of the ligand-induced responses, and PIAS3 inhibits the DNA binding of STAT3 (36). However, recent studies indicate that SOCS proteins do not inhibit constitutively active STAT3 (37). Although described as specific inhibitors of STAT proteins (36), the PIAS proteins have been shown now to act as transcriptional coactivators with steroid receptors (38) and appear to act as small ubiquitin-like modifier 1 ligases (39, 40) in the context of other transcription factors. More recent studies suggest that they have a more important role to play in steroid signaling than in JAK-STAT signaling (41). A poorly defined third mechanism of STAT3 inhibition is its dephosphorylation, through an unspecified STAT3-tyrosine-specific phosphatase (33). In light of the multiple physiological roles played by STAT3, no single mechanism is likely to account fully for the regulation of its activity.
In our attempts to define molecular mechanisms involved in IFN/RA-induced antitumor actions, we identified the gene product GRIM-19 (14). Although unremarkable from the point of its structural relationship to other apoptotic proteins, GRIM-19 caused apoptosis when overexpressed. Its nuclear and cytoplasmic distribution and the punctuate staining patterns observed in cells prompted us to suspect that GRIM-19 might interact with various protein(s) or protein complexes to regulate cellular responses. Indeed, a subsequent study (42) has shown that GRIM-19 is also a subunit of the mitochondrial NADH:ubiquinone oxidoreductase (respiratory complex I). More recently, we have shown that the viral oncoproteins vIRF1 of Kaposi's sarcoma-associated herpes virus and E6 of high-risk human papilloma viruses interact with GRIM-19 and down-regulate its apoptotic functions (43). In the current article, we show that GRIM-19 binds to STAT3 and inhibits transcription.
Because normal tyrosine and serine phosphorylation of STAT3 occurs even in the presence of overexpressed GRIM-19 (Fig. 4 A–C), receptor-proximal events involved in STAT3 activation are not interfered with. This characteristic distinguishes GRIM-19 from the SOCS proteins (35). Because there was no decrease in tyrosine or serine phosphorylation of STAT3 in GRIM-19-expressing cells, phosphatase activities are not likely to be involved in its anti-STAT3 action. EMSAs with nuclear extracts showed clearly that neither the DNA binding nor the ligand-induced nuclear translocation of STAT3 are inhibited in the presence of GRIM-19 (Fig. 4 D and H). In this respect, GRIM-19 differs from protein inhibitor of activated STAT3, which interferes with the DNA binding of STAT3 (36). Mutational analysis shows that GRIM-19 binding to STAT3 requires the TAD (Fig. 5). A STAT3 mutant defective in DNA binding still interacted with GRIM-19. Consistent with this finding, we observed an inhibition of STAT3-dependent transcription upon coexpression of GRIM-19. Conversely, STAT3-dependent transcription was significantly higher in cells expressing antisense GRIM-19, compared with the controls. Similarly, the STAT3-induced expression of cyclin B1 and cdc2 genes was inhibited in the presence of GRIM-19 (Fig. 3 J–L). Inhibition of these genes can suppress cell growth. STAT3 also promotes the expression of antiapoptotic mitochondrial regulators of permeability, such as Bcl-XL and BclII (44, 45). It is conceivable that down-regulation of these antiapoptotic proteins in conjunction with other proapoptotic regulators might further damage mitochondria, leading to disruption of oxidative phosphorylation, where GRIM-19 is present in complex I, and releases additional GRIM-19 into the cytoplasm, thus potentially amplifying apoptotic responses.
Remarkably, among several members of the STAT family, GRIM-19 binds only to STAT3 (Fig. 2 H–J). We have also shown that STAT1-dependent transcription is not inhibited by GRIM-19 (Fig. 3H). Because STAT1 is critical for IFN-induced responses and GRIM-19 is an IFN/RA-induced gene product, it would be counterproductive for GRIM-19 to inhibit its upstream regulator. In this respect GRIM-19 differs from the SOCS proteins, which inhibit their own inducers (35). Prolonged exposure of the GRIM-19 IP blots showed a weak interaction with STAT1α and STAT1β (data not shown), suggesting that the weak interaction of GRIM-19 occurs outside the TAD of STAT1. This weak interaction does not appear to be inhibitory and even seems to stimulate transcription. Lastly, the weak binding of GRIM-19 to STAT1α and STAT1β does not appear to be caused by the ability of STAT1 to form heterodimers with STAT3, because this interaction is also seen in STAT3–/– cells (data not shown). A previous report has shown no substantial difference in the transactivating capabilities of the STATs, because STAT1 functions can be driven equivalently by TADs of other STATs (46). However, with respect to inhibition, the STAT3 TAD differs from other STAT TADs on the basis of our results. Therefore, it is possible that specific inhibitors of other STAT TADs exist. However, thus far, we have not found any homologues of GRIM-19, but we cannot rule out the possibility of functional homologues. Because GRIM-19 homologues are present in all vertebrates, Drosophila, and Caenorhabditis elegans (14, 42), STAT inhibition by GRIM-19 may be a universal phenomenon. Lastly, a chimeric protein with the STAT3-TAD fused to the GAL4-DBD failed to bind to GRIM-19, suggesting that the TAD alone is insufficient for this interaction (data not shown). Therefore, in addition to the TAD, the primary high-affinity binding site, a secondary low-affinity site formed by the folding of STAT3 may be required for binding to GRIM-19. In the absence of the TAD, the secondary site itself may not bind GRIM-19 tightly. This possibility is hinted at by the S727A mutant, which retains residual binding to GRIM-19. Alternatively, another protein may mediate the interaction of GRIM-19 with the secondary site, which requires the engagement of GRIM-19 with the TAD.
While these results were being prepared for publication, a study appeared describing the anti-STAT3 activity of GRIM-19 (47). There are several major differences between our report and that of Lufei et al. (47), who have argued that GRIM-19 binds to the coiled-coil, DNA binding, and linker domains of STAT3. In contrast, our study shows that the STAT3 TAD is the primary target of GRIM-19. Conclusions drawn by Lufei et al. were based on STAT3 and GRIM-19 overexpression in COS-1 cells. Such experiments do not rule out the possibility that these interactions are mediated through the heterodimerization of mutant and endogenous STAT3. We have conducted the interaction studies in STAT3–/– cells, where the role of endogenous STAT3 in mediating GRIM-19 interactions can be excluded. That said, we have also obtained similar results with STAT3+/+ and HeLa cells (data not shown), indicating that the behavior of STAT3–/– cells is not idiosyncratic. Similarly, GRIM-19 exerted negative effects on epidermal growth factor-induced gene expression through STAT3 (data not shown), indicating that its effects are not inducer-specific. Because STAT3 phosphorylation, nuclear localization, and DNA binding are not different in GRIM-19-overexpressing cells (Fig. 4), we believe that the failure of STAT3 to translocate to the nucleus, as reported by Lufei et al., appears be an artifact of overexpression. We have shown clearly that S727 of STAT3 plays an important role in GRIM-19 binding because a mutation of this residue nearly eliminates binding (Fig. 6).
Our study has identified a cell growth inhibitory mechanism used by biologic therapeutic agents. Our results may also relate to the anti-STAT3 effects of IFN-α observed in some clinical studies (48). STAT3 joins a group of IFN-inhibited pro-growth transcription factors, which until now comprised c-myc (49) and E2F (50), although very different mechanisms of inhibition are involved in each case.
Acknowledgments
We thank Robert Schreiber for the STAT1–/– mouse embryonic fibroblasts. This work was supported by National Institutes of Health Grants CA78282 and CA71401 (to D.V.K.) and a Ministero dell'Istruzione dell'Università e della Ricerca COFIN grant (to V.P.).
Abbreviations: DBD, DNA binding domain; EMSA, electrophoretic mobility-shift assay; GRIM, gene associated with retinoid-IFN-induced mortality; IP, immunoprecipitation; pIRE, palindromic IFN-response element; RA, all-trans retinoic acid; SOCS, suppressor of cytokine signaling; STAT, signal transducer and activator of transcription; TAD, transactivation domain; TK, thymidine kinase.
References
- 1.Kimchi, A. (1998) Biochim. Biophys. Acta 1377, F13–F33. [DOI] [PubMed] [Google Scholar]
- 2.Kalvakolanu, D. V. (2000) Histol. Histopathol. 15, 523–537. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan, D. H., Shankaran, V., Dighe, A. S., Stockert, E., Aguet, M., Old, L. J. & Schreiber, R. D. (1998) Proc. Natl. Acad. Sci. USA 95, 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ikeda, H., Old, L. J. & Schreiber, R. D. (2002) Cytokine Growth Factor Rev. 13, 95–109. [DOI] [PubMed] [Google Scholar]
- 5.Willman, C. L., Sever, C. E., Pallavicini, M. G., Harada, H., Tanaka, N., Slovak, M. L., Yamamoto, H., Harada, K., Meeker, T. C., List, A. F. & Taniguchi, T. (1993) Science 259, 968–971. [DOI] [PubMed] [Google Scholar]
- 6.Holtschke, T., Lohler, J., Kanno, Y., Fehr, T., Giese, N., Rosenbauer, F., Lou, J., Knobeloch, K. P., Gabriele, L., Waring, J., et al. (1996) Cell 87, 307–317. [DOI] [PubMed] [Google Scholar]
- 7.Choubey, D., Li, S. J., Datta, B., Gutterman, J. U. & Lengyel, P. (1996) EMBO J. 15, 5668–5678. [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, C. J., Wang, H. & Lengyel, P. (1999) EMBO J. 18, 2845–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindner, D. J., Kalvakolanu, D. V. & Borden, E. C. (1997) Semin. Oncol. 24, S9-99–S9-104. [PubMed] [Google Scholar]
- 10.Moore, D. M., Kalvakolanu, D. V., Lippman, S. M., Kavanagh, J. J., Hong, W. K., Borden, E. C., Paredes-Espinoza, M. & Krakoff, I. H. (1994) Semin. Hematol. 31, 31–37. [PubMed] [Google Scholar]
- 11.Lindner, D. J., Borden, E. C. & Kalvakolanu, D. V. (1997) Clin. Cancer Res. 3, 931–937. [PubMed] [Google Scholar]
- 12.Hofmann, E. R., Boyanapalli, M., Lindner, D. J., Weihua, X., Hassel, B. A., Jagus, R., Gutierrez, P. L. & Kalvakolanu, D. V. (1998) Mol. Cell. Biol. 18, 6493–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deiss, L. P. & Kimchi, A. (1991) Science 252, 117–120. [DOI] [PubMed] [Google Scholar]
- 14.Angell, J. E., Lindner, D. J., Shapiro, P. S., Hofmann, E. R. & Kalvakolanu, D. V. (2000) J. Biol. Chem. 275, 33416–33426. [DOI] [PubMed] [Google Scholar]
- 15.Buettner, R., Mora, L. B. & Jove, R. (2002) Clin. Cancer Res. 8, 945–954. [PubMed] [Google Scholar]
- 16.Bromberg, J. F., Horvath, C. M., Besser, D., Lathem, W. W. & Darnell, J. E., Jr. (1998) Mol. Cell. Biol. 18, 2553–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, J., Angell, J. E., Zhang, J., Ma, X., Seo, T., Raha, A., Hayashi, J., Choe, J. & Kalvakolanu, D. V. (2002) J. Interferon Cytokine Res. 22, 1017–1026. [DOI] [PubMed] [Google Scholar]
- 18.Costa-Pereira, A. P., Tininini, S., Strobl, B., Alonzi, T., Schlaak, J. F., Is'harc, H., Gesualdo, I., Newman, S. J., Kerr, I. M. & Poli, V. (2002) Proc. Natl. Acad. Sci. USA 99, 8043–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meraz, M. A., White, J. M., Sheehan, K. C., Bach, E. A., Rodig, S. J., Dighe, A. S., Kaplan, D. H., Riley, J. K., Greenlund, A. C., Campbell, D., et al. (1996) Cell 84, 431–442. [DOI] [PubMed] [Google Scholar]
- 20.Weihua, X., Lindner, D. J. & Kalvakolanu, D. V. (1997) Proc. Natl. Acad. Sci. USA 94, 7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., Frisch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 22.Maity, A., McKenna, W. G. & Muschel, R. J. (1995) EMBO J. 14, 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa, Y., Iwase, S., Terui, Y., Kikuchi, J., Sakai, T., Nakamura, M., Kitagawa, S. & Kitagawa, M. (1996) J. Biol. Chem. 271, 28469–28477. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi, M., Seki, N., Ozaki, T., Kato, M., Kuno, T., Nakagawa, T., Watanabe, K., Miyazaki, K., Ohira, M., Hayashi, S., et al. (2002) Cancer Res. 62, 2203–2209. [PubMed] [Google Scholar]
- 25.Akimoto, M., Hangai, M., Okazaki, K., Kogishi, J., Honda, Y. & Kaneda, Y. (1998) Exp. Eye Res. 67, 395–401. [DOI] [PubMed] [Google Scholar]
- 26.Katula, K. S., Wright, K. L., Paul, H., Surman, D. R., Nuckolls, F. J., Smith, J. W., Ting, J. P., Yates, J. & Cogswell, J. P. (1997) Cell Growth Differ. 8, 811–820. [PubMed] [Google Scholar]
- 27.Taylor, W. R., Schonthal, A. H., Galante, J. & Stark, G. R. (2001) J. Biol. Chem. 276, 1998–2006. [DOI] [PubMed] [Google Scholar]
- 28.Yu, C. L., Meyer, D. J., Campbell, G. S., Larner, A. C., Carter-Su, C., Schwartz, J. & Jove, R. (1995) Science 269, 81–83. [DOI] [PubMed] [Google Scholar]
- 29.Muller, M., Laxton, C., Briscoe, J., Schindler, C., Improta, T., Darnell, J. E., Jr., Stark, G. R. & Kerr, I. M. (1993) EMBO J. 12, 4221–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, X., Blenis, J., Li, H. C., Schindler, C. & Chen-Kiang, S. (1995) Science 267, 1990–1994. [DOI] [PubMed] [Google Scholar]
- 31.Akira, S., Nishio, Y., Inoue, M., Wang, X. J., Wei, S., Matsusaka, T., Yoshida, K., Sudo, T., Naruto, M. & Kishimoto, T. (1994) Cell 77, 63–71. [DOI] [PubMed] [Google Scholar]
- 32.Zhong, Z., Wen, Z. & Darnell, J. E., Jr. (1994) Science 264, 95–98. [DOI] [PubMed] [Google Scholar]
- 33.Levy, D. E. & Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662. [DOI] [PubMed] [Google Scholar]
- 34.Bromberg, J. (2002) J. Clin. Invest. 109, 1139–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Starr, R. & Hilton, D. J. (1999) BioEssays 21, 47–52. [DOI] [PubMed] [Google Scholar]
- 36.Chung, C. D., Liao, J., Liu, B., Rao, X., Jay, P., Berta, P. & Shuai, K. (1997) Science 278, 1803–1805. [DOI] [PubMed] [Google Scholar]
- 37.Iwamoto, T., Senga, T., Naito, Y., Matsuda, S., Miyake, Y., Yoshimura, A. & Hamaguchi, M. (2000) Oncogene 19, 4795–4801. [DOI] [PubMed] [Google Scholar]
- 38.Junicho, A., Matsuda, T., Yamamoto, T., Kishi, H., Korkmaz, K., Saatcioglu, F., Fuse, H. & Muraguchi, A. (2000) Biochem. Biophys. Res. Commun. 278, 9–13. [DOI] [PubMed] [Google Scholar]
- 39.Jackson, P. K. (2001) Genes Dev. 15, 3053–3058. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt, D. & Muller, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2872–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kotaja, N., Aittomaki, S., Silvennoinen, O., Palvimo, J. J. & Janne, O. A. (2000) Mol. Endocrinol. 14, 1986–2000. [DOI] [PubMed] [Google Scholar]
- 42.Fearnley, I. M., Carroll, J., Shannon, R. J., Runswick, M. J., Walker, J. E. & Hirst, J. (2001) J. Biol. Chem. 276, 38345–38348. [DOI] [PubMed] [Google Scholar]
- 43.Seo, T., Lee, D., Shim, Y. S., Angell, J. E., Chidambaram, N. V., Kalvakolanu, D. V. & Choe, J. (2002) J. Virol. 76, 8797–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bromberg, J. F., Wrzeszczynska, M. H., Devgan, G., Zhao, Y., Pestell, R. G., Albanese, C. & Darnell, J. E., Jr. (1999) Cell 98, 295–303. [DOI] [PubMed] [Google Scholar]
- 45.Grad, J. M., Zeng, X. R. & Boise, L. H. (2000) Curr. Opin. Oncol. 12, 543–549. [DOI] [PubMed] [Google Scholar]
- 46.Shen, Y. & Darnell, J. E., Jr. (2001) J. Virol. 75, 2627–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lufei, C., Ma, J., Huang, G., Zhang, T., Novotny-Diermayr, V., Ong, C. T. & Cao, X. (2003) EMBO J. 22, 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkwood, J. M., Farkas, D. L., Chakraborty, A., Dyer, K. F., Tweardy, D. J., Abernethy, J. L., Edington, H. D., Donnelly, S. S. & Becker, D. (1999) Mol. Med. 5, 11–20. [PMC free article] [PubMed] [Google Scholar]
- 49.Raveh, T., Hovanessian, A. G., Meurs, E. F., Sonenberg, N. & Kimchi, A. (1996) J. Biol. Chem. 271, 25479–25484. [DOI] [PubMed] [Google Scholar]
- 50.Melamed, D., Tiefenbrun, N., Yarden, A. & Kimchi, A. (1993) Mol. Cell. Biol. 13, 5255–5265. [DOI] [PMC free article] [PubMed] [Google Scholar]