Abstract
Sox9 has essential roles in endochondral bone formation during axial and appendicular skeletogenesis. Sox9 is also expressed in neural crest cells, but its function in neural crest remains largely unknown. Because many craniofacial skeletal elements are derived from cranial neural crest (CNC) cells, we asked whether deletion of Sox9 in CNC cells by using the Cre recombinase/loxP recombination system would affect craniofacial development. Inactivation of Sox9 in neural crest resulted in a complete absence of cartilages and endochondral bones derived from the CNC. In contrast, all of the mesodermal skeletal elements and intramembranous bones were essentially conserved. The migration and the localization of Sox9-null mutant CNC cells were normal. Indeed, the size of branchial arches and the frontonasal mass of mutant embryos was comparable to that of WT embryos, and the pattern of expression of Ap2, a marker of migrating CNC cells, was normal. Moreover, in mouse embryo chimeras Sox9-null mutant cells migrated to their correct location in endochondral skeletal elements; however, Sox9-null CNC cells were unable to contribute chondrogenic mesenchymal condensations. In mutant embryos, ectopic expression of osteoblast marker genes, such as Runx2, Osterix, and Col1a1, was found in the locations where the nasal cartilages exist in WT embryos. These results indicate that inactivation of Sox9 causes CNC cells to lose their chondrogenic potential. We hypothesize that these cells change their cell fate and acquire the ability to differentiate into osteoblasts. We conclude that Sox9 is required for the determination of the chondrogenic lineage in CNC cells.
The vertebrate neural crest is a population of multipotential stem cells that are precursors of several different cell lineages. The branchial arches and frontonasal mass are populated by cranial neural crest (CNC) cells, which originate in the rhombomeres from the lateral ridges of the neural plate, delaminate, and migrate along defined pathways. These cells then differentiate into a variety of cell types, including chondrocytes, osteoblasts, and odontoblasts of the teeth (1, 2). Previous in vivo studies using techniques of laser ablation, generation of chicken/quail chimeras (3), and cell labeling with a vital dye (4, 5) resulted in a precise fate mapping of CNC cells. Although several transcription factors have been suggested to play important roles in establishing cell lineages (6–8), the molecular mechanisms underlying the specification of cell lineages and the determination of cell fate in CNC cells remain unclear.
The components of the craniofacial skeleton originate from either somitomere or the neural crest, except for a part of the occipital bone. Each bone forms through one of two distinct modes, endochondral or intramembranous bone formation. In endochondral bone formation, condensed mesenchymal cells differentiate into chondrocytes, which subsequently proliferate, hypertrophy, and then calcify their extracellular matrix. At the same time, cells in the perichondrium surrounding the cartilage anlagen differentiate into preosteoblasts and then functional osteoblasts, which replace the calcified cartilage by a bone matrix. In intramembranous ossification, the cells in mesenchymal condensations differentiate directly into osteoblasts and lay down osteoid tissue. CNC cells contribute to both pathways during craniofacial development, but the lack of genetic markers hampers a comprehensive analysis of the molecular basis of these cell lineages.
Sox9, a member of the Sox family of transcription factors, has been shown to regulate both chondrogenesis and sex determination. During embryogenesis, Sox9 is expressed in all chondroprogenitors and differentiated chondrocytes except hypertrophic chondrocytes (9, 10), and it plays essential roles in sequential steps of endochondral bone formation (11). Mutations in one allele of Sox9 in humans result in campomelic dysplasia (CD), a skeletal dysplasia syndrome characterized by sex reversal and skeletal malformations of endochondral bones (12, 13). Sox9 heterozygous mutant mice exhibit abnormalities of craniofacial skeletal elements derived from CNC cells that resemble those of CD patients (14). Indeed, the fact that Sox9 is expressed in CNC cells raises the possibility that Sox9 may have a function in these cells.
In this study, we conditionally inactivated the Sox9 gene specifically in neural crest cells by using the Cre recombinase/loxP (Cre/loxP) recombination system with mice harboring the Wnt1-Cre transgene. Sox9 conditional null mutant embryos had no endochondral skeletal elements derived from CNC cells. However, intramembranous ossification occurred normally, including formation of the mandible. In addition, our experiments suggest that Sox9-null mutant CNC-derived cells change their chondrogenic cell fate into an osteoblast lineage.
Materials and Methods
Generation of Sox9 Mutant Mice. Mice carrying Sox9flox,a Sox9 allele in which the DNA segment that includes exons 2 and 3 was flanked by loxP sites (Sox9flox/WT), were a generous gift from Andreas Schedl and Marie-Christine Chaboissier (University of Nice, Nice, France) (11). Chimeras derived from embryonic stem (ES) cells with lacZ genes inserted in both Sox9 null alleles and Sox9 heterozygotes were generated as described (14, 15). The Wnt1-Cre transgenic line was used to produce neural crest-specific inactivation of Sox9 (16). In a first cross, Wnt1-Cre transgenic mice were mated with Sox9flox/WT mice. The offspring inheriting Wnt1-Cre and a floxed allele were then mated with Sox9flox/WT mice to obtain embryos harboring the Wnt1-Cre transgene together with two Sox9 floxed alleles. Embryos were genotyped by PCR on DNA obtained from the yolk sacs or skin by using the following primer pairs for Cre and Sox9flox: 5′-TCCAATTTACTGACCGTACACCAA-3′ and 5′-CCTGATCCTGGCAATTTCGGCTA-3′, and 5′-GTCAAGCGACCCATGAACGC-3′ and 5′-TGGTAATGAGTCATACACAGTAC-3′, respectively.
Embryonic Analysis. For histological analyses, embryos were fixed with 4% paraformaldehyde, embedded in paraffin, sectioned to 7 μm, and stained with either alcian blue and nuclear fast red or von Kossa reaction and nuclear fast red. Skeletal preparations were performed on E18.5 embryos as described (17). Immunohistochemical staining of Sox9 was performed by using peroxidase chromogens (Zymed)/TrueBlue substrate (Kirkegaard & Perry Laboratories) with rabbit polyclonal anti-Sox9 antibody (1:150). Apoptosis was visualized on paraffin sections by terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling (TUNEL) analysis, using an ApopTag Plus Peroxidase In Situ Apoptosis Detection Kit (Intergen, Purchase, NY) and following the manufacturer's protocols. RNA in situ hybridization analyses were carried out as described (18). The probes were as described (10, 19, 20). Pictures of hybridization signals were taken with a red filter and superimposed with blue fluorescence images of cell nuclei stained with the Hoechst 33258 dye.
Results
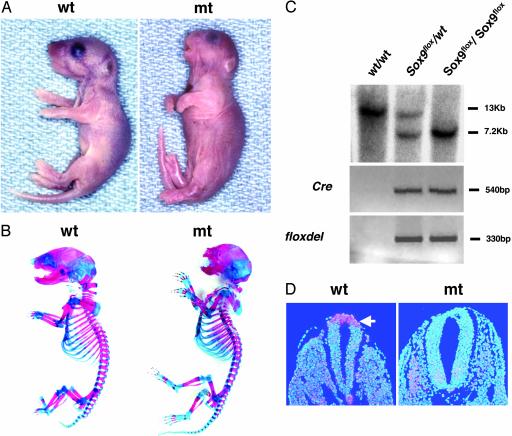
Generation of Sox9 Conditional Null Mice. To inactivate Sox9 in neural crest cells, compound heterozygotes carrying both a Sox9 floxed allele and the Wnt1-Cre transgene were generated by using the mating scheme described in Materials and Methods. Most Sox9flox/WT heterozygotes that harbored the Wnt1-Cre transgene were viable and developed into fertile mice with almost normal lifespans, although they had small cleft secondary palates and a mildly hypoplastic craniofacial skeleton (data not shown). In embryos harboring the Wnt1-Cre transgene, Cre is expressed starting at E8.5, and its expression is exclusively limited to the dorsal midline of the central nervous system (16). In the Sox9flox/flox; Wnt1-Cre embryos, Cre-mediated conversion of Sox9flox to Sox9floxdel, the deleted allele of Sox9flox, occurred only in embryos positive for Wnt1-Cre, indicating that Cre is capable of recombining loxP sites within the Sox9 gene in vivo (Fig. 1C). In situ hybridization of E9.5 and E13.5 showed a complete absence of Sox9 RNA in the neural crest and the frontonasal regions, respectively, indicating that inactivation of Sox9 was complete (Fig. 1D; see also Fig. 3B). Sox9 protein was not detectable in the frontonasal regions of E13.5 mutants either (see Fig. 3B).
Fig. 1.
Conditional inactivation of the Sox9 gene by using the Wnt1-Cre transgene. (A) Gross appearance of newborn mice. (B) Skeletons of newborn mice stained by alcian blue followed by alizarin red. (C) Southern blot analysis of fetal genomic DNA and PCR genotyping of Cre transgenes and floxdel alleles. Genomic DNA isolated from the skin or the yolk sac of WT, Sox9flox/WT; Wnt1-Cre, and Sox9flox/flox; Wnt1-Cre was digested with BamHI and then hybridized with a 3′ probe (11). The WT and floxed alleles were detected as 13-kb and 7.2-kb fragments, respectively. (D) Expression of Sox9 mRNA in the dorsal neural tube and neural crest of E9.5 WT and mutant embryos. Sox9 mRNA was not detected in mutants. The arrow indicates the dorsal neural tube and neural crest.
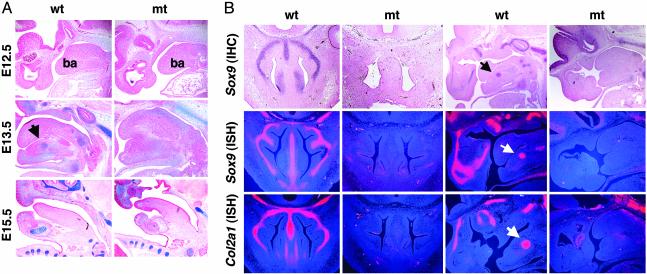
Fig. 3.
Histological analysis in Sox9flox/flox; Wnt1-Cre mice. (A) Alcian blue and nuclear fast red staining of E12.5, E13.5, and E15.5 WT and mutant (mt) embryos. ba, branchial arch 1. (B) Expression of Sox9 protein (IHC, immunohistochemistry) and Sox9 and Col2a1 mRNA (ISH, in situ hybridization) in E13.5 WT and mutant embryos. The arrows indicate Meckel's cartilage.
Complete Absence of Endochondral Craniofacial Skeletal Elements Derived from CNC in the Sox9flox/flox; Wnt1-Cre Mutant Mice. The Sox9 conditional null mutants resulting from expression of the Wnt1-Cre transgene, which were recovered with the expected Mendelian frequency, died in the immediate postnatal period from respiratory distress because of a large cleft secondary palate. Newborn mutants exhibited obvious craniofacial deformities characterized by a domed skull, a short snout, and short mandibles. In contrast, no abnormalities were observed in the trunk and the limbs (Fig. 1 A and B).
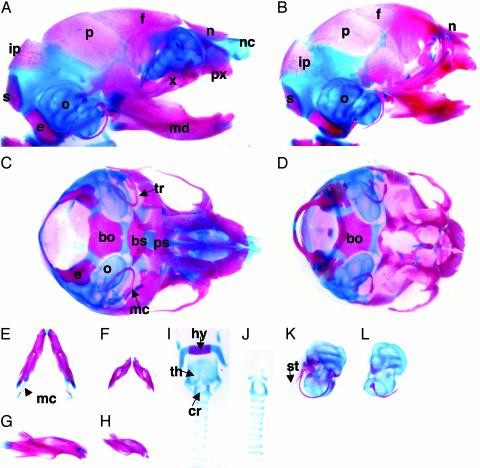
Analysis of the cranial base of skeletal preparations of E18.5 mutants showed that all of the cartilages and endochondral bones in the prechordal region, such as basisphenoid and presphenoid, were missing. However, no elements of the caudal part of the skull base, including basioccipital, exooccipital, and supraoccipital, were affected. Among the first branchial arch-derived skeletal elements of the mutants, the intramembranous bones, such as the maxilla, premaxilla, zygomatic bone, squamous portion of the temporal bone, pterygoid, nasal, lacrimal, and tympanic bones, were conserved. In contrast, the endochondral bones and cartilages, such as malleus and incus, and the nasal capsule were totally absent (Fig. 2 A–D, K, and L). In the mutants, Meckel's cartilage was also totally absent, and the mandibles were much smaller than those of the WT embryos (Fig. 2 E–H). All of the skeletal elements derived from the second and third branchial arches, such as the stapes, the lesser horns, and the body of the hyoid, and the styloid process were missing in the mutant embryos. Among the laryngeal cartilages, the thyroid cartilage was deleted, whereas the elements derived from more caudal branchial arches, including the arytenoid, the cricoid, and the tracheal rings, were conserved, indicating that CNC cells are the dominant contributors to the first four branchial arches, but not to more caudal branchial arches (Fig. 2 I and J).
Fig. 2.
Skeletal preparations of E18.5 WT (A, C, E, G, I, and K) and Sox9flox/flox; Wnt1-Cre (B, D, F, H, J, and L) embryos stained with alcian blue and alizarin red; lateral (A and B) and basal (C and D) views. The mandibles were removed to enhance the view of the cranial base. The basal (E and F) and lateral (G and H) views of the mandibles showed that, in the mutants, the morphology was conserved, but they were smaller than those in WT embryos. The hyoid bone, the laryngeal cartilages (I and J), the otic capsules, and middle ear elements (K and L) were dissected out. nc, nasal capsule; n, nasal; f, frontal; p, parietal; ip, interparietal; s, supraoccipital; e, exooccipital; o, otic capsule; md, mandible; x, maxilla; px, premaxilla; bs, basisphenoid; bo, basioccipital; ps, presphenoid; mc, Meckel's cartilage; tr, tympanic ring; hy, hyoid body; th, thyroid cartilage; cr, cricoid; st, styloid process.
Histological analysis showed no difference in the size of the branchial arches between WT and mutant embryos at E12.5 (Fig. 3A). In E13.5 WT embryos, the chondrogenic condensed mesenchymal cells differentiated into chondrocytes and formed Meckel's cartilage, nasal cartilage, and the cartilages of the skull base (Fig. 3). In E15.5 embryos, Meckel's cartilage develops and bone formation takes place in the mandible and the skull base. In contrast, in E13.5 mutant embryos, no discernible chondrogenic mesenchymal condensations were seen, and Meckel's cartilage, nasal cartilage, and bones in the precordal part of the skull base were not detectable (see Figs. 3A and 5A). Furthermore, in E15.5 mutant embryos the mandible and maxilla, which are formed by intramembranous ossification, showed bone trabeculae comparable with those of WT embryos (see Fig. 5B).
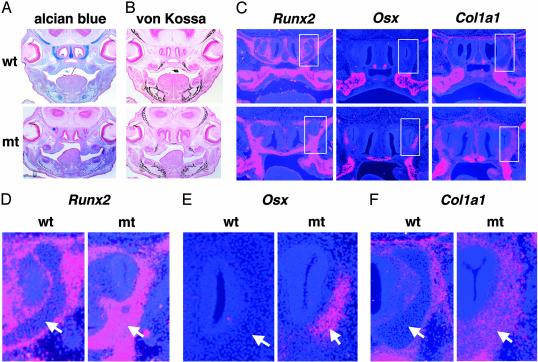
Fig. 5.
Expression of osteoblast markers in E15.5 Sox9flox/flox; Wnt1-Cre mice. (A) Alcian blue and nuclear fast red staining showed lack of cartilage formation. (B) Staining by von Kossa's method visualized a normal pattern of mineral deposition in intramembranous bones of WT and mutant (mt) embryos. (C) Ectopic expression of Runx2, Osx, and Col1a1 was detectable in mutant embryos. Boxed regions in C are shown at a high magnification (×200) in D–F. The arrows indicate cartilages in WT embryos and ectopic expression of osteoblast marker genes in equivalent locations in the mutants.
To characterize the complete defect in CNC-derived endochondral bone formation in the mutant embryos, we analyzed the expression of Col2A1, an early marker of chondrogenic cells, by in situ hybridization. In E13.5 WT embryos, Col2a1 is expressed in the skull base, nasal region, and Meckel's cartilage. In the mutant embryos, expression of Col2a1 was essentially abolished in these regions (Fig. 3B). Thus we concluded that no chondrocytes were present in the regions of the mutant embryos where endochondral bone formation derived from CNC cells occurs in WT embryos.
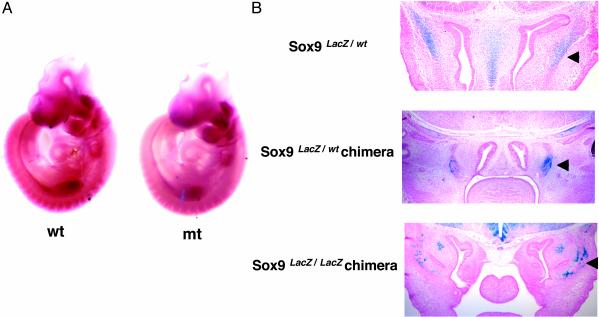
Sox9-Null CNC Cells Migrate Normally to Their Target Locations and Survive in These Regions. CNC cells migrate along specific pathways to the branchial arches, where they differentiate to give rise to craniofacial bones and cartilages (5, 16, 21). To determine whether the craniofacial skeletal defects in Sox9 mutants could result from aberrant migration of CNC cells into their targets, expression of Ap2, which encodes a transcription factor essential for survival of migratory CNC cells (19, 22, 23), was analyzed by whole-mount in situ hybridization (Fig. 4A). In E9.5 Sox9 conditional null mutant embryos, expression of Ap2 was detectable in streaks extending from the rhombomeres to the branchial arches and frontonasal mass, and was comparable with that of WT embryos. Furthermore, we examined the localization of LacZ-positive cells in Sox9lacZ/WT heterozygous embryos, in chimeric embryos derived from Sox9lacZ/WT ES cells, and in chimeras derived from Sox9lacZ/lacZ-null mutant ES cells (Fig. 4B). In heterozygous embryos, expression of lacZ reproduced expression of Sox9. Thus the lacZ sequence present in the Sox9/lacZ allele enabled the visualization of Sox9 expression by 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) staining (15). In E12.5 chimeras, the overall pattern of lacZ expression of Sox9-null mutant cells was comparable with that of heterozygous Sox9 mutant cells. Our results indicated that Sox9-null mutant CNC cells migrated to their normal craniofacial target regions.
Fig. 4.
Distribution of Sox9-null mutant cells. (A) Whole-mount RNA in situ hybridization of E9.5 WT and mutant (mt) embryos. Normal expression of Ap2 revealed normal migration of CNC cells in the mutants. (B) Transverse sections of whole-mount 5-bromo-4-chloro-3-indolylβ-d-galactoside (X-Gal) staining of Sox9 heterozygous (Sox9lacZ/WT), Sox9 heterozygous chimeras (Sox9lacZ/WT chimera), and Sox9-null chimeras (Sox9 lacZ/lacZ chimera) at 12.5 days postcoitum. The arrowheads indicate X-Gal-positive cells in chondrogenic mesenchymal condensations and cartilages.
The severe malformations and the arrest of outgrowth in craniofacial development of the mutant embryos could have been caused by apoptosis within the population of migratory CNC cells. To test this hypothesis, we performed a terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling assay (TUNEL). No apoptotic cells were detected in the branchial arches or the skull base of E13.5 mutant or WT embryos (data not shown). In E15.5 mutant embryos, only a few apoptotic cells appeared in the areas where condensed mesenchymal cells differentiate and form cartilages in WT embryos (data not shown). This result suggests that most of the Sox9-null mutant CNC cells survived after migration.
Ectopic Expression of Osteoblast Markers in the Sox9flox/flox; Wnt1-Cre Mutant Embryos. We asked whether the Sox9-null mutant CNC cells had adopted a different cell fate, given that endochondral bone formation derived from CNC cells was completely missing in the Sox9flox/flox; Wnt1-Cre mutant embryos, even though Sox9-null mutant CNC cells were present and specified. In the nasal region of WT embryos, well-differentiated cartilages but not mineralized bones were seen, whereas no cartilages were detectable in this region in the mutant embryos (Fig. 5 A and B). In situ hybridization analysis showed that in the nasal regions of E15.5 WT embryos, Runx2 and Col1a1, early markers of osteoblast differentiation, were expressed in the perichondrium of the cartilage primordia of the nasal turbinate. In WT embryos, Osterix (Osx), which encodes a transcription factor specifically expressed in osteoblast (20), was expressed only in the vomer and the ossification centers within the maxilla; both of these skeletal elements are intramembranous bones derived from CNC cells. In contrast, the expression domains of Runx2 and Col1a1 in the mutant embryos were expanded and much wider than those in WT embryos. Furthermore, expression of Osx was detectable in locations of the nasal turbinate that are cartilaginous in WT embryos (Fig. 5 C–F). Because Sox9-null cells in chimeras migrated in the nasal areas that were positive for Runx2, Osx, and Col1a1, these results strongly suggest that Sox9-null mutant CNC cells, which could not differentiate into chondrocytes, acquired a cell fate characterized by the expression of a series of genes that are typical of osteoblasts.
Discussion
Sox9 Is Essential for CNC-Derived Endochondral Bone Formation. CNC cells that migrate from the posterior midbrain and hindbrain rhombomeres into the branchial arches give rise to the craniofacial skeleton (16). In mouse embryo chimeras derived from Sox9-null ES cells, mutant cells were excluded from mesenchymal condensations and did not express chondrocyte-specific markers, such as Col2a1, Col11a2, and Aggrecan (15). Furthermore, inactivation of Sox9 in mouse limb buds before the onset of chondrogenic mesenchymal condensations by using the Cre/loxP recombination system resulted in a complete absence of mesenchymal condensations and subsequent cartilage and bone formation in the mutant limb buds. In these mutant limb buds, expression of Sox5 and Sox6, which are needed for overt chondrocyte differentiation (24), and that of Runx2, which is needed for osteoblast differentiation (25, 26), were abolished. Thus during limb endochondral bone formation in vivo, Sox9 is required for both chondrogenesis and osteogenesis (11, 15). During the establishment of CNC cells, expression of Sox9 is first detected in the lateral edges of the neural tube in E8.0 embryos, is down-regulated in migratory CNC cells, and becomes up-regulated again, first in branchial arches and later in chondrogenic condensed mesenchymal cells and chondrocytes (9, 10).
To further understand the role of Sox9 in CNC cells, we inactivated the Sox9 gene in Wnt1-expressing CNC cells, which include all precursors of the craniofacial skeleton (16). The Wnt1 promoter that was used in these experiments is active specifically in the neural plate, in the dorsal neural tube, and in the early migratory neural crest population (16, 27). Expression of Wnt1 is extinguished as the crest cells migrate away from the neural tube, and Wnt1 is not expressed at any other time or at any other place except in the midbrain. Taking advantage of the tissue and developmental stage-specific expression of the Wnt1 gene, Sox9 can be deleted in CNC cells before or at the onset of migration.
Inactivation of Sox9 in these cells resulted in a complete deletion of CNC-derived endochondral bones and cartilages. The absence of Sox9 in CNC did not affect any intramembranous bone formation. The complete lack of these endochondral bones and cartilages was attributable to the failure of mesenchymal condensations and cell differentiation of postmigrating CNC cells into chondrocytes and osteoblasts. Indeed, based on the pattern of expression of Ap2, the mutant CNC cells migrated normally into branchial arches and the frontonasal mass. Furthermore, in chimeras derived from Sox9-null ES cells, Sox9-null mutant cells were present in the cellular locations where endochondral bone formation takes place in the WT embryos.
Cell-fate mapping of CNC-derived craniofacial skeletal elements has been well described. However, there are still conflicting views as to the origin of some skeletal components (28). Taking advantage of the complete absence of CNC-derived endochondral bone formation in the Sox9flox/flox; Wnt1-Cre mutant embryos, we can clarify what the dominant contribution of CNC cells is to some skeletal components. Recent studies have suggested that the ventrally emigrating neural tube (VENT) cells, in addition to the CNC cells, contribute to the formation of Meckel's cartilage (29). In the Sox9flox/flox; Wnt1-Cre mutant embryos, Meckel's cartilage was entirely missing. In addition, the Wnt1 promoter is not expressed in the VENT cells. Furthermore, a recent study using morpholino antisense oligonucleotides showed that depletion of Sox9 protein in developing Xenopus embryos caused a complete loss of Meckel's cartilage (30). These results provide evidence that CNC cells have an essential role in the specification of the cells of Meckel's cartilage.
It has been proposed that the fourth and sixth arches originate from the lateral plate mesoderm (31). However, a recent study that examined the fate of LacZ-positive cells in embryos harboring both the Wnt1-Cre transgene and the Rosa26 reporter allele showed that mesenchymal cells in the fourth and sixth arches were derived from the neural crest (32, 33). In our mutant embryos, the fourth arch thyroid cartilage was entirely absent, strongly suggesting that it originates from CNC cells. In contrast, the cricoid cartilage, which is derived from the sixth arch, was present in the mutant, suggesting that it is not derived from the neural crest.
Sox9 Is Needed for the Specification of Both the Chondrocyte and Osteoblast Lineages in CNC-Derived Endochondral Bones. Previous fate-mapping and trace-cell-mapping studies suggested that the neural crest cells are preprogrammed before their migration to form specific embryonic structures and are unable to alter their cell fate (3, 34). However, recent studies argue against this fixed cell autonomy model and have provided evidence that CNC cells are plastic and are capable of respecification in response to distinct environmental signals (35). Neural crest cells are multipotent stem cells that give rise to various cell lineages. A number of signaling molecules, including Wnts, transforming growth factor-β, and Notch, promote the specification of particular cell fates in neural crest cells (36–40). Other experiments have shown that osteoblastic cells from embryonic calvarial bones have the potential to differentiate into chondrocytes in vitro (41–43), suggesting that there are bipotential progenitor cells in these tissues derived from CNC cells, and that there is a degree of plasticity in the cell fate determination of these cells.
However, the genetic pathways controlling the specification of osteoblast and chondrocyte lineages from CNC cells remain unknown. Recent gene targeting studies revealed that Msx1; Msx2 double-knockout mice completely lack calvarial ossification, whereas the chondrocranium forms normally (44). Thus it is possible that Msx1 and Msx2 are involved in the specification of the osteoblast lineage, at least in some CNC cells. Msx2 and Sox9 are coexpressed in early migrating CNC cells, and Msx2 was shown to inhibit the chondrogenic differentiation of these cells (45). In addition, we have recently reported that Osx, a zinc finger-containing transcription factor specifically expressed in all developing bones, is required for osteoblast differentiation (20). In Osx-null mice, in which no bone formation occurs, Osx mutant CNC cells migrate normally into intramembranous and endochondral skeletal elements; but these cells, which are blocked from differentiating into osteoblasts, express chondrocyte marker genes, including Sox9, Sox5, and Col2a1. The implication of these observations is that intrinsic genetic pathways involved in the specification of the osteoblast lineage inhibit the specification of the chondrocyte lineage in CNC cells. In the present study, we showed that inactivation of Sox9 in CNC cells resulted in a complete defect of CNC-derived endochondral bone formation, but that intramembranous bones formed normally. In addition, our experiments strongly suggest that Sox9-null CNC-derived cells, which were blocked from differentiating into chondrocytes, expressed instead osteoblast marker genes, including Runx2, Osx, and Col1a1, suggesting that Sox9-null CNC derived-cells were respecified into an osteoblast lineage.
Although Sox9 is required in CNC cells for both the chondrocyte and osteoblast lineages destined for endochondral bone formation, it is not needed for cells of the osteoblast lineage in intramembranous skeletal elements derived from the neural crest. But the cells of the osteoblast lineage that are destined for intramembranous bone formation also have the potential to become chondrocytes when Sox9 is present (20).
Overall, our previous (20) and our present experiments provide evidence supporting the hypothesis that CNC cells contain common precursor cells with the potential to differentiate into either osteoblasts or chondrocytes during both endochondral and intramembranous bone formation. Sox9 is needed for the specification of an osteochondroprogenitor of both the chondrocyte and osteoblast lineages in endochondral bone formation. We speculate that, in chondrocyte precursor cells, Sox9 is a negative regulator of Runx2 and Osx and thus prevents these cells from choosing an osteoblast lineage.
Acknowledgments
We thank Andreas Schedl and Marie-Christine Chaboissier for Sox9flox mice, Zhaoping Zhang and Heidi Eberspaecher for valuable help, Kazuhisa Nakashima and Richard Behringer for discussions, and Janie Finch for editorial assistance. This work was funded by National Institutes of Health Grant PO1 AR42919, The G. Harold and Leila Y. Mathers Charitable Foundation (B.d.C.), and National Institutes of Health Cancer Center Support Grant CA16672 for DNA sequence analysis.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CNC, cranial neural crest; ES, embryonic stem; En, embryonic day n.
References
- 1.Bronner-Fraser, M. (1995) Exp. Cell Res. 218, 405–417. [DOI] [PubMed] [Google Scholar]
- 2.Hall, B. K. (1999) The Neural Crest in Development and Evolution (Springer, New York).
- 3.Noden, D. (1983) Dev. Biol. 96, 144–165. [DOI] [PubMed] [Google Scholar]
- 4.Serbedzija, G. N., Bronner-Fraser, M. & Fraser, S. E. (1989) Development (Cambridge, U.K.) 106, 809–816. [DOI] [PubMed] [Google Scholar]
- 5.Serbedzija, G. N., Bronner-Fraser, M. & Fraser, S. E. (1992) Development (Cambridge, U.K.) 116, 297–307. [DOI] [PubMed] [Google Scholar]
- 6.Martinsen, B. J. & Bronner-Fraser, M. (1998) Science 281, 988–991. [DOI] [PubMed] [Google Scholar]
- 7.Lister, J. A., Robertson, C. P., Lepage, T., Johnson, S. L. & Raible, D. W. (1999) Development (Cambridge, U.K.) 126, 3757–3767. [DOI] [PubMed] [Google Scholar]
- 8.Paratore, C., Goerich, D. E., Suter, U., Wegner, M. & Sommer, L. (2001) Development (Cambridge, U.K.) 128, 3949–3961. [DOI] [PubMed] [Google Scholar]
- 9.Ng, L. J., Wheatley, S., Muscat, G. E., Conway-Campbell, J., Bowles, J., Wright, E., Bell, D. M., Tam, P. P., Cheah, K. S. & Koopman, P. (1997) Dev. Biol. 183, 108–121. [DOI] [PubMed] [Google Scholar]
- 10.Zhao, Q., Eberspaecher, H., Lefebvre, V. & de Crombrugghe, B. (1997) Dev. Dyn. 209, 377–386. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama, H., Chaboissier, M. C., Martin, J. F., Schedl, A. & de Crombrugghe, B. (2002) Genes Dev. 16, 2813–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, J. W., Dominguez-Steglich, M. A., Guioli, S., Kowk, G., Weller, P. A., Stevanovic, M., Weissenbach, J., Mansour, S., Young, I. D., Goodfellow, P. N., et al. (1994) Nature 372, 525–530. [DOI] [PubMed] [Google Scholar]
- 13.Wagner, T., Wirth, J., Meyer, J., Zabel, B., Held, M., Zimmer, J., Pasantes, J., Bricarelli, F. D., Keutel, J., Hustert, E., et al. (1994) Cell 79, 1111–1120. [DOI] [PubMed] [Google Scholar]
- 14.Bi, W., Huang, W., Whitworth, D. J., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. (2001) Proc. Natl. Acad. Sci. USA 98, 6698–6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bi, W., Deng, J. M., Zhang, Z., Behringer, R. R. & de Crombrugghe, B. (1999) Nat. Genet. 22, 85–89. [DOI] [PubMed] [Google Scholar]
- 16.Chai, Y., Jiang, X., Ito, Y., Bringas, P., Jr., Han, J., Rowitch, D. H., Soriano, P., McMahon, A. P. & Sucov, H. M. (2000) Development (Cambridge, U.K.) 127, 1671–1679. [DOI] [PubMed] [Google Scholar]
- 17.Hogan, B., Beddington, R., Costantini, F. & Lucy, E. (1994) Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY).
- 18.Albrecht, U., Eichele, G., Helms, J. A. & Lu, H. C. (1997) in Molecular and Cellular Methods in Developmental Toxicology, ed. Daston, G. P. (CRC, Boca Raton, FL), pp. 23–48.
- 19.Mitchell, P. J., Timmons, P. M., Hebert, J. M., Rigby, P. W. J. & Tjian, R. (1991) Genes Dev. 5, 105–119. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima, K., Zhou, X., Kunkel, G., Zhang, Z., Deng, J. M., Behringer, R. R. & de Crombrugghe, B. (2002) Cell 108, 17–29. [DOI] [PubMed] [Google Scholar]
- 21.Tan, S. S. & Morriss-Kay, G. M. (1986) J. Embryol. Exp. Morphol. 98, 21–58. [PubMed] [Google Scholar]
- 22.Schorle, H., Meier, P., Buchert, M., Jaenisch, R. & Mitchell, P. J. (1996) Nature 381, 235–238. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, J., Hagopian-Donaldson, S., Serbedzija, G., Elsemore, J., Plehn-Dujowich, D., McMahon, A. P., Flavell, R. A. & Williams, T. (1996) Nature 381, 238–241. [DOI] [PubMed] [Google Scholar]
- 24.Smits, P., Li, P., Mandel, J., Zhang, Z., Deng, J. M., Behringer, R. R., de Crombrugghe, B. & Lefebvre, V. (2001) Dev. Cell 1, 277–290. [DOI] [PubMed] [Google Scholar]
- 25.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., et al. (1997) Cell 89, 755–764. [DOI] [PubMed] [Google Scholar]
- 26.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., et al. (1997) Cell 89, 765–771. [DOI] [PubMed] [Google Scholar]
- 27.Echelard, Y., Vassileva, G. & McMahon, A. P. (1994) Development (Cambridge, U.K.) 120, 2213–2224. [DOI] [PubMed] [Google Scholar]
- 28.Kontges, G. & Lumsden, A. (1996) Development (Cambridge, U.K.) 122, 3229–3242. [DOI] [PubMed] [Google Scholar]
- 29.Sohal, G. S., Ali, M. M., Ali, A. A. & Dai, D. (1999) Dev. Dyn. 216, 37–44. [DOI] [PubMed] [Google Scholar]
- 30.Spokony, R. F., Aoki, Y., Saint-Germain, N., Magner-Fink, E. & Saint-Jeannet, J. P. (2002) Development (Cambridge, U.K.) 129, 421–432. [DOI] [PubMed] [Google Scholar]
- 31.Kaufman, M. H. & Bard, J. B. L. (1999) The Anatomical Basis of Mouse Development (Academic, San Diego).
- 32.Soriano, P. (1999) Nat. Genet. 21, 70–71. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, X., Rowitch, D. H., Soriano, P., McMahon, A. P. & Sucov, H. M. (2000) Development (Cambridge, U.K.) 127, 1607–1616. [DOI] [PubMed] [Google Scholar]
- 34.Couly, G., Grapin-Botton, A., Coltey, P., Ruhin, B. & Le Douarin, N. M. (1998) Development (Cambridge, U.K.) 125, 3445–3459. [DOI] [PubMed] [Google Scholar]
- 35.Trainor, P. & Krumlauf, R. (2000) Nat. Cell. Biol. 2, 96–102. [DOI] [PubMed] [Google Scholar]
- 36.Shah, N. M., Marchionni, M. A., Isaacs, I., Stroobant, P. & Anderson, D. J. (1994) Cell 77, 349–360. [DOI] [PubMed] [Google Scholar]
- 37.Shah, N. M., Groves, A. K. & Anderson, D. J. (1996) Cell 85, 331–343. [DOI] [PubMed] [Google Scholar]
- 38.Dorsky, R. I., Moon, R. T. & Raible, D. W. (1998) Nature 396, 370–373. [DOI] [PubMed] [Google Scholar]
- 39.Morrison, S. J., Perez, S. E., Qiao, Z., Verdi, J. M., Hicks, C., Weinmaster, G. & Anderson, D. J. (2000) Cell 101, 499–510. [DOI] [PubMed] [Google Scholar]
- 40.Jin, E. J., Erickson, C. A., Takada, S. & Burrus, L. W. (2001) Dev. Biol. 233, 22–37. [DOI] [PubMed] [Google Scholar]
- 41.Grigoriadis, A. E., Heersche, J. N. & Aubin, J. E. (1990) Dev. Biol. 142, 313–318. [DOI] [PubMed] [Google Scholar]
- 42.Bernier, S. M. & Goltzman, D. (1993) J. Bone Miner. Res. 8, 475–484. [DOI] [PubMed] [Google Scholar]
- 43.Stringa, E. & Tuan, R. S. (1996) Anat. Embryol. 194, 427–437. [DOI] [PubMed] [Google Scholar]
- 44.Satokata, I., Ma, L., Ohshima, H., Bei, M., Woo, I., Nishizawa, K., Maeda, T., Takano, Y., Uchiyama, M., Heaney, S., et al. (2000) Nat. Genet. 24, 391–395. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi, K., Nuckolls, G. H., Takahashi, I., Nonaka, K., Nagata, M., Ikura, T., Slavkin, H. C. & Shum, L. (2001) Dev. Dyn. 222, 252–262. [DOI] [PubMed] [Google Scholar]