Abstract
T-box genes encode transcription factors that play critical roles in generating the vertebrate body plan. In many developmental fields, multiple T-box genes are expressed in overlapping domains, establishing broad regions in which different combinations of T-box genes are coexpressed. Here we demonstrate that three T-box genes expressed in the zebrafish mesoderm, no tail, spadetail, and tbx6, operate as a network of interacting genes to regulate region-specific gene expression and developmental fate. Loss-of-function and gain-of-function genetic analyses reveal three kinds of interactions among the T-box genes: combinatorial interactions that generate new regulatory functions, additive contributions to common developmental pathways, and competitive antagonism governing downstream gene expression. We propose that T-box genes, like Hox genes, often function within gene networks comprised of related family members.
T-box genes encode related transcription factors that regulate tissue specification, morphogenesis, and cell proliferation (1–3). In addition to tissue-specific roles, T-box genes govern regional identities within developmental fields (4–6). One puzzling aspect of T-box gene function is the recurrent finding that the primary cellular focus of the defect seen in a T-box mutant corresponds to only a limited portion of the expression domain of the mutated T-box gene (5). For example, whereas haploinsufficiency of TBX3 results in posterior forelimb deficiencies, the gene is expressed in the anterior and posterior margin of the developing hind limb and forelimb (7, 8). Similarly, although Brachyury orthologues are expressed throughout the nascent mesoderm in vertebrate embryos, loss of Brachyury function blocks differentiation of the notochord, a dorsal mesoderm tissue, but has only limited effects on the morphogenesis of the ventrolateral mesoderm, allowing differentiation of the full range of mesoderm cell types derived from this tissue (9–11). These findings implicate factors that modify T-box gene function so that individual T-box genes carry out different functions in different regions of their expression domain (12–15). Because multiple T-box genes are expressed in overlapping patterns in many developmental fields (7, 16–19), we hypothesized that T-box gene interactions contribute to regionalization of T-box gene function.
We examined individual and combined functions of three T-box genes, no tail (ntl), spadetail (spt), and tbx6, which are expressed exclusively in the developing zebrafish mesoderm (20–24). The three genes are expressed in broad domains that overlap and together mark all of the mesoderm (Fig. 1A). We demonstrate that ntl, the zebrafish orthologue of Brachyury, regulates different downstream targets in different portions of its uninterrupted expression domain. Because spt and tbx6 are coexpressed with ntl in selected regions of the mesoderm, we analyzed whether these genes interact with ntl to modify its function in a region-specific manner. Amacher et al. (25) recently showed that ntl and spt provide some overlapping functions in the mesoderm. In this report, we document three kinds of interactions among the three T-box genes: combinatorial interactions that generate new regulatory functions, additive contributions to common developmental pathways, and competitive antagonism governing downstream gene expression. Our findings demonstrate that T-box genes can perform multiple region-specific functions within a developmental field and indicate how loss of function of one T-box gene can alter the function of other T-box genes expressed in the same field.
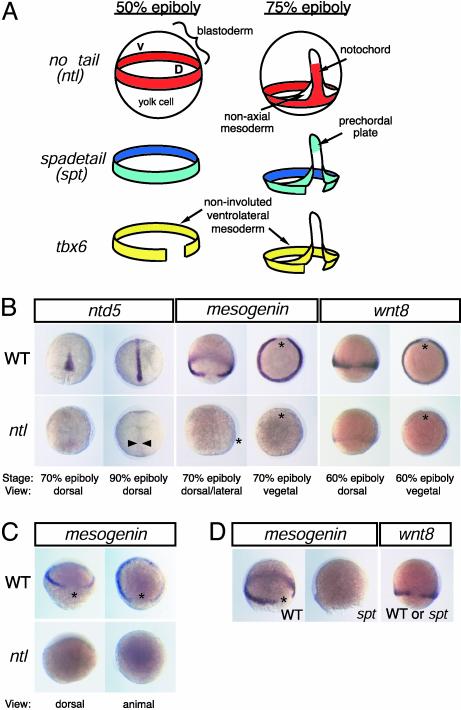
Fig. 1.
T-box genes have region-specific functions in the zebrafish mesoderm. (A) Schematic representation of ntl, spt, and tbx6 expression domains during gastrulation. (B) ntl regulates different target genes in the dorsal and ventrolateral mesoderm. ntd5, which encodes a product with Sushi/CCP domains found in adhesion and complement proteins (54), is expressed in the presumptive notochord of WT (Upper) but not of ntl mutant (Lower) embryos. wnt8 and mesogenin are expressed in the ventrolateral mesoderm in a ntl-dependent manner. (C) mesogenin expression is first detected at 30% epiboly (Upper), but it is not initiated in ntl embryos (Lower). (D) mesogenin is expressed in WT, but not spt mutant, embryos; wnt8 is expressed in both types of embryos. D or *, dorsal midline; V, ventral midline; arrowheads, axial mesoderm.
Materials and Methods
Isolation of ntl-Regulated Genes. A mesendoderm-specific cDNA library was constructed by subtractive hybridization (26, 27): cDNA from midgastrula embryos was depleted for sequences expressed in isolated animal cap tissue (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org). Independent transformants (1,728 in all) were arrayed and analyzed by differential hybridization screening. cDNA from WT and individually genotyped ntlb195 mutant 70% epiboly embryos was used to generate WT-ntl, ntl-WT-subtracted, and ntl-unsubtracted cDNA probes. Candidate ntl-dependent sequences hybridized only with WT-ntl probe. Whole-mount in situ hybridization was performed on WT and mutant sibling embryos to identify genes whose expression depended on ntl function.
Genetics and Genotype Analysis. Embryos from natural spawnings were raised at 28.5°C (28). WT embryos were from the AB* line. The sptb104, ntlb160, and ntlb195 alleles were used (11, 20, 24, 29). To analyze additive effects of mutant alleles on MyoD expression, genotyping with allele-specific primers was performed on tissue removed before in situ hybridization (see Supporting Materials and Methods).
Ectopic Gene Expression Experiments. Sequences encoding No Tail, the No Tail DNA-binding domain (DBD) (amino acids 1–229), Tbx6, the Tbx6 DBD (amino acids 1–240), the repressor domain of the Drosophila Engrailed protein [amino acids 2–299 (30)], or the activation domain of VP16 (amino acids 19–101) were cloned into the CS2+ and CS2+MT plasmids (31). Proteins were expressed ectopically after injection of in vitro-generated 5′-capped mRNA (mMESSAGE mMACHINE kit, Ambion, Austin, TX) along with fluorescent lineage tracer dye into one- to two-cell embryos. Normally cleaving embryos with widespread dye were selected for analysis. Myc-epitope-tagged protein was detected with the 9E10 antibody (Santa Cruz Biotechnology). To measure gene expression in animal caps, caps were removed at 3.5 h from injected embryos and incubated for 5 h at 28°C (21), and RNA expression was detected by RT-PCR and quantified (see Supporting Materials and Methods). Relative MyoD expression was determined as the ratio of MyoD PCR product to cytokeratin8 PCR product for each experimental condition.
Electrophoretic Mobility-Shift Assays. T-sites were synthesized, cloned into the SmaI site of pBluescript, purified such that all oligomers were ≈55 bp, and radiolabeled with [32P]dATP by using a modified Klenow fragment of DNA polymerase (Stratagene). Binding conditions are provided as Supporting Materials and Methods. Binding reaction products were resolved by gel electrophoresis and quantified after scanning of autoradiograms. Free DNA signal was used to determine fraction oligo bound. Curves represent best fits to the equation fraction oligo shifted = 1/[1 + (Kd/[P])]. R values for the binding curves are: Tbx6-myc-1/2T-site, 0.985; Tbx6-myc-palT-site, 0.975; No Tail-1/2T-site, 0.986; and No Tail-palT-site, 0.970.
Transactivation Assays. 293 cells were transfected by using Lipofectamine Plus reagent (Invitrogen). T-box transcription factors were expressed from CS2+ expression plasmids. Luciferase reporter plasmids contained three T-sites 55 bp upstream of the promoter of –36 PRL-luc (32), a pGL3-derived plasmid (Promega) containing 73 bp of the rat prolactin promoter TATA box region. Plasmid expressing Renilla luciferase constitutively was used as an internal reference to standardize transfection efficiency. Experiments were performed in triplicate and repeated at least twice.
Results
Region-Specific Functions of ntl in the Mesoderm. ntl, spt, and tbx6 are expressed exclusively in the developing zebrafish mesoderm (20, 21, 23). Overlapping expression of the three genes demarcates distinct regions of mesoderm identities (Fig. 1 A). Before gastrulation, at the time of mesoderm specification, the combined expression marks two domains: dorsal mesoderm, where ntl and spt are expressed without tbx6, and ventrolateral mesoderm, where all three genes are present. During gastrulation, the expression patterns of the genes are refined to generate four domains: anterior axial mesoderm, or prechordal plate, expresses only spt; posterior axial mesoderm, or presumptive notochord, expresses only ntl; nonaxial mesoderm generated by involution of the ventrolateral mesoderm expresses spt and tbx6 together; and noninvoluted ventrolateral mesoderm maintains expression of all three genes. We hypothesized that individual T-box genes have distinct functions in different portions of their expression domains and that the function of each T-box gene is defined in part by the combination of T-box genes with which it is coexpressed.
To define cellular regions of ntl function, we identified genes whose expression depends on ntl by hybridizing a mesendoderm-specific target cDNA library with a probe enriched for sequences expressed in WT, but not ntl mutant, gastrulae (see Materials and Methods). Two classes of genes were recovered (Fig. 1B). One class, exemplified by ntl-dependent gene 5 (ntd5), was expressed solely in the presumptive notochord. A second class, exemplified by mesogenin and wnt8 (33), was expressed in the noninvoluted ventrolateral mesoderm. Loss of expression of these genes was not secondary to loss of mesoderm tissue: other markers of axial and ventrolateral mesoderm continued to be expressed through midgastrula stages (data not shown and refs. 21, 34, and 35). Furthermore, mesogenin appears to be a proximal target of ntl, because its expression is detectable within 30 min of the onset of ntl transcription in 30% epiboly WT embryos and fails to be initiated in ntl mutants (Fig. 1C). Regardless of the mechanism by which the ntl-dependent genes are regulated, the genes serve as markers of cellular phenotype regulated by ntl. These results provide direct evidence that ntl regulates gene expression in the ventrolateral, as well as the dorsal, mesoderm and indicate that ntl has distinct region-specific functions in the mesoderm.
Combinatorial Interactions Between ntl and spt. mesogenin and wnt8 are transcribed only in the portion of the ntl expression domain where spt is also present, raising the possibility that spt modifies ntl function in the ventrolateral mesoderm. Analysis of spt mutant embryos indicated mesogenin is fully dependent on spt function (Fig. 1D), and, thus, both ntl and spt are required for initiating expression of mesogenin. Because mesogenin is not transcribed where each T-box is expressed alone, the regulation of mesogenin is a previously uncharacterized function that ntl and spt acquire when expressed together. spt is not the only interacting gene responsible for directing the ventral mesoderm activities of ntl, because wnt8 is not regulated by spt (Fig. 1D).
ntl and spt Contribute Additive Functions to Mesoderm Development and Gene Expression. Null mutations of ntl or spt are fully recessive mutations that primarily affect different regions of the mesoderm (11, 20, 24, 29, 36). Whereas ntl mutants lack notochord and posterior mesoderm, spt mutants produce notochord and generate posterior mesoderm in which both somites and notochord are formed in the tail. Conversely, spt, but not ntl, is required for formation of trunk somites. In spt mutants, somitic precursors fail to migrate properly and accumulate as undifferentiated mesoderm in the tail. The two T-box genes seem to contribute in a partially redundant manner to some aspects of mesoderm development, because double mutants exhibit a more severe phenotype than the simple addition of the defects present in ntl and spt mutants (25).
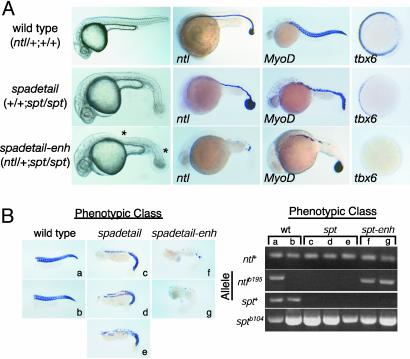
To identify pathways regulated in concert by both genes, we tested whether heterozygosity at one locus would enhance the mutant phenotype associated with loss of the other gene. Among progeny of matings between doubly heterozygous individuals, a previously uncharacterized phenotypic class appeared in addition to the expected WT, ntl, spt, and ntl; spt phenotypic classes (Table 1). The previously uncharacterized phenotypic class, here called spadetail-enhanced (spadetail-enh), was present in numbers consistent with the interpretation that it represented embryos homozygous for the spt mutation and heterozygous for the ntl mutation (Table 1). spadetail-enh embryos exhibited the surfeit of tail tissue characteristic of spt mutants, but they were distinguishable from the canonical spt phenotype in that they displayed a complete loss of somites in the tail and loss of axial tissue in the trunk (Fig. 2A). Simultaneous phenotypic and genotypic analyses performed on individual embryos demonstrated that the ntl mutation acted as a fully penetrant dominant enhancer of the spt phenotype (Fig. 2B).
Table 1. Gene interactions between ntl and spt.
|
Phenotypic class, %
|
|||||
|---|---|---|---|---|---|
| WT | ntl | spt | spadetail-enh | ntl; spt | |
| ntl/+; spt/+ × ntl/+; spt/+ | |||||
| Expected | 56.25 | 18.75 | 18.75 | 6.25 | |
| Observed (n = 842) | 56.3 | 18.3 | 6.4 | 12.7 | 6.3 |
| ntl/+; spt/+ × +/+; spt/+ | |||||
| Expected | 75 | 25 | |||
| Observed (n = 116) | 77.5 | 12.9 | 9.5 | ||
| ntl/+; spt/+ × ntl/+; +/+ | |||||
| Expected | 75 | 25 | |||
| Observed (n = 57) | 73.6 | 26.3 | |||
Progeny of indicated crosses were scored for morphology at day 1. “Expected” indicates percentage in each class if there is no interaction.
Fig. 2.
ntl and spt make additive contributions to dorsal and ventrolateral mesoderm development. (A) The recessive ntl mutation acts dominantly to enhance the spt mutant phenotype. Enhancement is evident by morphology at day 1 (Left); ntl expression at 20 h; MyoD expression at day 1; or tbx6 expression at 6 h (60% epiboly; animal pole views). spadetail-enh embryos lack somites in the tail and have a deficit of axial mesoderm (indicated by *). Phenotypic class (and genotype in parentheses) of embryos are indicated on the left. (B) The ntl/+; spt/spt genotype is responsible for the spadetail-enh (spt-enh) phenotype. Individual embryos (a–g) produced from ntl/+; spt/+ × spt/+ matings were collected at day 1 and were analyzed phenotypically for MyoD expression and genotypically by using an allele-specific PCR assay. WT embryos (a and b) harbored at least one WT allele of ntl and spt. All phenotypically spt embryos (c, d, and e) carried only mutant alleles of spt and WT alleles of ntl. All spadetail-enh embryos (f and g) carried only mutant alleles of spt and both WT and mutant alleles of ntl.
Mesoderm differentiation was analyzed in spadetail-enh embryos by using ntl expression as a marker of notochord development and MyoD expression as a marker of somite formation (23, 37). Although heterozygous ntl/+ embryos have a WT pattern of ntl and MyoD expression, the ntl mutation acts dominantly in a spt mutant background. spt mutants make a complete notochord marked by ntl expression, and they form MyoD-expressing somites in the tail and patches of MyoD-expressing cells in the trunk. In contrast, spadetail-enh mutants exhibit only discontinuous patches of ntl-expressing tissue in the midline and have an almost complete lack of MyoD-expressing tissue in the trunk and tail. Thus, loss of spt function sensitizes both the notochord and somite pathways to the level of ntl expression and indicates that the two T-box genes work together to promote development of both tissues.
To determine the relative quantitative contributions of ntl and spt to mesoderm development, we measured the effects of ntl and spt mutations on tbx6 expression (Fig. 2 A). Absence of spt function results in a significant but incomplete decrease in the expression of tbx6 in early gastrula embryos (20). Although loss of one or both copies of ntl in an otherwise WT genetic background does not have a detectable effect on early expression of tbx6 (21), loss of one copy of ntl in a spt mutant lowers tbx6 expression to undetectable levels. These results indicate that ntl and spt regulate a common set of genes in the mesoderm, contributing in an additive but unequal manner to their full expression.
Antagonism of Some ntl Functions by tbx6. ntl is required in the dorsal mesoderm to promote differentiation of the notochord and production of the MyoD-expressing adaxial cells that flank the presumptive notochord (Fig. 3 A and B) (11, 37). Given that the absence of ntl's dorsal-specific functions in the ventrolateral mesoderm correlates with the presence of tbx6, we asked whether tbx6 can antagonize the dorsal-specific functions of ntl (Fig. 3 A and B). Ectopic expression of a bona fide antagonist of No Tail, Ntl-EnR-myc, in which the DBD of No Tail was fused with the Engrailed repressor domain, suppressed posterior notochord and adaxial tissue development (10). Ectopic expression of Tbx6-myc or Tbx6 produced a similar range of phenotypes. Tbx6 acted locally to antagonize No Tail function as adaxial cells that lacked ectopic Tbx6-myc maintained normal MyoD expression (Fig. 3B). Expression of β-galactosidase or the Myc-epitope peptide had no effect on notochord development (data not shown). In sum, ectopic expression of tbx6 acts to antagonize ntl, resulting in a developmental syndrome expected from hypomorphic activity of ntl.
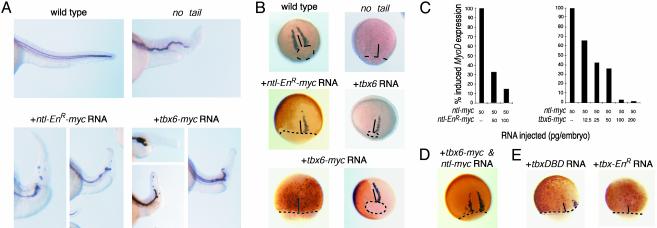
Fig. 3.
Tbx6 protein suppresses ntl-dependent functions. (A) Suppression of notochord development. collagen type 2A (col2A) expression in the floorplate and hypochord is used to outline the notochord in lateral views of the trunk/tail region of day-1 embryos. ntl mutant embryos lack differentiated notochord. Embryos that were injected at the one- to two-cell stage (+RNA) with 100 pg of ntl-EnR-myc or 100–200 pg of tbx6-myc RNA have reduced notochord development. The tail region is greatly reduced and either entirely lacks or contains isolated islands of col2A-expressing cells. The fraction of affected embryos depended on the amount of injected tbx6-myc RNA: 100 pg of tbx6-myc RNA yielded 37% (n = 46) with truncated notochord, 200 pg of tbx6-myc RNA yielded 57% (n = 46), and 400 pg of tbx6-myc RNA yielded 80% (n = 46). (B) Tbx6 suppresses MyoD expression in adaxial cells. WT MyoD expression in adaxial cells bordering the presumptive notochord of 80% epiboly embryos is absent in ntl mutant embryos. Adaxial MyoD expression (purple staining) is absent in embryos that express Ntl-EnR-myc or Tbx6-myc (brown staining) dorsally. Ectopic expression of WT Tbx6 also suppresses MyoD expression. Ninety-four percent (n = 66) of embryos injected with 100 pg of tbx6-myc RNA and 86% (n = 102) of embryos injected with 200 pg of tbx6 RNA exhibited suppression of MyoD expression. (C) Tbx6 blocks No Tail-dependent activation of MyoD transcription in animal caps. One- to two-cell embryos were injected with 50 pg of ntl-myc RNA along with either ntl-EnR-myc RNA or tbx6-myc RNA. Data indicate the amount of MyoD transcription relative to that caused by 50 pg of ntl-myc RNA. Replicate experiments yielded similar results. (D) Overexpression of No Tail counteracts the ability of Tbx6 to suppress MyoD expression in adaxial cells. Adaxial MyoD expression in an embryo injected at the one-cell stage with 100 pg of ntl-myc RNA and 200 pg of tbx6-myc RNA. Brown immunostaining indicates that ectopic proteins are expressed widely, including the dorsal mesoderm. (E) Transcriptionally inactive forms of Tbx6 can antagonize ntl. Ectopic expression of Tbx6DBD-myc or Tbx6-EnR-myc protein on the dorsal side of the embryo suppresses MyoD expression in adaxial cells. Suppression was observed in 18 of 20 embryos injected with 100 pg of tbx6DBD-myc RNA, and 42 of 46 embryos injected with 100 pg of tbx6-EnR-myc RNA. Dotted lines indicate blastoderm margins. Solid lines indicate dorsal midlines.
Ectopic expression of Brachyury proteins in ectodermal animal cap cells induces MyoD expression in a dose-dependent manner (38). It is likely that MyoD is regulated directly by No Tail in early zebrafish gastrulae, because No Tail protein is physically associated with chromatin containing the MyoD promoter at this stage (K.H., unpublished data). Coinjection of increasing amounts of ntl-EnR-myc RNA or tbx6-myc RNA along with a fixed amount of ntl-myc RNA inhibited No Tail-activated MyoD expression in a dose-dependent manner (Fig. 3C). Comparable amounts of tbx6-myc and ntl-EnR-myc RNA produced similar reductions in the amount of MyoD expression, indicating that the two proteins may have similar capacities to antagonize No Tail-dependent activation of gene expression.
Cell fate choices depend on the relative amount of ntl and tbx6 expression in individual embryonic cells. Whereas overexpression of either ntl or tbx6 perturbed MyoD expression in gastrula embryos in opposite ways, overexpression of both genes together could rescue a WT pattern of MyoD (Fig. 3D; Table 2). The finding that the suppression of MyoD expression in adaxial cells caused by ectopic tbx6 mRNA could be overcome by coexpression with ntl indicates that tbx6 generally does not suppress mesoderm formation; however, tbx6 and ntl can interact as competitive antagonists that govern mesoderm fate in the embryo. Furthermore, Tbx6 need not activate transcription to antagonize No Tail. Ectopic expression of Tbx6DBD or Tbx6DBD-EnR-myc, neither of which could promote expression of reporter genes that harbored Tbx6 binding sites (Fig. 4E and data not shown), suppressed the normal pattern of MyoD expression (Fig. 3E) and notochord development (data not shown).
Table 2. ntl and tbx6 act competitively to regulate MyoD expression in adaxial cells.
|
RNA injected, pg
|
MyoD expression pattern, %
|
|||
|---|---|---|---|---|
| ntl | tbx6 | Suppressed | WT | Ectopic |
| 50 | — | 0 | 7 (2/30) | 93 (28/30) |
| — | 200 | 90 (17/19) | 11 (2/19) | 0 |
| 50 | 200 | 48 (15/31) | 48 (15/31) | 3 (1/31) |
| 100 | 200 | 31 (10/32) | 56 (18/32) | 13 (4/32) |
Pattern of MyoD expression in embryos with ectopically expressed No Tail-myc, Tbx6-myc, or both proteins. Embryos were injected with RNA at the one-to two-cell stage and analyzed at 80% epiboly for MyoD expression by in situ hybridization. Only embryos with dorsal expression of ectopic proteins, detected immunohistochemically, were analyzed.
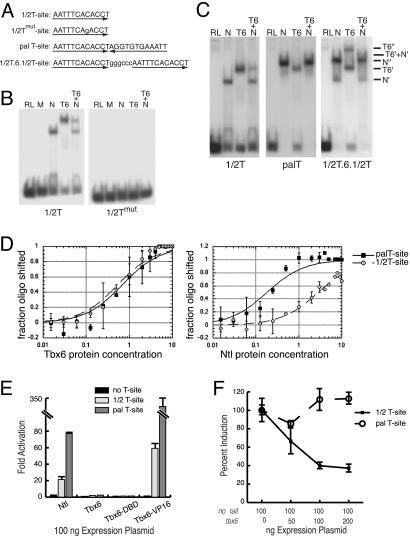
Fig. 4.
Tbx6 efficiently antagonizes ntl-dependent transcription at some T-sites. (A) Sequences of T-sites (1/2T-site motif is underlined) used. (B and C) Electrophoretic mobility-shift assay (EMSA) of T-box protein binding. (B) Protein–DNA complexes are formed on the WT 1/2T-site with No Tail (N) or Tbx6-myc (T6) but not with unrelated proteins (M, Myc-epitope peptide; RL, unprogrammed reticulocyte mixture). Protein–DNA complexes are not formed on the mutant 1/2T-site. (C) Tbx6-myc and No Tail each form single protein–DNA complexes (T6′ and N′) with the 1/2T-site. Tbx6-myc also binds the palT-site as a monomer, but No Tail binds this sequence as a homodimer (N′′). Incubation of the two types of target site with an identical mixture of the two proteins (T6+N) produces both No Tail–DNA and Tbx6–DNA complexes with the 1/2T-site (see also B), but it predominantly produces No Tail–DNA complexes with the palT-site. Protein binding to an oligomer with two separated 1/2T-sites (1/2T.6.1/2T) yields both one- and two-protein complexes, including mixed protein complexes (T6′+N′). (D) Tbx6 and No Tail have different relative affinities for 1/2T and palT sites. Increasing amounts of Tbx6-myc or No Tail protein were incubated with a constant amount of target T-sites. One unit of protein corresponds to 1 μl of programmed reticulocyte lysate. Binding reactions analyzed by EMSA are plotted as a function of the fraction of oligomer bound. Curves shown are fitted for bimolecular reactions. (E and F) Transcription activity of T-box proteins. (E) No Tail promotes expression of reporter genes that harbor either the 1/2T-site or palT-site. Neither Tbx6 nor Tbx6DBD is efficient at promoting T-site mediated expression. Tbx6DBD-VP16 promotes reporter gene expression in a T-site-dependent manner. (F) Cotransfection of tbx6 with ntl caused a substantial reduction in expression of luciferase from reporter genes that harbored 1/2T-sites but not from reporter genes that harbored palT-sites.
To explain how Tbx6 might selectively suppress dorsal-specific functions of No Tail, we hypothesized that Tbx6 can effectively antagonize No Tail at only some target sites. Other T-box proteins are known to have overlapping but nonidentical interactions with binding sites called T-sites (39). We measured the ability of No Tail and Tbx6 to bind to and promote transcription from different forms of the T-site (Fig. 4A) (40–42). No Tail and Tbx6-myc demonstrated similar sequence specificity: both bound the 1/2T-site as a monomer but failed to bind the 1/2Tmut-site, which harbored a single base substitution affecting a critical contact site for Brachyury proteins (41, 42) (Fig. 4 B and C). However, whereas Tbx6 bound both the 1/2T- and the palindromic palT-sites as a monomer, with identical affinity for the two kinds of sites, No Tail exhibited considerably enhanced affinity for the palT-site, on which it formed homodimers (Fig. 4 C and D). Consistent with these findings, incubation of 1/2T-site oligomer with a mixture of Tbx6 and No Tail yielded protein–DNA complexes containing both proteins (Fig. 4 B and C), but incubation of the palT-site oligomer with the same protein mixture yielded only No Tail homodimer protein–DNA complexes (Fig. 4C). Heterodimer protein–DNA complexes were not detected in these experiments, although mixed protein complexes could have been detected (T6′+N′ in Fig. 4C).
Tbx6 can compete effectively with No Tail to regulate T-site-dependent transcription. Whereas No Tail efficiently promoted expression of reporter genes that harbored either the palT- or 1/2T-site, Tbx6 failed to promote expression at these sites even though the Tbx6DBD is capable of recognizing T-sites in cells, indicated by the finding that Tbx6DBD-VP16 activated reporter gene expression (Fig. 4E). Coexpression of Tbx6 with No Tail at ratios ≤1:1 led to a substantial reduction of No Tail-dependent expression from the 1/2T-site reporter (Fig. 4F). In contrast, expression of the two proteins at similar ratios had no measurable effect on No Tail-dependent expression of the palT reporter gene. Thus, in cells expressing the two proteins, Tbx6 can effectively inhibit No Tail-dependent transcription at promoters that harbor some, but not all, kinds of T-sites.
Discussion
T-Box Genes Function as an Interacting Network. ntl, spt, and tbx6 function as interacting members of a network that directs mesoderm gene expression and developmental fate in the zebrafish. The interactions are not solely additive in that coexpression of ntl and spt brings about new functions and tbx6 can suppress some ntl functions. Because the interactions determine the function of a participating T-box gene, the developmental role of any one T-box gene may vary from cell to cell, depending on the expression of other family members. The existence of these interactions has two implications for the analysis of T-box gene function: (i) individual T-box genes may have multiple region-specific functions within a single developmental field; and (ii) loss-of-function of one T-box gene is likely to alter the function of a second T-box gene expressed in the same morphogenetic field.
As T-box genes are expressed in overlapping patterns in many developmental fields in both vertebrate and invertebrate embryos (5, 43), we propose that the formation of interacting networks is integral to the mode of action of T-box genes, as it is for Hox gene function (44). Overlapping expression of interacting T-box genes contributes to the subdivision of the zebrafish mesoderm into smaller regional elements with distinct gene expression patterns. The use of overlapping interactive genes is a resilient strategy for mediating positional information to effect regionalization of a large field. Even if embryo-to-embryo variability alters the exact breadth of a T-box expression domain, cells are never left without identity, and neighbor relationships among tissue precursors are preserved.
Interactions Between T-Box Genes. Our results show that pairs of T-box genes can interact combinatorially, additively, or antagonistically; however, the data do not reveal the mechanistic basis of these interactions. Combinatorial interactions are demonstrated by the finding that coexpression of ntl and spt is required for mesogenin transcription. Given the very short delay between the onset of ntl expression and the onset of mesogenin expression, we propose that No Tail and Spadetail interact directly, perhaps forming a heterodimeric transcription factor with previously uncharacterized specificity. Other T-box proteins form homodimers or heterodimers with partners of a different transcription factor family (13, 41, 42). Alternatively, the combined presence of No Tail and Spadetail may determine the cofactors accessible to each, thereby affecting the target specificity of each T-box protein (45).
Amacher et al. (25) initially demonstrated that ntl and spt contribute in an additive way to mesoderm development. Our finding that the loss of a single copy of ntl acts dominantly to enhance the spt phenotype indicates that both genes contribute to muscle and notochord development. Furthermore, the studies of tbx6 regulation indicate the two T-box genes have unequal and additive effects on expression of some downstream genes, an interaction observed between some pairs of paralogous Hox genes (46).
Finally, Tbx6 can act as a competitive inhibitor of some No Tail functions. Experiments in embryos and tissue culture indicate that cells can measure the relative abundance of the two proteins. Tbx6 need not be the only factor that suppresses ntl function ventrally, as our preliminary experiments indicate that embryos treated with tbx6 antisense morpholino oligonucleotides exhibit only mild axial defects. Our studies show that differences among promoter binding sites may explain how Tbx6 can competitively inhibit expression of some, but not all, target genes regulated by No Tail. We have used the 1/2T- and palT-sites to model potential interactions between Tbx6 and No Tail, realizing that the promoter sequences mediating No Tail transcription in vivo are likely to be more complex. Both the sequence of T-sites (39, 47) and the presence of additional sequences that recruit cofactors (13, 14) might affect the strength of No Tail binding to target promoters and thus affect the relative affinity of Tbx6 and No Tail for targets.
Gene Families and Gene Networks. Gene duplication events are thought to lead to diversification of gene functions and thus to contribute to biological diversity (48–50). One widely recognized mechanism for increasing the roles of an ancestral gene is through diversification of the biochemical functions and/or cellular expression patterns of individual homologous family members (51–53). Here we illustrate how interactions between homologues effectively increase diversity of gene function. The existence of interacting networks of Hox and T-box genes suggests that a recurrent mechanism for increasing diversity of gene function may involve combinatorial interactions among members of a transcription factor family.
Supplementary Material
Acknowledgments
We thank G. Walter and S. Johnson for excellent technical support. S. Amacher and B. Draper graciously shared unpublished information. This work was supported by National Institutes of Health Grant HD37572 (to D.J.G.) and by predoctoral fellowships from the Huntsman Cancer Institute (to L.M.G.) and the National Institutes of Health (to B.H. and L.M.G.).
Abbreviation: DBD, DNA binding domain.
Data deposition: The sequences reported in this article have been deposited in the GenBank database [accession nos. AY150226 (mesogenin) and AY150227 (ntd5)].
References
- 1.Jacobs, J. J., Keblusek, P., Robanus-Maandag, E., Kristel, P., Lingbeek, M., Nederlof, P. M., van Welsem, T., van de Vijver, M. J., Koh, E. Y., Daley, G. Q. & van Lohuizen, M. (2000) Nat. Genet. 26, 291–299. [DOI] [PubMed] [Google Scholar]
- 2.Papaioannou, V. E. (2001) Int. Rev. Cytol. 207, 1–70. [DOI] [PubMed] [Google Scholar]
- 3.Smith, J. (1999) Trends Genet. 15, 154–158. [DOI] [PubMed] [Google Scholar]
- 4.Ruvinsky, I. & Gibson-Brown, J. J. (2000) Development (Cambridge, U.K.) 127, 5233–5244. [DOI] [PubMed] [Google Scholar]
- 5.Wilson, V. & Conlon, F. L. (2002) Genome Biol. 3, reviews3008.1–3008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koshiba-Takeuchi, K., Takeuchi, J. K., Matsumoto, K., Momose, T., Uno, K., Hoepker, V., Ogura, K., Takahashi, N., Nakamura, H., Yasuda, K. & Ogura, T. (2000) Science 287, 134–137. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, D. L., Garvey, N., Hancock, S., Alexiou, M., Agulnik, S. I., Gibson-Brown, J. J., Cebra-Thomas, J., Bollag, R. J., Silver, L. M. & Papaioannou, V. E. (1996) Dev. Dyn. 206, 379–390. [DOI] [PubMed] [Google Scholar]
- 8.Bamshad, M., Lin, R. C., Law, D. J., Watkins, W. C., Krakowiak, P. A., Moore, M. E., Franceschini, P., Lala, R., Holmes, L. B., Gebuhr, T. C., et al. (1997) Nat. Genet. 16, 311–315. [DOI] [PubMed] [Google Scholar]
- 9.Chesley, P. (1935) J. Exp. Zool. 70, 429–459. [Google Scholar]
- 10.Conlon, F. L., Sedgwick, S. G., Weston, K. M. & Smith, J. C. (1996) Development (Cambridge, U.K.) 122, 2427–2435. [DOI] [PubMed] [Google Scholar]
- 11.Halpern, M. E., Ho, R. K., Walker, C. & Kimmel, C. B. (1993) Cell 75, 99–111. [PubMed] [Google Scholar]
- 12.Artinger, M., Blitz, I., Inoue, K., Tran, U. & Cho, K. W. (1997) Mech. Dev. 65, 187–196. [DOI] [PubMed] [Google Scholar]
- 13.Hiroi, Y., Kudoh, S., Monzen, K., Ikeda, Y., Yazaki, Y., Nagai, R. & Komuro, I. (2001) Nat. Genet. 28, 276–280. [DOI] [PubMed] [Google Scholar]
- 14.Lamolet, B., Pulichino, A. M., Lamonerie, T., Gauthier, Y., Brue, T., Enjalbert, A. & Drouin, J. (2001) Cell 104, 849–859. [DOI] [PubMed] [Google Scholar]
- 15.O'Reilly, M. A., Smith, J. C. & Cunliffe, V. (1995) Development (Cambridge, U.K.) 121, 1351–1359. [DOI] [PubMed] [Google Scholar]
- 16.Ruvinsky, I., Oates, A. C., Silver, L. M. & Ho, R. K. (2000) Dev. Genes Evol. 210, 82–91. [DOI] [PubMed] [Google Scholar]
- 17.Mione, M., Shanmugalingam, S., Kimelman, D. & Griffin, K. (2001) Mech. Dev. 100, 93–97. [DOI] [PubMed] [Google Scholar]
- 18.Dheen, T., Sleptsova-Friedrich, I., Xu, Y., Clark, M., Lehrach, H., Gong, Z. & Korzh, V. (1999) Development (Cambridge, U.K.) 126, 2703–2713. [DOI] [PubMed] [Google Scholar]
- 19.Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E. & Rubin, G. M. (2002) Genome Biol. 3, research0088.1–0088.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin, K. J., Amacher, S. L., Kimmel, C. B. & Kimelman, D. (1998) Development (Cambridge, U.K.) 125, 3379–3388. [DOI] [PubMed] [Google Scholar]
- 21.Hug, B., Walter, V. & Grunwald, D. J. (1997) Dev. Biol. 183, 61–73. [DOI] [PubMed] [Google Scholar]
- 22.Ruvinsky, I., Silver, L. M. & Ho, R. K. (1998) Dev. Genes Evol. 208, 94–99. [DOI] [PubMed] [Google Scholar]
- 23.Schulte-Merker, S., Ho, R. K., Herrmann, B. G. & Nusslein-Volhard, C. (1992) Development (Cambridge, U.K.) 116, 1021–1032. [DOI] [PubMed] [Google Scholar]
- 24.Schulte-Merker, S., van Eeden, F. J., Halpern, M. E., Kimmel, C. B. & Nusslein-Volhard, C. (1994) Development (Cambridge, U.K.) 120, 1009–1015. [DOI] [PubMed] [Google Scholar]
- 25.Amacher, S. L., Draper, B. W., Summers, B. R. & Kimmel, C. B. (2002) Development (Cambridge, U.K.) 129, 3311–3323. [DOI] [PubMed] [Google Scholar]
- 26.Patel, M. & Sive, H. L. (1996) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Wiley, New York).
- 27.Shimono, A. & Behringer, R. R. (2000) Methods Mol. Biol. 136, 333–344. [DOI] [PubMed] [Google Scholar]
- 28.Westerfield, M. (1993) The Zebrafish Book: Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) rerio (Univ. of Oregon Press, Eugene).
- 29.Kimmel, C. B., Kane, D. A., Walker, C., Warga, R. M. & Rothman, M. B. (1989) Nature 337, 358–362. [DOI] [PubMed] [Google Scholar]
- 30.Hollemann, T., Bellefroid, E. & Pieler, T. (1998) Development (Cambridge, U.K.) 125, 2425–2432. [DOI] [PubMed] [Google Scholar]
- 31.Turner, D. L. & Weintraub, H. (1994) Genes Dev. 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- 32.Thorburn, J., Xu, S. & Thorburn, A. (1997) EMBO J. 16, 1888–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lekven, A. C., Thorpe, C. J., Waxman, J. S. & Moon, R. T. (2001) Dev. Cell 1, 103–114. [DOI] [PubMed] [Google Scholar]
- 34.Melby, A. E., Kimelman, D. & Kimmel, C. B. (1997) Dev. Dyn. 209, 156–165. [DOI] [PubMed] [Google Scholar]
- 35.Talbot, W. S., Trevarrow, B., Halpern, M. E., Melby, A. E., Farr, G., Postlethwait, J. H., Jowett, T., Kimmel, C. B. & Kimelman, D. (1995) Nature 378, 150–157. [DOI] [PubMed] [Google Scholar]
- 36.Ho, R. K. & Kane, D. A. (1990) Nature 348, 728–730. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg, E. S., Allende, M. L., Kelly, C. S., Abdelhamid, A., Murakami, T., Andermann, P., Doerre, O. G., Grunwald, D. J. & Riggleman, B. (1996) Development (Cambridge, U.K.) 122, 271–280. [DOI] [PubMed] [Google Scholar]
- 38.Cunliffe, V. & Smith, J. C. (1992) Nature 358, 427–430. [DOI] [PubMed] [Google Scholar]
- 39.Conlon, F. L., Fairclough, L., Price, B. M., Casey, E. S. & Smith, J. C. (2001) Development (Cambridge, U.K.) 128, 3749–3758. [DOI] [PubMed] [Google Scholar]
- 40.Casey, E. S., O'Reilly, M. A., Conlon, F. L. & Smith, J. C. (1998) Development (Cambridge, U.K.) 125, 3887–3894. [DOI] [PubMed] [Google Scholar]
- 41.Kispert, A. & Herrmann, B. G. (1993) EMBO J. 12, 3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller, C. W. & Herrmann, B. G. (1997) Nature 389, 884–888. [DOI] [PubMed] [Google Scholar]
- 43.Papaioannou, V. E. & Silver, L. M. (1998) BioEssays 20, 9–19. [DOI] [PubMed] [Google Scholar]
- 44.Lewis, E. B. (1978) Nature 276, 565–570. [DOI] [PubMed] [Google Scholar]
- 45.Mann, R. S. & Morata, G. (2000) Annu. Rev. Cell Dev. Biol. 16, 243–271. [DOI] [PubMed] [Google Scholar]
- 46.Greer, J. M., Puetz, J., Thomas, K. R. & Capecchi, M. R. (2000) Nature 403, 661–665. [DOI] [PubMed] [Google Scholar]
- 47.Tada, M. & Smith, J. C. (2001) Dev. Growth Differ. 43, 1–11. [DOI] [PubMed] [Google Scholar]
- 48.Cooke, J., Nowak, M. A., Boerlijst, M. & Maynard-Smith, J. (1997) Trends Genet. 13, 360–364. [DOI] [PubMed] [Google Scholar]
- 49.Duboule, D. & Wilkins, A. S. (1998) Trends Genet. 14, 54–59. [DOI] [PubMed] [Google Scholar]
- 50.Ohno, S. (1970) Evolution by Gene Duplication (Springer, Heidelberg).
- 51.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klinghoffer, R. A., Mueting-Nelsen, P. F., Faerman, A., Shani, M. & Soriano, P. (2001) Mol. Cell 7, 343–354. [DOI] [PubMed] [Google Scholar]
- 53.Szebenyi, G. & Fallon, J. F. (1999) Int. Rev. Cytol. 185, 45–106. [DOI] [PubMed] [Google Scholar]
- 54.Bork, P., Downing, A. K., Kieffer, B. & Campbell, I. D. (1996) Q. Rev. Biophys. 29, 119–167. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.