Abstract
Two approaches based on hybridization of viral probes with oligonucleotide microarrays were developed for rapid analysis of genetic variations during microevolution of RNA viruses. Microarray analysis of viral recombination and microarray for resequencing and heterogeneity analysis were able to generate instant genetic maps of vaccine-derived polioviruses (VDPVs) and reveal the degree of their evolutionary divergence. Unlike conventional methods based on cDNA sequencing and restriction fragment length polymorphism, the microarray approaches are better suited for analysis of heterogeneous populations and mixtures of different strains. The microarray hybridization profile is very sensitive to the cumulative presence of small quantities of different mutations, including those that cannot be revealed by sequencing, making this approach useful for characterization of profiles of nucleotide sequence diversity in viral populations. By using these methods, we identified a type-3 VDPV isolated from a healthy person and missed by conventional methods of screening. The mutational profile of the polio strain was consistent with >1 yr of circulation in human population and was highly virulent in transgenic mice, confirming the ability of VDPV to persist in communities despite high levels of immunity. The proposed methods for fine genotyping of heterogeneous viral populations can also have utility for a variety of other applications in studies of genetic changes in viruses, bacteria, and genes of higher organisms.
Keywords: molecular evolution, recombination, population heterogeneity, mutation screening, live vaccine
Genetic instability and plasticity of genomes are inherent properties of many RNA viruses with many profound implications for their replication, evolution, and pathogenesis (1–4). They include accumulation of point mutations and intermolecular recombination, resulting in exchanges of homologous genomic segments. Although few mutations make viruses more viable, the increased mutational burden often results in reduction of viral fitness (5–7). Recombination that occurs in some viruses results in exchange of functional domains and may restore the overall fitness of the virus (8). These concurrent processes serve as a continuous source of new viral variants. This selection is driven by changes in replication conditions or simply occurs by random sampling (bottlenecking). The evolution of dynamic swarms of viral mutants plays an important role in pathogenesis of chronic viral infections such as HIV, hepatitis C virus, and many others (4, 9, 10) and may occur in response to drug treatment or make immunization inefficient.
The evolution of RNA viruses is influenced by numerous factors related to conditions of viral replication. Analysis of the genomic structure of continuously emerging viral variants may reveal the nature of selective forces, helping to predict and possibly control the direction of evolution. Despite the fundamental role this dynamic polymorphism plays in processes having important practical implications, current experimental methods for its analysis do not easily reveal the full picture. First, any protocol that includes cDNA cloning is inadequate unless it involves generation of a large representative subset of independent clones. Recently, we proposed an alternative approach for genetic engineering of RNA viruses based on full-length PCR amplification of viral cDNA, which allowed us to maintain and manipulate viral heterogeneity in vitro (11). Next, methods for evaluation of genomic heterogeneity of RNA viruses are generally very labor-intensive and poorly adapted for high-throughput analysis. In addition to the most informative but tedious sequencing of a large number of viral clones, these methods include PCR–restriction fragment length polymorphism (RFLP) (12), mutant analysis by PCR, and restriction enzyme cleavage (13). They are either focused on one or few nucleotides in viral genome or are insufficiently sensitive or precise.
Areas of research requiring methods devoid of these shortcomings include screening for emerging viruses, monitoring of viral evolution in different natural hosts, and safety of live viral vaccines. Oral polio vaccine (OPV) (14) consists of live attenuated strains of poliovirus and is among the most widely used and successful vaccines in the history of medicine. It possesses an inherent genetic instability and has been implicated in rare cases of vaccine-associated paralytic poliomyelitis, caused by vaccine-derived strains with increased virulence having point mutations; a large number of them are intertypic recombinants (15, 16). It has been traditionally assumed that even though the revertant polioviruses possess higher neurovirulence, they cannot circulate and therefore are rapidly cleared from human populations (17–19). Recent outbreaks of poliomyelitis in Egypt (20), the Philippines (21), the Dominican Republic and Haiti (22), and Madagascar (23) were caused by vaccine-derived poliovirus (VDPV) strains that fully regained their virulence and ability to circulate in a human population. This raises serious questions about the strategy of polio eradication that can be answered only by conducting a large-scale worldwide screening of poliovirus isolates to uncover the reasons and mechanisms of the emergence of newly virulent VDPVs.
Hybridization with microarrays of immobilized DNA or oligonucleotides that contain thousands of individual probes appears to be suitable for parallel analysis of a large number of markers. Recently, this approach was used for genotyping of viruses, bacteria, and bacterial factors (24–28). The methods were based on either short highly specific oligonucleotide probes (oligoprobes) or longer degenerate probes that covered a wide range of viral diversity. In this study, we have explored the utility of microarray techniques for analysis of the polymorphous poliovirus genomes and characterization of individual and unique viral stocks. This was made possible by a combination of full-length PCR amplification of viral cDNA, with a large number of oligoprobes used to systematically interrogate the entire viral genome. The methods described here allowed us to obtain instant genetic maps of recombinant strains and to determine evolutionary divergence and mutational profiles of individual viral stocks.
Materials and Methods
Virus Isolation and Preliminary Characterization. The origins of the VDPV strains studied are given in Table 1. Virus isolation from stool samples was done in RD cells by standard methods (29). Attenuated poliovirus Sabin 1 was passaged with high multiplicity in Vero cells at 37°C (30), and the viruses were serotyped in microneutralization tests with type-specific sera (29). Sequencing of the PCR-amplified viral cDNA products was performed by using an ABI Prism 310 Genetic Analyzer and d-Rhodamine DNA sequencing kits (Applied Biosystems). RFLP analysis was done as described (12).
Table 1. Poliovirus isolates studied.
| Isolate no. | Date of isolation | Geographic location | Diagnosis |
|---|---|---|---|
| 14 | October 23, 1999 | Perm, Russia | Poliomyelitis |
| 11264 | October 24, 1999 | Perm, Russia | Healthy |
| 15763 | June 7, 2001 | Tjumen, Russia | Poliomyelitis |
| 9457 | February 16, 1999 | Azerbaijan | Acute flaccid paralysis |
| 14829 | February 18, 2001 | Buryat Republic | Poliomyelitis |
Test for Neurovirulence. TgPVR-21 transgenic mice (31) were purchased at the Central Institute of Experimental Animals in Tokyo, intraspinally inoculated (32) with five doses (10-fold dilutions) of poliovirus to determine 50% of the paralytic dose, and observed for signs of paralysis during the 14 days after inoculation.
Design and Fabrication of Microchips. Microarrays for resequencing and sequence heterogeneity (MARSH) assay. Three sets of oligonucleotides (Tm = 47°C; see Table 3, which is published as supporting information on the PNAS web site, www.pnas.org) overlapping at half length, matching genomic sequences of three OPV strains, and covering the virion protein 1 (VP1) coding region were synthesized. Each oligoprobe contained an aminolink group at the 5′ end for immobilization and was purified after automated synthesis as described (25). Microchips were printed on 3 × 1-in sialylated (aldehyde-coated) glass slides (Cell Associates, The Sea Ranch, CA) by using contact microspotting robot (Cartesian Technologies, Ann Arbor, MI) equipped with a microspotting pin CMP-7 (ArrayIt, Sunnyvale, CA) as described (28). Each oligoprobe was spotted four times within a single microarray, and four MARSH chips were printed on one glass slide.
Microarray analysis of viral recombination (MAVR) assay. Genotype-specific oligoprobes for identification of Sabin strains (GenBank accession nos. AY184219–AY184221) were selected by using custom oligoscan software (unpublished data), and were spaced ≈150 bases from each other (see Table 4, which is published as supporting information on the PNAS web site). They had a moderate GC content (Tm between 41 and 57°C) and were printed in three rows according to their location in the genome. Five individual microarrays for MAVR analysis were spotted on each slide.
Preparation of Samples for Hybridization. Primers matching conserved regions of the 5′ and 3′ nontranslated regions were used for full-length PCR amplification of viral cDNA performed with the Perkin–Elmer XL-PCR kit as described (11). The forward primer contained biotin at the 5′ end, which allowed us to separate strands of PCR products by using streptavidin-coated magnetic beads (GenoVision, Oslo) as described (25). The VP1 coding region was PCR amplified with universal poliovirus primers. The reverse primer contained a T7 RNA polymerase promoter, and PCR products were transcribed in vitro by using the MEGAscript T7 RNA polymerase kit (Ambion, Austin, TX). Single-stranded DNA or RNA (0.1–0.5 μg) was labeled by using the Cy5 or Cy3 Micromax ASAP RNA Labeling Kit (Perkin–Elmer) and purified by using CENTRI-SEP (Princeton Separations, Adelphia, NJ) spin columns.
Microarray Hybridization, Scanning, and Data Analysis. Immediately before hybridization, fluorescently labeled samples were dried, reconstituted in ASAP Hybridization Buffer III (Perkin–Elmer), and denatured for 1 min at 95°C. The final concentration of each probe in the hybridization solution did not exceed 0.2 μM. Five-microliter aliquots were applied to the microarray area and covered with an individual plastic coverslip. Hybridization was performed in the incubation chamber (ArrayIt) for at least 30 min at 45°C and then washed as described (28). Microchip images were taken by using a confocal fluorescent scanner ScanArray 5000 (GSI Lumonics, Billerica, MA) equipped with green and red HeNe lasers (543 and 632 nm for excitation of Cy3 and Cy5, respectively). MARSH images were then analyzed by using quantarray software (Packard). Data obtained from four replicates of each oligoprobe were cleaned up by “median filtration” (the highest and the lowest values were dropped, and the remaining data points were averaged). The values from each microarray element were normalized by the total signal from the entire array. The normalized signals from the reference array were then divided by the respective signals from the test array and the results expressed as a ratio.
Results
Accumulation of point mutations and recombination are the two major mechanisms of poliovirus evolution. They were studied by MARSH and MAVR assays, respectively.
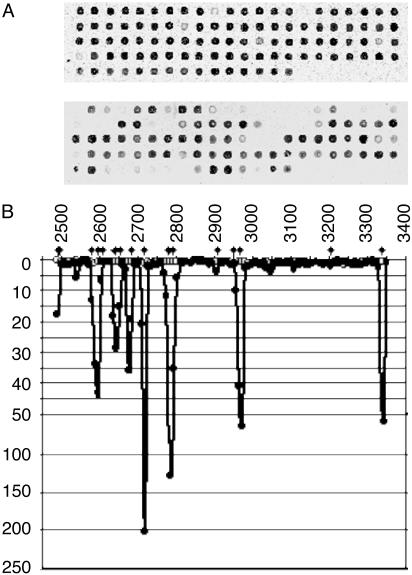
MARSH Assay. To screen for emerging point mutations, we have created a microchip containing oligonucleotides overlapping at half-length covering the region coding for VP1, the most variable capsid protein of poliovirus. Each microarray contained 102–103 (depending on the serotype of poliovirus) oligoprobes printed in quadruplicates to assess reproducibility of results. Fig. 1A shows the results of analysis of VDPV strain 11264 isolated from the contact of vaccine-associated paralytic poliomyelitis case. The small size of oligonucleotides (14–24 nt) ensured that their binding was critically affected by even a single-nucleotide mismatch. Relative fluorescence intensity produced by individual oligoprobes depends on their structure and varied ≈50-fold. Therefore, two microarrays were simultaneously hybridized with fluorescently labeled RNA samples prepared from the reference Sabin strain and from a test strain. The fluorescent signal from each spot in the reference microarray was divided by the respective signal in the test array. Mutations in the test sample resulted in a decreased binding to some oligoprobes and thus higher ratio (Fig. 1B). The peak size varied, reflecting the nature of different mutations and their number and location within oligoprobes. Mutations in the middle of short oligonucleotides tended to produce larger peaks (for example, the peaks with ratio close to 1:150 and 1:200; Fig. 1B). Mutations closer to the ends of oligoprobes produced smaller peaks. For instance, peaks attributed to mutations at nucleotides 2908 and 3202 are hardly visible at the scale used in Fig. 1B. However, the ratio values for these oligoprobes (3.5 and 2.0) significantly differed from the values for the homogeneous sample (0.9 and 1.3, respectively). On the other hand, we observed peaks at locations where no mutation could be detected by sequencing (e.g., the peak around nucleotide 2550 shown in Fig. 1B). Closer inspection of raw sequencing data revealed the presence of more than one nucleotide per site in this region, suggesting that MARSH can detect mutations present at a low level (sequence heterogeneities).
Fig. 1.
MARSH analysis of strain 11264. (A) Hybridization patterns of reference Sabin 3 (Upper) and test 11264 (Lower) fluorescent RNA samples with MARSH arrays. In each case, only one quadruplicate array is shown. (B) The ratio of signals from two MARSH chips hybridized with Sabin 3 reference is shown with open circles. The ratio of signals obtained with the Sabin 3 reference and strain 11264 is shown by filled circles. x axis, the nucleotide coordinates on poliovirus genome; y axis, ratio values. Diamonds show the location of mutations determined by sequencing.
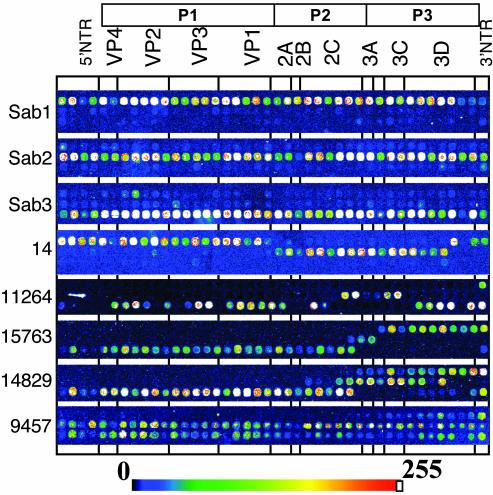
MAVR Assay. The microarray contained 42 oligoprobes specific for each of the three Sabin strains spaced by ≈150 nt along the entire genome and printed on the microchip in three rows according to their location in the genome. Fig. 2 shows the results of hybridization of Sabin and VDPV strains with this microchip. Most recombinant strains contained more than one crossover point [Fig. 2 (14, 11264, 15763, 14829)], and in some cases there was an indication of the presence of more than one genotype in a sample [Fig. 2 (11264, 14829, 9457)]. The recombination patterns were consistent with the results of RFLP analysis of these strains (data not shown) but are more detailed and their interpretation more straightforward. For example, MAVR analysis identified the genomic recombination schemes of strains 15763 and 14829 (Fig. 2) including the presence of more than one type of recombinant in the latter, whereas RFLP failed to detect the mixture. In addition, the HinfI and RsaI restriction patterns of the 3AC and 3D-1 regions of strain 15763 differed from those of Sabin strains. Although the intensity of fluorescent signals produced by individual spots varied, identification of the origin of individual parts of viral genomes in most cases was unambiguous.
Fig. 2.
MAVR analysis of OPV and VDPV strains.
Type-3 Highly Divergent VDPV. Strain 14 of type-1 poliovirus was the causative agent of poliomyelitis in an unvaccinated 7-mo-old infant and was shown to have circulated in human population for ≈2 yr (33). As part of the epidemiological investigation of this case, two strains (11264 and 11265) were isolated from healthy contacts and shown by neutralization to be serotype-3 viruses and thus, at first, were considered unrelated to the original case. Indeed, on MARSH analysis, isolate 11265 turned out to be a slightly evolved type-3 VDPV (data not shown) and might have been a result of recent vaccination. However, MARSH analysis of the VP1-coding region of the isolate 11264 showed a significant number of mutations (Fig. 1). This was confirmed by sequencing that revealed 17 mutations in the VP1-coding region, consistent with ≈18-mo circulation, the time assessed based on roughly linear rate of fixation of mutations in naturally evolving polioviruses (33).
MAVR analysis of isolate 11264 showed that it was a type-3/type-2/type-3 recombinant (Fig. 2). Isolate 14 from the original vaccine-associated paralytic poliomyelitis case was a type-1/type-2/type-1 recombinant, sharing type-2 segments in the region coding for nonstructural proteins (Fig. 2). Comparison of type-2 sequences of these strains revealed that 5 of 11 mutations in the shared region were common to both isolates, suggesting that type-2 regions might be related. Many of the common mutations were clustered in a short region (nucleotides 4958–5283) that appears to be a recombination hot spot [nucleotides 4923–4952 (15, 16, 34–37)], further supporting the notion that both isolates may have had a common type-2 VDPV ancestor.
Even though isolate 11264 came from a healthy child, it has a very high degree of divergence from its vaccine progenitor, suggesting that it might have regained neurovirulent properties. To check this, we tested it in transgenic mice susceptible to poliovirus and found that it was almost as virulent as a wild-type Leon/37 virus (Table 2).
Table 2. TgPVR21 mouse neurovirlence test with type-3 polioviruses.
|
Paralyzed mice (in groups of 10)
|
|||||||
|---|---|---|---|---|---|---|---|
|
Dose (log10 TCID50)
|
|||||||
| Virus strain | 1 | 2 | 3 | 4 | 5 | 6 | PD50 (log10 TCID50) |
| Leon/37 | 6 | 10 | 10 | 10 | 10 | nd | <1.0 |
| 11264 | 5 | 10 | 10 | 10 | 10 | 10 | 1 |
| Sabin 3 | nd | 1 | 1 | 6 | 10 | 10 | 3.2 |
TCID50, tissue culture 50% infective dose; PD50, 50% of the paralytic dose.
Screening for Mutations in High-Passage Stocks of Attenuated Poliovirus. Passaging in vitro and in vivo tends to result in accumulation of random mutations in viral stocks, leading to increased heterogeneity of viral populations. To see whether the MARSH method is sensitive enough to detect small differences in mutational profiles of serially passaged viral stocks, we compared profiles of Sabin 1 virus at passage levels from 1 to 10 in Vero cells. Fig. 3A shows two big peaks on the profile of passage 9 (purple curve). Sequencing of VP1 coding region revealed the presence of a G→A substitution at the nucleotide 2975 (Val-166→Met) and a visible heterogeneity (mix of A, T, and G) at position 2772 (Fig. 3B, passage 9), matching the two peaks. MARSH analysis and sequencing of passage levels 3, 4, and 6 showed that the size of these peaks increased with passages, reflecting gradual accumulation of mutations (Fig. 3). However, we also observed few peaks with ratio close to or more than 10 on the profile of passage 9 (e.g., the peaks in the regions of nucleotides 2500–2600 or 3100–3350 shown in Fig. 3A) that did not match mutations detectable by direct sequencing. These orphan peaks on the mutational profile can be explained by cumulative effect of several mutations within a genomic region matching one oligoprobe, if each of these mutations were present at a low level, undetectable by conventional sequencing techniques. Interestingly, there were regions of many contiguous oligoprobes where viral RNA was stable and consistently showed the ratio close to 1 in all passages (Fig. 3A; genomic regions corresponding to ≈nucleotides 2650–2750 and 3000–3100). This demonstrates that MARSH analysis can reveal both conserved and variable regions in RNA viral genomes.
Fig. 3.
Mutational profiles of the VP1 coding region of Sabin 1 viral stocks obtained at different passage levels. (A) The ratio of signals obtained from hybridization of MARSH with reference sample (first passage of Sabin 1) and other passages: passage 3, yellow; passage 4, green; passage 6, blue; passage 9, red. x and y axes, see legend to Fig. 1. (B) The raw sequencing data of VP1 genomic regions (nucleotides 2767–2777 and 2970–2980) of Sabin 1 passages 3, 4, 6, and 9. Nucleotides 2772 and 2975 are underlined.
Discussion
Two microarray methods have been described here for the analysis of genetic diversity of RNA viruses at the level of genomic recombination and at the level of nucleotide sequence heterogeneity. Together, these two approaches produce a snapshot of genomic organization of the entire viral population without resorting to more laborious techniques. This simple analysis can be completed within a few hours simultaneously for a large number of viral isolates. The method does not require cloning of the test samples, thus preserving the natural composition of viral populations. Use of these methods opens the possibility of a large-scale full-genome screening of viral isolates needed for thorough epidemiological surveillance, vaccine quality control, and analysis of genetic changes in response to drug treatment.
Use of oligonucleotide microchips for screening of random mutations is based on their ability to distinguish single-nucleotide polymorphisms (25, 38–40). However, not all mutations are expected to produce the same effect on binding of oligoprobes. For instance, the strongest peaks were produced by mutations affecting the center of an oligoprobe, and the shortest probes produced better discrimination. Because the oligoprobes overlap and on average each site in the sequence matches two oligoprobes, the chances of a mutation to be missed is low and can be further decreased by a bigger overlap.
The MARSH approach resembles resequencing by hybridization with Affymetrix microchips containing all possible oligonucleotides present in a gene of interest (41). Affymetrix chips provide a complete coverage of the gene under study with oligonucleotide probes and can also be used for mutational screening. However, our system has important advantages. Ninety-six to 98% of the probes produced by photolithographic oligonucleotide synthesis have sequences different from the intended one, which results in lower hybridization specificity and makes it difficult to quantitatively assess sequence heterogeneities. In contrast, the presynthesized oligonucleotides are significantly more homogeneous and produce better discrimination. For instance, even single-point mutations often produce ratios 1:100 and more (Fig. 1), meaning that they disrupt binding of oligoprobes by >99% and also that cross-hybridization may be <1%. The second more practical advantage is that, unlike Affymetrix microchips that are produced by using highly sophisticated industrial equipment, our microchips can be easily created and modified as needed in virtually any laboratory, whereas any modification of photolithographically produced chips can be done only by the manufacturer.
Unlike direct sequencing, the MARSH method can determine only an approximate location of mutations within a single oligoprobe. However, this limitation also has an advantage, because it allows several closely located mutations present on different RNA molecules to be detected even if each of them is present at a relatively low level undetectable by conventional sequencing methods. This cumulative effect of small quantities of mutations may provide a tool for profiling minor genetic heterogeneity in a gene or even entire genome. For instance, we observed a difference in oligonucleotide-binding profiles of a high-passage stock of attenuated poliovirus (Fig. 3), suggesting the presence of multiple but low-sequence heterogeneities. For some probes, the ratio was extremely high, even though the level of heterogeneity detectable by the conventional sequencing method was moderate. This ability to detect minor sequence diversity could be used for identification of genome regions with different stability in individual stocks of virus grown under differing conditions. Comparison of mutational profiles of poliovirus stocks at different passage levels revealed “hot-spot” regions in VP1-coding regions, in which mutations accumulated much faster. On the other hand, there were rather long segments of the gene with no detectable heterogeneity even after nine passages. Thus, in addition to “hot-spots,” the MARSH approach can discover “cold spots” of higher-than-average genetic stability. These conserved parts of the VP1-coding region deserve special attention, and it was previously noted that they contain an unusual concentration of rarely used codons, possibly controlling the ribosome elongation rate (7).
One obvious application of this approach could be quality control and consistency monitoring of live viral vaccines production. Our previous studies demonstrating a correlation between minor sequence heterogeneity and neurovirulence of OPV (13, 42–44) and plasticity of the genome of attenuated mumps virus (45, 46) have led us to propose a molecular consistency approach to monitoring production of live viral vaccines (47). Mutant analysis by PCR and the restriction enzyme cleavage method we proposed for this purpose were accepted by the World Health Organization as a standard quality control test of OPV (48). MARSH analysis can be the ultimate tool to determine complete mutational profiles of individual vaccine stocks rather than to monitor quantities of individual mutations. Another area with a critical need to distinguish viral stocks, differing only in its fine mutational composition, is monitoring of the microevolution of viruses causing chronic infections, whose natural populations are notoriously heterogeneous, e.g., hepatitis C virus and HIV. The MARSH method could be used to reveal mutations responsible for emergence of drug resistance and other important properties of the viruses.
The recombination chip is, in essence, an extension of genotype-specific oligonucleotide microchips previously used for identification of different viruses and bacteria (24, 25). Location of spots within the poliovirus MAVR chips resulted in a hybridization image that graphically revealed recombination patterns and crossover regions. One potential caveat of MAVR analysis is that it can positively identify only regions derived from strains represented on the microchip. If one recombination partner is unknown, the microchip will reveal a “gap” or an irregular pattern, calling for nucleotide sequencing as a tool of last resort. Alternatively, conserved oligoprobes with broader specificity could be included into MAVR chips to tentatively identify the origin of “orphan” genomic segments.
Even though MAVR mapping of poliovirus genomes is less detailed than that made by complete nucleotide sequencing, its extraordinary throughput makes it a reasonable compromise. RFLP also used for this purpose is more time-consuming and less informative than MAVR. One unique advantage of MAVR analysis is that it allows us to genotype naturally heterogeneous populations. For instance, 14829 in Fig. 2 shows that the strain is a mixture of at least two and probably more different types of recombinants. In combination with PCR amplification directly from clinical specimens, this opens the possibility of studying natural heterogeneity of viral populations in vivo.
Despite the ≈50-yr history of OPV use, highly divergent VDPVs were discovered only recently, primarily because thousands of isolates were screened as part of a worldwide polio eradication campaign. The utility of microarray techniques for this purpose was demonstrated by identification of the VDPV strain with divergence consistent with its ≈1.5-yr persistence in the human population. It was isolated from a healthy individual in contact with a case of paralytic poliomyelitis caused by the long-circulating VDPV strain described recently (33). Even though it was isolated from a healthy child, its neurovirulence was comparable to that of wild-type poliovirus (Table 2). Previously undescribed circulation of type-3 highly divergent VDPV showed that all three Sabin strains can revert to both neurovirulence and transmissibility. The primary case was caused by a recombinant between Sabin 1- and 2-related viruses, whereas the “contact” strain was classified antigenically as type-3 poliovirus and at first was dismissed as unrelated. MAVR analysis revealed that it was a recombinant between types 3 and 2. Type-2-derived parts of these viruses shared 38% of mutations, suggesting that they may have had a recent common ancestor.
Isolation of these two VDPV strains in the same orphanage leads us to an alarming suggestion that this cryptic circulation of highly divergent polioviruses may not be uncommon, regardless of the level of population immunity. A plausible reason why these strains did not cause outbreaks may be just the high vaccination coverage in this particular community. In countries with an undervaccinated population, such VDPVs present a greater risk and may cause outbreaks (20–23), confirming that maintaining of high vaccination coverage is important even after eradication of wild-type polioviruses. Occurrence of highly divergent VDPVs capable of causing paralytic disease blurs the line between “attenuated” and “wild-type” polioviruses even further, to the point where these definitions no longer can be used for practical decisions in the eradication campaign. Can the region where Sabin-derived polioviruses circulate and cause paralytic disease be called free from wild-type polio? It is apparent that the polio eradication campaign encountered a host of very difficult questions at the time when the final goal appears within reach. Answers to some of these questions can be obtained by an extensive worldwide screening of VDPVs, and the proposed methods can play an important role in this.
It is clear that the approaches described here can be used not only for poliovirus but for screening for point mutations and heterogeneity in other viruses as well as in genes of other organisms. Currently we are working on using the MARSH method for analysis of the dynamics of heterogeneous viral populations in a number of other systems, as well as for single-nucleotide polymorphism screening in human genes.
Supplementary Material
Acknowledgments
We thank O. E. Ivanova and T. P. Eremeeva for providing epidemiological information. We also express our gratitude to G. Y. Lipskaya for help in the coordination of this study. This work was supported in part by grants from the Defense Advanced Research Project Agency, the National Vaccine Program Office, and the World Health Organization.
Abbreviations: MARSH, microarrays for resequencing and sequence heterogeneity; MAVR, microarray analysis of viral recombination; RFLP, restriction fragment length polymorphism; VDPV, vaccine-derived poliovirus; OPV, oral polio vaccine; VP1, virion protein 1.
References
- 1.Domingo, E., Martinez-Salas, E., Sobrino, F., de la Torre, J. C., Portela, A., Ortin, J., Lopez-Galindez, C., Perez-Brena, P., Villanueva, N., Najera, R., et al. (1985) Gene 40, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Domingo, E. & Holland, J. J. (1997) Annu. Rev. Microbiol. 51, 151–178. [DOI] [PubMed] [Google Scholar]
- 3.Domingo, E., Escarmis, C., Menendez-Arias, C. & Holland, J. J. (1999) in Origin and Evolution of Viruses, eds. Domingo, E., Webster, R. & Holland, J. J. (Academic, San Diego), pp. 141–161.
- 4.Sala, M. & Wain-Hobson, S. (2000) J. Mol. Evol. 51, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duarte, E., Clarke, D., Moya, A., Domingo, E. & Holland, J. (1992) Proc. Natl. Acad. Sci. USA 89, 6015–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, D. K., Duarte, E. A., Moya, A., Elena, S. F., Domingo, E. & Holland, J. (1993) J. Virol. 67, 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavrilin, G. V., Cherkasova, E. A., Lipskaya, G. Y., Kew, O. M. & Agol, V. I. (2000) J. Virol. 74, 7381–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agol, V. I. (2002) in The Molecular Biology of Picornaviruses, eds. Semler, B. L. & Wimmer, E. (Am. Soc. Microbiol., Washington, DC), pp. 269–284.
- 9.Yoneyama, T., Yoshida, H., Shimizu, H., Yoshii, K., Nagata, N., Kew, O. & Miyamura, T. (2001) Dev. Biol. 105, 93–98. [PubMed] [Google Scholar]
- 10.Woolhouse, M. E., Webster, J. P., Domingo, E., Charlesworth, B. & Levin, B. R. (2002) Nat. Genet. 32, 569–577. [DOI] [PubMed] [Google Scholar]
- 11.Chumakov, K. M. (1996) J. Virol. 70, 7331–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balanant, J., Guillot, S., Candrea, A., Delpeyroux, F. & Crainic, R. (1991) Virology 184, 645–654. [DOI] [PubMed] [Google Scholar]
- 13.Chumakov, K. M., Powers, L. B., Noonan, K. E., Roninson, I. B. & Levenbook, I. S. (1991) Proc. Natl. Acad. Sci. USA 88, 199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sabin, A. B. (1955) Ann. N.Y. Acad. Sci. 61, 924–938. [DOI] [PubMed] [Google Scholar]
- 15.Lipskaya, G. Y., Muzychenko, A. R., Kutitova, O. K., Maslova, S. V., Equestre, M., Drozdov, S. G., Perez-Bercoff, R. P. & Agol, V. I. (1991) J. Med. Virol. 35, 290–296. [DOI] [PubMed] [Google Scholar]
- 16.Driesel, G., Diedrich, S., Kunkel, U. & Schreier, E. (1995) Eur. J. Epidemiol. 11, 647–654. [DOI] [PubMed] [Google Scholar]
- 17.Fine, P. E. & Carneiro, I. A. (1999) Am. J. Epidemiol. 150, 1001–1021. [DOI] [PubMed] [Google Scholar]
- 18.Wood, D. J., Sutter, R. W. & Dowdle, W. R. (2000) Bull. World Health Org. 78, 347–357. [PMC free article] [PubMed] [Google Scholar]
- 19.Wood, D. J. (2001) Dev. Biol. 105, 69–72. [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention (2001) Morbid. Mortal. Wkly. Rep. 50, 41.–42, 51. [Google Scholar]
- 21.Centers for Disease Control and Prevention (2001) Morbid. Mortal. Wkly. Rep. 50, 874–875. [Google Scholar]
- 22.Kew, O., Morris-Glasgow, V., Landaverde, M., Burns, C., Shaw, J., Garib, Z., Andre, J., Blackman, E., Freeman, C. J., Jorba, J., et al. (2002) Science 296, 356–359. [DOI] [PubMed] [Google Scholar]
- 23.Centers for Disease Control and Prevention (2002) Wkly. Epidemiol. Rec. 77, 241–242.12149772 [Google Scholar]
- 24.Chizhikov, V., Rasooly, A., Chumakov, K. & Levy, D. D. (2001) Appl. Environ. Microbiol. 67, 3258–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chizhikov, V., Wagner, M., Ivshina, A., Hoshino, Y., Kapikian, A. Z. & Chumakov, K. (2002) J. Clin. Microbiol. 40, 2398–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, D., Coscoy, L., Zylberberg, M., Avila, P. C., Boushey, H. A., Ganem, D. & DeRisi, J. L. (2002) Proc. Natl. Acad. Sci. USA 99, 15687–15692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapa, S., Mikheev, M., Shchelkunov, S., Mikhailovich, V., Sobolev, A., Blinov, V., Babkin, I., Guskov, A., Sokunova, E., Zasedatelev, A., et al. (2002) J. Clin. Microbiol. 40, 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volokhov, D., Rasooly, A., Chumakov, K. & Chizhikov, V. (2002) J. Clin. Microbiol. 40, 4720–4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization (1997) Manual for the Virologic Investigation of Poliomyelitis (World Health Organization, Geneva).
- 30.Rezapkin, G. V., Chumakov, K. M., Lu, Z., Ran, Y., Dragunsky, E. M. & Levenbook, I. S. (1994) Virology 202, 370–378. [DOI] [PubMed] [Google Scholar]
- 31.Koike, S., Taya, C., Kurata, T., Abe, S., Ise, I., Yonekawa, H. & Nomoto, A. (1991) Proc. Natl. Acad. Sci. USA 88, 951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dragunsky, E., Chernokhvostova, Y., Taffs, R., Chumakov, K., Gardner, D., Asher, D., Nomura, T., Hioki, K. & Levenbook, I. (1997) Vaccine 15, 1863–1866. [DOI] [PubMed] [Google Scholar]
- 33.Cherkasova, E. A., Korotkova, E. A., Yakovenko, M. L., Ivanova, O. E., Eremeeva, T. P., Chumakov, K. M. & Agol, V. I. (2002) J. Virol. 76, 6791–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuervo, N. S., Guillot, S., Romanenkova, N., Combiescu, M., Aubert-Combiescu, A., Seghier, M., Caro, V., Crainic, R. & Delpeyroux, F. (2001) J. Virol. 75, 5740–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cammack, N., Phillips, A., Dunn, G., Patel, V. & Minor, P. D. (1988) Virology 167, 507–514. [PubMed] [Google Scholar]
- 36.Minor, P. D., John, A., Ferguson, M. & Icenogle, J. P. (1986) J. Gen. Virol. 67, 693–706. [DOI] [PubMed] [Google Scholar]
- 37.King, A. M. Q. (1988) in RNA Genetics, eds. Domingo, E., Holland, J. J. & Ahlquist, P. (CRC, Boca Raton, FL), pp. 149–165.
- 38.Hacia, J. G. (1999) Nat. Genet. 21, 42–47. [DOI] [PubMed] [Google Scholar]
- 39.Hacia, J. G., Fan, J. B., Ryder, O., Jin, L., Edgemon, K., Ghandour, G., Mayer, R. A., Sun, B., Hsie, L., Robbins, C. M., et al. (1999) Nat. Genet. 22, 164–167. [DOI] [PubMed] [Google Scholar]
- 40.Raitio, M., Lindroos, K., Laukkanen, M., Pastinen, T., Sistonen, P., Sajantila, A. & Syvanen, A. C. (2001) Genome Res. 11, 471–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ahrendt, S. A., Halachmi, S., Chow, J. T., Wu, L., Halachmi, N., Yang, S. C., Wehage, S., Jen, J. & Sidransky, D. (1999) Proc. Natl. Acad. Sci. USA 96, 7382–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezapkin, G. V., Norwood, L. P., Taffs, R. E., Dragunsky, E. M., Levenbook, I. S. & Chumakov, K. M. (1995) Virology 211, 377–384. [DOI] [PubMed] [Google Scholar]
- 43.Rezapkin, G. V., Alexander, W., Dragunsky, E., Parker, M., Pomeroy, K., Asher, D. M. & Chumakov, K. M. (1998) Virology 245, 183–187. [DOI] [PubMed] [Google Scholar]
- 44.Rezapkin, G. V., Fan, L., Asher, D. M., Fibi, M. R., Dragunsky, E. M. & Chumakov, K. M. (1999) Virology 258, 152–160. [DOI] [PubMed] [Google Scholar]
- 45.Amexis, G., Oeth, P., Abel, K., Ivshina, A., Pelloquin, F., Cantor, C. R., Braun, A. & Chumakov, K. (2001) Proc. Natl. Acad. Sci. USA 98, 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amexis, G., Rubin, S., Chizhikov, V., Pelloquin, F., Carbone, K. & Chumakov, K. (2002) Virology 300, 171–179. [DOI] [PubMed] [Google Scholar]
- 47.Chumakov, K. M. (1999) Dev. Biol. Stand. 100, 61–74. [PubMed] [Google Scholar]
- 48.World Health Organization (2002) WHO Tech. Rep. Ser. 904, 31–93. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.