Abstract
Formation of the vascular system requires differentiation and patterning of endothelial and smooth muscle cells (SMCs). Although much attention has focused on development of the vascular endothelial network, the mechanisms that control vascular SMC development are largely unknown. Myocardin is a smooth and cardiac muscle-specific transcriptional coactivator of serum response factor, a ubiquitous transcription factor implicated in smooth muscle gene expression. When expressed ectopically in nonmuscle cells, myocardin can induce smooth muscle differentiation by its association with serum response factor. Here we report that mouse embryos homozygous for a myocardin loss-of-function mutation die by embryonic day 10.5 and show no evidence of vascular SMC differentiation. Myocardin is the only transcription factor known to be necessary and sufficient for vascular SMC differentiation.
The cardiovascular system is the first organ system to form and function during embryogenesis. Vascular development begins with the organization of endothelial cells into a primitive vascular plexus that becomes progressively remodeled to ultimately form a complex vascular network (1). Smooth muscle cells (SMCs) are recruited to the endothelial vasculature and ensheathe it, providing support and contractility to the vascular system. Several peptide growth factors and their tyrosine kinase receptors have been shown to play key roles in assembly and patterning of the endothelial vasculature and recruitment of SMCs. In contrast, little is known of the transcriptional events responsible for development of vascular SMCs in vivo.
Vascular SMCs are derived from a variety of embryonic progenitors, including lateral mesoderm, cranial mesenchyme, and the neural crest (2, 3). Differentiation of SMCs is triggered by extracellular cues and is accompanied by the transcriptional activation of an array of smooth muscle (SM) genes whose products confer the unique contractile, morphological, and structural properties that distinguish them from other muscle cell types. The mcm1, Agamous, Deficiens, serum response factor (SRF) box transcription factor plays a critical role in SM gene activation. SRF binds to a DNA sequence known as a CArG box, which is required for the expression of virtually every SM gene analyzed to date (4–10). The importance of SRF for SM gene expression has also been suggested by the finding that a dominant-negative SRF mutant can block SM differentiation in epicardial explant cultures (11). However, the role of SRF in SM development in vivo has been clouded by the fact that SRF knockout mice die during gastrulation from a lack of mesoderm well before vascular development is initiated (12). Moreover, because SRF is expressed throughout the embryo, it alone cannot account for the specificity of SM gene transcription. Thus, it has been proposed that SRF controls SM genes by recruiting cell type-specific cofactors. Indeed, the transcriptional activity of SRF in cultured cells can be modulated by its association with positive and negative cofactors and by extracellular signaling, but there is little or no direct evidence for the involvement of either of these types of mechanisms in the control of SM genes by SRF in vivo (13).
The SAP domain transcriptional coactivator, myocardin, is an extraordinarily powerful SRF cofactor expressed specifically in smooth and cardiac muscle cells (14). Myocardin selectively activates smooth and cardiac muscle promoters by its interaction with SRF. Expression of a dominant-negative myocardin mutant in Xenopus embryos blocks heart formation, suggesting that myocardin cooperates with SRF to activate cardiac gene expression (14). Interestingly, when expressed in nonmuscle cells in vitro, myocardin activates smooth but not cardiac muscle gene expression (15–18). Two myocardin-related transcription factors (MRTFs) also associate with SRF but are expressed in more widespread patterns during embryogenesis than myocardin (19).
To determine the function of myocardin during embryogenesis, we targeted the mouse myocardin gene by homologous recombination. Here we show that mice lacking myocardin die by embryonic day (E) 10.5 from a complete absence of vascular SMCs. In contrast, cardiac development occurs normally in myocardin mutant embryos. The avascular phenotype of myocardin mutant mice, combined with previous studies demonstrating that myocardin can activate expression of SM genes in nonmuscle cells (15–18), demonstrates that myocardin is a master regulator of SM development both sufficient and necessary for SMC differentiation.
Methods
Generation of Myocardin Mutant Mice. The gene structure of myocardin has been described (19). A myocardin-targeting vector was constructed to delete exons 8 and 9 by using a pN-Z-TK2 vector, which contains a nuclear LacZ (nLacZ) cassette and a neomycin-resistance gene under the control of the RNA polymerase II promoter and two herpes simplex virus thymidine kinase (TK) gene cassettes (a generous gift of R. Palmiter, University of Washington, Seattle). Genomic DNA flanking myocardin exons 8 and 9 was PCR amplified from a mouse 129SvEv genomic DNA library (Stratagene) and inserted into the targeting vector as short and long arms, respectively. The targeting vector was electroporated into 129 SvEv-derived ES cells, and selection was performed with G-418 and FIAU, respectively. Four hundred ES cell clones were isolated and analyzed by Southern blotting for homologous recombination. Three clones with a disrupted myocardin gene were injected into 3.5-day mouse C57BL/6 blastocysts, and the resulting chimeric male mice were bred to C57BL/6 females to achieve germline transmission of the mutant allele.
RT-PCR. Total RNA was purified from tissues with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For RT-PCR, total RNA was used as a template for reverse transcriptase and random hexamer primers. Primer sequences are available on request.
Immunostaining and Histology. Whole-mount staining for plateletendothelial cell adhesion molecule (PECAM) and SM α-actin was performed by using anti-mouse PECAM mAb MEC13.3 (Pharmigen) and SM α-actin Ab (clone 1A4, Sigma), as described (20). Embryos for histology were fixed in 4% paraformaldehyde, sectioned, and processed for in situ hybridization by using standard procedures (21).
SM22-lacZ Transgenic Mice. Staining of transgenic embryos for lacZ expression was performed as described (22). The SM22-lacZ transgene contained the 1,343-bp SM22 promoter linked to lacZ (22). The transgene was introduced into the myocardin mutant background by interbreeding the appropriate strains of mice.
Results
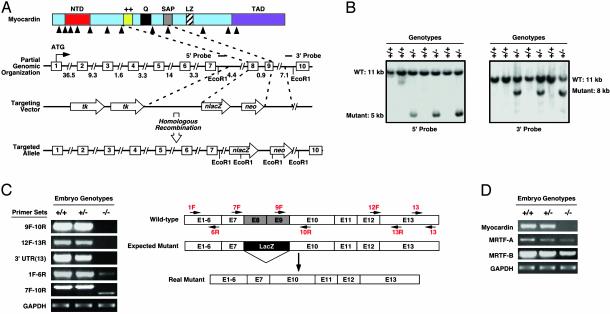
Generation of Myocardin Knockout Mice. To determine the function of myocardin during mouse development, we generated myocardin-deficient mice by targeted disruption of the myocardin gene. The protein-coding region of the mouse myocardin gene encompasses 13 exons and spans ≈93 kb of genomic DNA (Fig. 1A). Exons 8 and 9, which encode the basic domain, glutaminerich domain, and part of the SAP domain (19), were replaced with a promoterless lacZ gene and a neomycin-resistance gene (Fig. 1 A). The basic and glutamine-rich domains are required for interaction with SRF. Deletion of these domains abolishes all myogenic activity of myocardin (14, 18). Thus, this mutation inactivates the gene.
Fig. 1.
Generation of myocardin mutant mice. (A) Targeting strategy. Homologous recombination resulted in deletion of exons 8 and 9 and insertion of lacZ and neomyocin-resistance cassettes. The positions of 5′ and 3′ probes used for Southern analysis in B are shown. Intron junctions within the coding region are shown by arrowheads beneath the schematized protein. Exons are shown in boxes, and sizes of introns are indicated. (B) Southern blot analysis. Genomic DNA from ES cell clones was isolated from tail biopsies and analyzed by Southern blot with 5′ and 3′ probes after digestion with EcoRI. The positions of WT and mutant bands are shown. (C) Analysis of myocardin transcripts by RT-PCR. RNA was isolated from hearts of WT and myocardin mutant embryos at E9.5 and analyzed by RT-PCR by using different pairs of primers, as shown to the left of each panel. Genotypes are shown at the top. A schematic of exons (E) with positions of primers is shown at the right. Transcripts for GAPDH were detected as a control for RNA loading and integrity. In the targeted allele, exon 7 is spliced to exon 10. (D) RNA was isolated from hearts of WT and myocardin mutant embryos at E9.5 and analyzed for myocardin, MRTF-A, and MRTF-B transcripts by RT-PCR.
The targeting vector was electroporated into 129SV/Ev ES cells and targeted clones were identified by Southern blot analysis of genomic DNA (Fig. 1B). ES cells heterozygous for the targeted myocardin gene were injected into C57BL/6 blastocysts to generate chimeric mice, which transmitted the mutant allele through the germ line.
Mice heterozygous for the mutant myocardin allele were viable, fertile, and phenotypically normal. Genotyping of off-spring from heterozygous intercrosses in the isogenic 129 background or in a 129/C57BL/6 mixed genetic background yielded WT and myocardin+/– mice in an approximate 1:2 ratio but no myocardin–/– mice, indicating that the homozygous mutation resulted in embryonic lethality. Analysis of the genotypes of embryos from timed matings showed Mendelian ratios up to E10.5 but no live homozygous mutants at later developmental time points.
To confirm the gene-targeting event, we performed RT-PCR analysis of mRNA from the hearts of WT and mutant embryos at E9.5, using primers representing exon sequences within and surrounding the deleted region of the gene (Fig. 1C). These assays revealed that exons 8 and 9 of myocardin were replaced with the LacZ-Neo cassette, as expected, and that this mutation resulted in alternative splicing of the targeted allele, such that exon 7 was spliced to exon 10. This was further confirmed by sequence analysis of RT-PCR products. Because of such alternative splicing, the lacZ gene was not expressed in mutant mice (data not shown). Notably, the truncated transcript generated from the mutant allele was expressed at a much lower level than the WT myocardin transcript, presumably because of instability of the mutant transcript (Fig. 1C).
Because myocardin shares extensive amino acid homology with MRTF-A and MRTF-B, which can also act as SRF cofactors (19), we assayed their expression by RT-PCR of RNA from hearts of E9.5 embryos. As shown in Fig. 1D, both transcripts were readily detectable in WT and mutant hearts, and neither was up-regulated in the absence of myocardin.
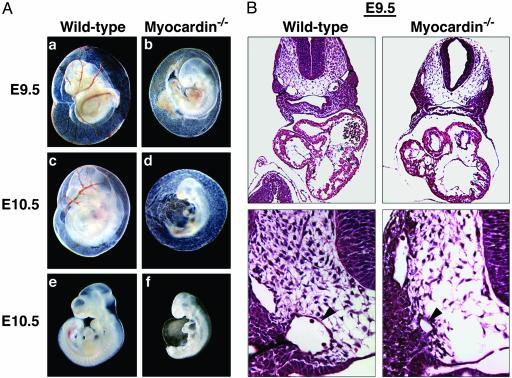
Lethal Vascular Abnormalities in Myocardin Mutant Embryos. Analysis of myocardin–/– embryos obtained from timed matings revealed no abnormalities before E8.0, and most embryos appeared to develop normally up to E8.5 (data not shown). However, homozygous mutant embryos could be readily identified at E9.5 by their pale yolk sacs, which lacked blood vessels (Fig. 2Aa–d). Homozygous mutant embryos also showed growth retardation and delayed development at E9.5. The gross morphology of the hearts in mutant embryos appeared normal with completed rightward looping and normal chamber formation.
Fig. 2.
(A) Vascular abnormalities in myocardin mutant embryos. Shown are WT and mutant yolk sacs and embryos at E9.5 and E10.5. In e and f, the yolk sacs were removed from the embryos shown in c and d. (B) Hematoxylin/eosin sections of WT and mutant embryos at E9.5. (Lower) High magnifications of the region with the dorsal aorta, indicated by an arrowhead.
Histological analysis of transverse sections of mutant embryos at E9.5 confirmed the normal appearance of the atrial and ventricular chambers but revealed severe vascular defects in which the dorsal aortae were clearly underdeveloped (Fig. 2B). However, the anterior cardinal veins appeared normal in mutant embryos at this stage. It is noteworthy that SM marker genes are not yet activated in the cardinal veins of WT embryos, whereas differentiated SMCs are present in the dorsal aortae at this stage. By E10.5, mutant embryos were severely delayed developmentally and pericardial effusion was often observed, indicative of cardiovascular insufficiency (Fig. 2 A e and f).
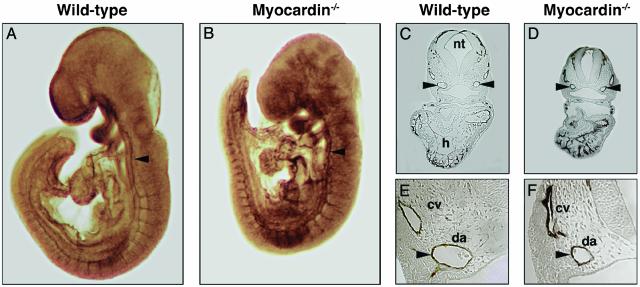
Normal Endothelial Cell Differentiation and Organization in Myocardin–/– Embryos. Vascular development initiates around E7.5 in the mouse with the differentiation and migration of endothelial progenitor cells (1–3). To visualize the embryonic vasculature, we performed whole-mount Ab staining for PECAM-1, an endothelial marker (23). At E8.5, PECAM staining of the newly formed vasculature was indistinguishable in WT and myocardin mutant embryos (data not shown). At E9.5, differentiated endothelial cells were properly positioned in mutant embryos, even though mutant embryos began to show growth retardation by this stage (Fig. 3 A and B). Notably, the complexity and patterning of the cranial vasculature, the intersomitic vasculature, and the dorsal aorta as revealed by PECAM-1 staining appeared similar in WT and myocardin mutant embryos. Transverse sections clearly showed the presence of PECAM-1-positive endothelial cells in the dorsal aorta and cardinal veins of both WT and mutant embryos, although the dorsal aorta in mutant embryos was smaller than normal (Fig. 3 C–F). Similarly, PECAM staining was detected in the endocardial layer of the heart of WT and myocardin mutant embryos. These findings indicated that vascular endothelial cell differentiation and organization were unaffected by the myocardin mutation.
Fig. 3.
Endothelial cell patterning detected by PECAM staining is unperturbed in myocardin mutant embryos. (A and B) WT and myocardin mutant embryos at E9.5 stained for PECAM. The arrowhead points to the dorsal aorta. (C–F) Histological sections of embryos stained for PECAM (C and E, WT; D and F, myocardin–/–). E and F show high magnifications of the region of the dorsal aorta, indicated by arrowheads. cv, cardinal vein; da, dorsal aorta; h, heart; nt, neural tube.
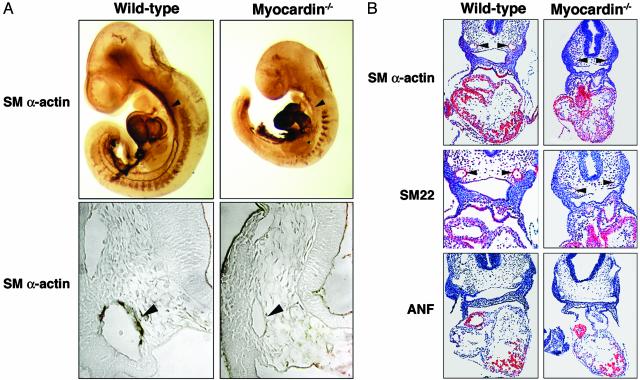
Defects in Vascular SMC Differentiation in Myocardin–/– Embryos. To determine whether the vascular abnormalities in myocardin mutant embryos resulted from a defect in SMC differentiation, we stained E9.5 embryos with an Ab to SM α-actin. As shown in Fig. 4A, SM α-actin-positive SMCs were present in the dorsal aorta and cardinal veins of WT embryos. However, no such SM α-actin-positive cells were detected in the vasculature of myocardin–/– embryos. Comparison of transverse sections of SM α-actin-stained embryos revealed that SMCs were missing from the dorsal aortae of myocardin–/– embryos (Fig. 4A). Unexpectedly, however, there was no decrease in the expression of SM α-actin in myocardin–/– hearts.
Fig. 4.
Lack of expression of SM markers in myocardin mutant embryos. (A) WT and myocardin mutant embryos at E9.5 stained for SM α-actin. The arrowhead points to the dorsal aorta. (B) Detection of smooth and cardiac muscle transcripts by in situ hybridization to E9.5 embryo sections. Silver grains are shown pseudocolored red. Arrowheads point to the dorsal aortae.
We further examined the expression of SM genes by in situ hybridization to embryo sections at E9.5 (Fig. 4B). The SM22 and SM α-actin genes are direct target genes of SRF and are induced by myocardin in transfected fibroblasts (5–7, 14, 18). Transcripts for both genes were expressed in the heart and developing vasculature of WT embryos (Fig. 4B). In contrast, neither transcript was detected in the vasculature of myocardin–/– embryos, although normal expression was detected in the hearts of mutant embryos. The atrial natriuretic factor (ANF) gene, a cardiac-specific target of myocardin, was also expressed normally in mutant embryos. These findings demonstrated that vascular SMC differentiation was specifically disrupted in myocardin–/– embryos, which is the likely cause of vascular abnormalities and embryonic lethality.
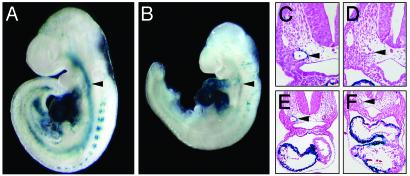
Expression of SM22-lacZ in Myocardin–/– Embryos. The SM22 promoter contains two CArG boxes that are required for expression in smooth, cardiac, and skeletal muscle cells at E9.5 and for transactivation of the promoter by myocardin in vitro (4, 5, 14). The finding that the endogenous SM22 gene was expressed in the heart of myocardin–/– embryos raised the question whether this promoter region was regulated by myocardin/SRF in vivo or whether another regulatory region was able to sustain the expression of SM22 in the heart of mutant embryos. We therefore introduced an SM22-lacZ transgene into the myocardin mutant background by breeding to the corresponding transgenic line. As shown in Fig. 5, SM22-lacZ expression was specifically ablated in the dorsal aortae of homozygous mutant embryos, but not in the heart or somites, confirming that myocardin is required specifically for vascular SM gene expression.
Fig. 5.
Ablation of SM expression of SM22-lacZ in myocardin mutant embryos. (A and B) WT and myocardin–/– mutant embryos harboring an SM22-lacZ transgene were stained for lacZ expression at E9.5. Arrowheads point to the dorsal aortae. (C–F) Sections of WT and mutant embryos from A and B counterstained with light eosin (C and E, WT; D and F, myocardin–/–). E and F show low magnifications to include the heart. Arrowheads point to the dorsal aortae.
Discussion
The phenotype of myocardin mutant mice reveals an essential role of myocardin in differentiation of vascular SMCs. These findings coupled with previous studies demonstrating the ability of myocardin to induce SM gene expression in nonmuscle cells (15–18) establish myocardin as the only transcription factor known to be both necessary and sufficient for SM differentiation.
The Role of Myocardin in SM Development. SRF has been reported to activate SM genes in transfection assays by recruiting several other cofactors. For example, the homeodomain protein Mhox and two A/T-rich DNA-binding proteins referred to as MRFα and β have been proposed as SM-restricted activators of SRF target genes (24, 25). The combination of GATA6 and LIM domain proteins of the cysteine-rich lim-only protein family has also been reported to be sufficient and necessary for SM gene expression in transfected cells, apparently by enhancing SRF DNA binding (26). However, it remains unclear which, if any, of these purported SRF cofactors are essential for SM gene expression in vivo or whether there is so much redundancy among SRF cofactors that no single cofactor is indispensable for SM gene activation.
The abnormalities in vascular development in myocardin mutant embryos are highly specific to SMCs and occur in the absence of associated cardiac abnormalities, which contrasts with numerous other mouse mutants in which vascular demise is secondary to cardiac dysfunction. The complete block of SM development in myocardin–/– embryos demonstrates that myocardin is an essential activator of the SM differentiation program in vivo and that no other factor can substitute for this promyogenic function, despite numerous reports that other transcription factors can cooperate with SRF to stimulate SM gene expression in vitro. These findings also demonstrate that SRF alone is incapable of activating SM target genes in vivo without recruiting myocardin.
Myocardin and Cardiac Gene Expression. Based on the absence of cardiac gene expression in Xenopus embryos expressing a dominant-negative myocardin mutant, and the ability of myocardin to activate cardiac gene promoters in transfection assays (14), we anticipated that myocardin would be required for heart development in the mouse. Nevertheless, we detected no abnormalities in cardiac morphogenesis or gene expression in myocardin mutant embryos. Even direct target genes of myocardin, such as SM22, SM α-actin, and ANF were expressed normally in the heart.
How can these findings be explained? We favor the possibility that MRTF-A or -B, which are expressed in the developing heart (19), or other cardiac transcription factors, may substitute for myocardin at this early stage, and such redundancy is lacking in the SM lineage. It is interesting to note that the early heart tube resembles a vessel and expresses many SM genes, which are later down-regulated. We propose that myocardin controls an early muscle regulatory program shared by the smooth and cardiac muscle lineages and that cardiac muscle cells possess additional myogenic regulators that modify this program. Combining the myocardin mutation with mutations in MRTF genes or other cardiac transcription factors should further illuminate the potential role of myocardin in the developing heart.
Acknowledgments
We thank David Sutcliffe for histology, Alisha Tizenor and Stephen Johnson for assistance with graphics, Jennifer Page for editorial assistance, and Rhonda Bassel-Duby for comments on the paper. E.N.O. was supported by grants from the National Institutes of Health, the Donald W. Reynolds Foundation, and the McGowan Foundation. D.-Z.W. was supported by the Muscular Dystrophy Association and a start-up fund from the University of North Carolina.
Abbreviations: SM, smooth muscle; SMC, SM cell; PECAM, platelet-endothelial cell adhesion molecule; En, embryonic day n; SRF, serum response factor; MRTF, myocardin-related transcription factor.
References
- 1.Conway, E. M., Collen, D. & Carmeliet, P. (2001) Cardiovasc. Res. 49, 507–521. [DOI] [PubMed] [Google Scholar]
- 2.Owens, G. K. (1995) Physiol. Rev. 75, 487–517. [DOI] [PubMed] [Google Scholar]
- 3.Majesky, M. W. (2003) Curr. Atheroscler. Rep. 5, 208–213. [DOI] [PubMed] [Google Scholar]
- 4.Li, L., Liu, Z., Mercer, B., Overbeek, P. & Olson, E. N. (1997) Dev. Biol. 187, 311–321. [DOI] [PubMed] [Google Scholar]
- 5.Kim, S., Ip, H. S., Lu, M. M., Clendenin, C. & Parmacek, M. S. (1997) Mol. Cell. Biol. 17, 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manabe, I. & Owens, G. K. (2001) J. Clin. Invest. 107, 823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack, C. P. & Owens, G. K. (1999) Circ. Res. 84, 852–861. [DOI] [PubMed] [Google Scholar]
- 8.Miano, J. M., Carlson, M. J., Spencer, J. A. & Misra, R. P. (2000) J. Biol. Chem. 275, 9814–9822. [DOI] [PubMed] [Google Scholar]
- 9.Lilly, B., Olson, E. N. & Beckerle, M. C. (2001) Dev. Biol. 240, 531–547. [DOI] [PubMed] [Google Scholar]
- 10.Herring, B. P. & Smith, A. F. (1997) Am. J. Physiol. 272, C1394–C1404. [DOI] [PubMed] [Google Scholar]
- 11.Landerholm, T. E., Dong, X. R., Lu, J., Belaguli, N. S., Schwartz, R. J. & Majesky, M. W. (1999) Development (Cambridge, U.K.) 126, 2053–2062. [DOI] [PubMed] [Google Scholar]
- 12.Arsenian, S., Weinhold, B., Oelgeschlager, M., Ruther, U. & Nordheim, A. (1998) EMBO J. 17, 6289–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Treisman, R. (1994) Curr. Opin. Genet. Dev. 4, 96–101. [DOI] [PubMed] [Google Scholar]
- 14.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851–862. [DOI] [PubMed] [Google Scholar]
- 15.Chen, J., Kitchen, C. M., Streb, J. W. & Miano, J. M. (2002) J. Mol. Cell. Cardiol. 34, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 16.Du, K. L., Ip, H. S., Li, J., Chen, M., Dandre, F., Yu, W., Lu, M. M., Owens, G. K. & Parmacek, M. S. (2003) Mol. Cell. Biol. 23, 2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida, T., Sinha, S., Dandre, F., Wamhoff, B. R., Hoofnagle, M. H., Kremer, B. E., Wang, D. Z., Olson, E. N. & Owens, G. K. (2003) Circ. Res. 92, 856–864. [DOI] [PubMed] [Google Scholar]
- 18.Wang, Z., Wang, D. Z., Pipes, G. C. T. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 7129–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, D. Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., Richardson, J. A., Nordheim, A. & Olson, E. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Q., Lu, J., Yanagisawa, H., Webb, R., Lyons, G. E., Richardson, J. A. & Olson, E. N. (1998) Development (Cambridge, U.K.) 125, 4565–4574. [DOI] [PubMed] [Google Scholar]
- 21.Shelton, J. M., Lee, M. H., Richardson, J. A. & Patel, S. B. (2000) J. Lipid Res. 41, 532–537. [PubMed] [Google Scholar]
- 22.Li, L., Miano, J. M., Mercer, B. & Olson, E. N. (1996) J. Cell Biol. 132, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin, H. S., Shen, H. M., Yan, H. C., DeLisser, H. M., Chung, A., Mickanin, C., Trask, T., Kirschbaum, N. E., Newman, P. J., Albelda, S. M., et al. (1994) Development (Cambridge, U.K.) 120, 2539–2553. [DOI] [PubMed] [Google Scholar]
- 24.Hautmann, M. B., Thompson, M. M., Swartz, E. A., Olson, E. N. & Owens, G. K. (1997) Circ. Res. 81, 600–610. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe, M., Layne, M. D., Hsieh, C. M., Maemura, K., Gray, S., Lee, M. E. & Jain, M. K. (2002) Circ. Res. 91, 382–389. [DOI] [PubMed] [Google Scholar]
- 26.Chang, D. F., Belaguli, N. S., Iyer, D., Roberts, W. B., Wu, S. P., Dong, X. R., Marx, J. G., Moore, M. S., Beckerle, M. C., Majesky, M. W., et al. (2003) Dev. Cell 4, 107–118. [DOI] [PubMed] [Google Scholar]