Abstract
We demonstrate the use of a chromosomal walk (or “tiling path”) printed as DNA microarrays for mapping protein–DNA interactions across large regions of contiguous genomic DNA in Drosophila melanogaster. Microarrays were constructed with genomic DNA fragments 430–920 bp in length, covering 2.9 million base pairs of the Adh–cactus region of chromosome 2 and 85,000 base pairs of the 82F region of chromosome 3. We performed DNA localization mapping for the heterochromatin protein HP1 and for the sequence-specific GAGA transcription factor, producing a comprehensive, high-resolution map of in vivo protein–DNA interactions throughout these regions of the Drosophila genome.
The Drosophila model has served as a guidepost for working out the molecular genetics of gene expression regulation in a developmental context, and the availability of the complete Drosophila melanogaster genome sequence has presented a new challenge: to systematically decode the genome into regulatory networks that direct complex developmental processes. This decoding will be accelerated through the use of gene expression and protein–DNA interaction data to map transcriptional regulatory networks on a genome-wide scale (1), but the appropriate technologies must be developed for such mapping to be efficient and comprehensive. DNA microarrays have been used to study gene expression patterns genome-wide during developmental processes in Drosophila, Caenorhabditis elegans, and mouse (2–7). Methodologies to map protein–DNA interactions using cDNA microarrays have also been developed in Drosophila (8). However, the most extensive protein–DNA interaction mapping has been carried out in yeast, where DNA microarrays containing all intergenic regions of the genome have been used to systematically identify the binding sites of transcription factors (9–11). In these studies, chromatin immunoprecipitation (ChIP) was used to isolate protein–DNA complexes, and the resulting purified DNA was labeled and hybridized to the intergenic DNA microarrays. Recently, ChIP has been used with human DNA microarrays to identify binding sites of GATA-1 in the 75-kb sequence of the β-globin locus and binding sites of E2F in promoters of genes expressed during cell cycle entry (12–14). The results from yeast demonstrate that intergenic arrays can be extremely valuable for the study of transcriptional regulatory networks, and the results from human show that, in principle, the technology can be applied to study complex genetic loci.
Here we demonstrate the use of genomic DNA tiling path microarrays to map protein–DNA interactions at high resolution along large segments of genomic DNA from D. melanogaster. We used DNA microarrays tiled across two genomic regions: 2.9 Mbp of Adh–cactus region on chromosome 2 and 85 kb of 82F region on chromosome 3. These arrays allowed us to assay protein–DNA interactions in coding and noncoding genomic sequence that contains at least 220 genes (15–18). The arrays were composed of overlapping fragments with sizes of 850–920 bp each across the Adh–cactus region and 430–500 bp each across the 82F region. To map protein–DNA interactions, we used the DamID chromatin profiling technique (8, 19). This technique involves in vivo expression of a trace amount of a chromatin protein of interest fused to Escherichia coli DNA adenine methyltransferase (Dam). As a result, DNA in the target loci of the chromatin protein is preferentially methylated by the tethered Dam. Subsequently, methylated DNA fragments are purified, labeled with a fluorescent dye, and hybridized to a microarray. To correct for unspecific binding of Dam and local differences in DNA accessibility, methylated DNA fragments of control cells transfected with Dam alone are labeled with a different fluorescent dye and cohybridized. The obtained ratio of fluorescent dyes reflects the extent of protein binding to the probed DNA sequence (8).
We performed high-resolution binding site mapping of a sequence-specific DNA-binding factor, GAF (20), and the heterochromatin protein HP1 (21). Binding profiles of both proteins have previously been determined in a study using cDNA arrays containing ≈300 cDNA fragments (8). Only binding sites in the immediate vicinity of transcribed regions can be detected by using cDNA arrays. However, localization of chromatin-associated proteins is often distant from transcribed regions. Here we demonstrate that genomic tiling path arrays can be used for comprehensive and high-resolution mapping of chromatin-associated proteins in the Drosophila genome. We discovered dozens of new GAF-binding sites in the 3 Mb of genomic DNA surveyed, and we were able to initially map these sites to a few hundred base pairs in most cases. The use of computational sequence analysis methods allowed many sites of chromosomal association to be pinpointed to within several nucleotides. Furthermore, ChIP analyses verified several randomly selected sites identified through this analysis, providing validation by using an independent method for direct mapping of GAF–DNA interactions. In addition to the high-resolution mapping of GAF protein, we found new patterns of HP1 association with transposable elements throughout this region of the genome.
Materials and Methods
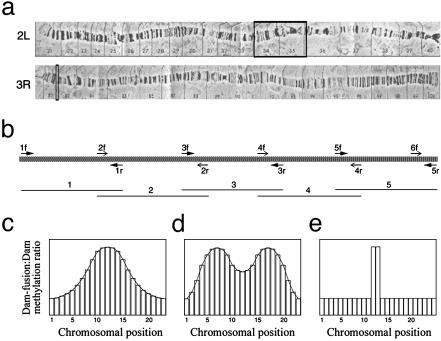
DNA Array Construction. To create the initial set of test arrays reported in this study, primers were designed to amplify 3,648 fragments representing the 2.9-Mb Adh–cactus region (on chromosome 2L) and 192 fragments covering 85 kb of the 82F region (on chromosome 3R) (Fig. 1 a and b). The primer3 program was used (http://www-genome.wi.mit.edu/genome_software/other/primer3.html). For the Adh–cactus region, each fragment was between 850 and 920 bp in size with a 25- to 200-bp overlap between neighboring fragments. Overlapping DNA segments were extracted from the Adh–cactus region sequence contig by using a perl program. PCR primer lengths varied between 21 and 23 nucleotides, and the melting temperature was set to 60°C in the PRIMER 3software. For the 82F region, each fragment is 430–500 bp in size with a 20- to 100-bp overlap between neighboring fragments. Fragments were amplified from whole genomic DNA of D. melanogaster y; cn bw sp strain in a 96-well format. The reaction mixture to produce each amplicon contained 50 ng of Drosophila genomic DNA, 1 μM forward primer, 1 μM reverse primer, 1× Taq Gold Buffer (Applied Biosystems), 2 mM MgCl2, 0.25 mM dATP, 0.25 mM dTTP, 0.25 mM dCTP, 0.25 mM dGTP, and 0.02 unit AmpliTaq Gold enzyme (Applied Biosystems). The following PCR protocol was used: an initial denaturation step for 9 min at 95°C followed by 40 cycles of denaturation for 30 sec at 95°C, annealing for 30 sec at 60°C, and elongation for 60 sec at 72°C. PCR products were run on agarose gels to confirm amplification success. See Supporting Methods and Table 3, which are published as supporting information on the PNAS web site, www.pnas.org, for a complete description of PCR results. The arrays used in this study were built based on the initial Adh–cactus and 82F sequences that were available when we began the project (16), before the version 1 release of the Drosophila genome sequence (22). PCR products were spotted on polylysine coated glass slides by using an Omnigrid arrayer.
Fig. 1.
Tiling path construction for Adh–cactus and 82F chromosomal regions, and theoretical distributions for tiling path microarray results for a protein–DNA interaction experiment. (a) Polytene chromosomes 2L and 3R, with regions used for microarray construction indicated by square black boxes. (b) Design of tiling path. The thick stippled line represents genomic template DNA, arrows indicate forward (f) and reverse (r) primers, and thin lines below represent amplified overlapping DNA fragments printed on tiling path arrays. (c) A monotonic binding profile expected if binding occurs in DNA sequences at a single chromosomal site. (d) A multimodal binding profile expected if binding occurs in DNA sequences at multiple chromosomal sites in close proximity to one another. (e) Binding profile expected in the case of cross-hybridization to DNA elsewhere in the genome that is either near a binding site(s) or is itself bound.
DamID. The DamID procedure was performed in Drosophila Kc167 cells as described (8, 19), except that methylated DNA fragments were not obtained by DpnI digestion and subsequent sucrose gradient centrifugation, but selectively amplified by PCR.
Genomic DNA isolated from Kc167 cells transfected with Dam or a Dam-fusion protein was isolated as described (8). In brief, ≈108 cells from one 10-cm plate were collected, pelleted, and resuspended in 1 ml of ice-cold T10E10 (10 mM Tris·HCl,pH7.5/10 mM EDTA). One milliliter of freshly prepared TENSK buffer [100 mM NaCl/0.5% SDS/200 μl of Proteinase K (Roche Molecular Biochemicals) in T10E10] was added and mixed by inversion. After incubation for 2 h at 55°C, 2.0 ml of buffer-saturated phenol/chloroform/isoamylalcohol was added, followed by mixing by inversion and spinning for 10 min at 3.5 krpm. The water phase was transferred to 2.0 ml of isopropanol and 0.2 ml of 3 M sodium acetate (pH 5.2), and mixed; the DNA was recovered by spooling on a yellow tip, completely dissolved in 0.3 ml of T10E10 with 2 μg of DNase-free RNase (Roche Molecular Biochemicals), and incubated at 37°C for at least 1 h. Next, 0.3 ml of TENSK was added, followed by incubation for 30 min at 55°C. A second phenolchloroform extraction followed, after which the water phase was transferred to 0.6 ml of isopropanol and 60 μl of 3 M sodium acetate (pH 5.2). The solution was mixed by inversion and the DNA precipitate was recovered, rinsed in 70% ethanol, and dissolved in 50 μl of T10E10 by incubation at 37°C for several hours.
For selective PCR amplication of methylated DNA fragments, 40 μg of the isolated genomic DNA was digested for 16 h at 37°C with 40 units of DpnI (New England Biolabs) in the presence of 12.5 ng of DNase-free RNase A (Roche Molecular Biochemicals) in a total volume of 50 μl of buffer 4 (New England Biolabs). After inactivation of DpnI at 80°C for 20 min, 4 μg of the DpnI-digested genomic DNA was ligated to 40 pmol of a double-stranded unphosphorylated adaptor (top strand: 5′-CTAATACGACTCACTATAGGGCAGCGTGGTCGCGGCCGAGGA-3′, bottom strand: 5′-TCCTCGGCCG-3′) for 2 h at 16°C with 5 units of T4-Ligase (Roche Molecular Biochemicals) in a total volume of 20 μl of ligation buffer. To prevent amplification of DNA fragments containing unmethylated GATCs, 1 μg of the adaptor-ligated DNA was cut with 2 units of DpnII (New England Biolabs) for1hat37°C in a total volume of 20 μl of DpnII buffer. Next, amplification was performed by using 0.5 μg of DpnII-cut DNA, 1 μl of Advantage cDNA PCR polymerase mix (CLONTECH), 10 nmol of each dATP, dCTP, dGTP, and dTTP, and 62.5 pmol of primer (5′-GGTCGCGGCCGAGGATC-3′) in 50 μl total volume of Advantage PCR buffer, under the following cycling conditions: activation of the polymerase and nick translation for 10 min at 68°C, followed by one cycle of 1 min at 94°C, 5 min at 65°C and 15 min at 68°C; 3 cycles of 1 min at 94°C, 1 min at 65°C and 10 min at 68°C; and 14 cycles of 1 min at 94°C, 1 min at 65°C and 2 min at 68°C. The PCR products were purified by using the QIAquick PCR purification kit (Qiagen) and labeled with Cy3 or Cy5 as described (8).
Finally, labeled experimental (Dam–protein fusion) and reference (Dam) DNA samples were mixed and hybridized to microarrays in 3× SSC (450 mM sodium chloride/45 mM sodium citrate, pH 7.0) supplemented with 0.22% SDS, 20 μg of poly(dA–dT), 100 μg of yeast tRNA, and 25 μg of unlabeled DpnI-digested plasmid encoding the fusion protein used for transfection. After a 15-min incubation at 42°C, hybridization was performed at 63°C for 16 h, followed by a sequential washing at room temperature in 1.14× SSC plus 0.0285% SDS, 1.14× SSC, 0.228× SSC, and 0.057× SSC. Immediately after washing, arrays were spun dry at 1,000 × g for 5 min in a table-top centrifuge.
Motif Analysis. Consensus binding motifs were inferred from the complete set of binding log-ratios by using three different algorithms: the motif-based linear regression method REDUCE, which exploits the correlation between the occurrence of sequence motifs near exons of genes and the expression of those exons (23), the method proposed by Keles et al. (24), which is conceptually similar to REDUCE, but uses a different motif selection scheme, and the MDscan method, which uses a modified Gibbs sampling strategy to search for common patterns in the segments with high binding ratios (25).
ChIP of GAF Binding Fragments. ChIP was performed by using formaldehyde cross-linking, and by using anti-GAF antibody with chromatin extracts of both Kc cells and Drosophila embryos as described (26). Primers were designed to amplify five GAF-binding fragments identified with DamID and seven fragments that did not show any GAF binding in the DamID experiments. However, all fragments with GAGAG sites were selected, regardless of whether they were positive for GAF binding in the DamID assay. PCR products were run on an agarose gel (1.4%) and transferred to a nylon membrane for Southern blot analysis. Blots were hybridized either with a probe made from a mock immunoprecipitation (IP) sample or with a probe from GAF ChIP. Hybridized membrane was then subjected to a 24-h exposure in a phosphorimager cassette, and results were quantified as presented in Table 2. Tested fragments were scored as “ChIP positive” if the ratio of mock IP to GAF IP was ≥2.0 in ChIP with embryo chromatin extracts. Our positive controls (Fab7, Mcp, and bxd from the Bithorax complex regulatory region) were enriched, although the enrichment value in Kc167 cells is not as high as usually found in embryos (26). We therefore lowered the criteria for enrichment in GAF ChIP for Kc cells to 1.5-fold. ChIP experiments were performed in duplicate.
Table 2. Independent verification of GAF binding sites using ChIP.
| Dam positive; |
Enrichment in ChIP
|
|||||
|---|---|---|---|---|---|---|
| ChIP positive (Kc167); ChIP positive (embryo) | Position start | Position stop | No. of GAGAG motifs | GAF binding signal from DamID | Kc cells | Embryo |
| +;+;+ | 2L:14119949 | 2L:14121575 | 8 | 4.3 | 1.5 | 2.7 |
| +;+;+ | 2L:14879240 | 2L:14880923 | 17 | 4.8 | 1.6 | 4.4 |
| +;+;+ | 2L:15147642 | 2L:15149729 | 5 | 3.2 | 5.2 | 2.0 |
| +;+;+ | 2L:16099455 | 2L:16101408 | 30 | 5.3 | 14.9 | 3.9 |
| +;+;+ | 2L:16130105 | 2L:16131818 | 14 | 2.6 | 1.5 | 4.3 |
| -;-;- | 2L:14003030 | 2L:14005454 | 11 | 1.0 | 0.82 | 0.9 |
| -;-;- | 2L:14213274 | 2L:14215052 | 9 | 1.0 | 0.93 | 1.2 |
| -;-;(+/-) | 2L:14638083 | 2L:14639717 | 5 | 0.95 | 0.53 | 1.4 |
| -;-;+ | 2L:15375422 | 2L:15377025 | 12 | 0.95 | 1.0 | 3.2 |
| -;-;- | 2L:14348388 | 2L:14350038 | 3 | 0.95 | 0.74 | 1.2 |
| -;-;- | 2L:14913451 | 2L:14915108 | 2 | 0.93 | 0.54 | 0.9 |
| -;-;(+/-) | 2L:15275807 | 2L:15277498 | 6 | 0.95 | 0.26 | 1.5 |
Results from GAF DamID in Kc167 cells, GAF ChIP in Kc167 cell extracts, and GAF ChIP in embryo extracts.
Results
Binding Site Profiling Using Tiling Path DNA Microarrays. We designed DNA microarrays containing contiguous sequences from two different chromosomes. A total of 2.9 Mb of the chromosomal sequences were from the well-studied Adh–cactus region of chromosome 2L (Fig. 1a), which has been the focus of intensive genetic analyses and annotation efforts (16, 17). The remainder of the sequences was from 85 kb of the 82F region of chromosome 3R (Fig. 1a) near the L82 gene, which has a complex structure that has been carefully mapped (18). These arrays contained overlapping genomic DNA fragments (Fig. 1b), allowing comprehensive mapping of contacts between chromatin proteins and chromosomes.
To begin, we consider the characteristic patterns of microarray data expected when these tiling path microarrays are used with the Dam ID technique, which compares genomic methylation patterns in the presence of a Dam-fusion protein to background methylation from expression of Dam alone (19). In the simplest case, the association of a Dam-fusion protein would occur at a single point along the chromosome. At that point, the signal ratio from a DamID experiment (Dam-fusion protein/Dam alone) would be high. One expects that targeted methylation levels of DNA in either direction from that point will progressively decrease proportional to distance, with a concomitant decease in the signal ratio. The quantitative result from the microarray experiment will accordingly be represented as a curve with its maximum over the point (Fig. 1c). This curve would be expected to be monotonic if GATC sequences targeted by Dam are randomly distributed around the focal point of the DNA–protein interaction. Multiple binding sites in a region may produce bimodal or other complex distributions (Fig. 1d). It is also important to consider that for protein–DNA binding assays using microarrays, repetitive DNA associated with the assayed protein in one part of the genome may cross-hybridize with DNA from another region printed on the microarray. At the place where sequence identity is lost between the DNA tiling path elements and the cross-hybridizing sequences from a remote genomic location(s), signal intensity would be expected to drop off sharply and no curve is expected outside the cross-hybridizing sequences, whereas inside the cross-hybridizing sequences, one expects either no curve at all (Fig. 1e) or a curve that reflects real binding to the remote sequences. We determined the actual distributions of signal intensities by using two Dam-fusion proteins, GAF-Dam and Dam-HP1. We refer to these data as GAF or HP1 binding profiles.
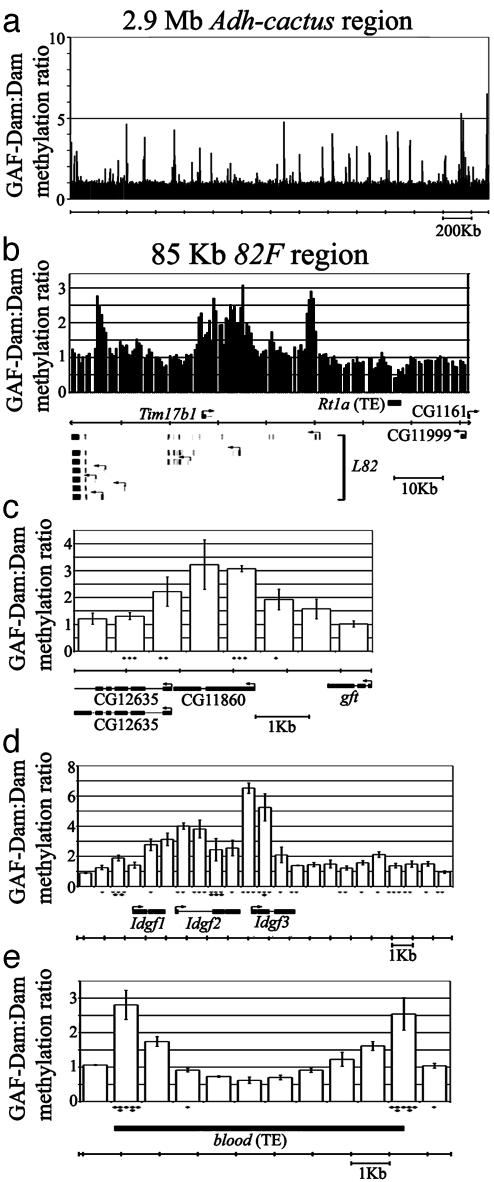
GAF Binding Profiles. We used a local linear weighted regression method that is more sensitive than a standard t test to identify 169 genomic DNA fragments with significantly elevated GAF-Dam/Dam methylation ratios (see Supporting Methods and ref. 27). These fragments congregated into 46 chromosomal areas (groups of adjacent fragments) (Table 4, which is published as supporting information on the PNAS web site). Because the affinity of GAF binding may be reflected in the microarray measurements, we imposed an additional criterion of a threshold cut-off to divide the 169 significant fragments into a set that shows a >2-fold differential (“high binding ratio”) and a set that does not (“low binding ratio”) (all ratios >2 also were significant by using a standard t test with P < 0.025; see Supporting Methods). We found 54 fragments in 23 areas in the 2.9-Mb Adh–cactus region, and 10 fragments in three areas in the 82F region (26 areas total) that showed high GAF binding ratios (Table 4, Fig. 2). Most of the 26 areas display a GAF-binding profile consistent with direct associations between chromosomal DNA fragments and GAF. Among the 23 areas with high GAF binding ratios in the Adh–cactus region, 15 display monotonic binding profiles (Fig. 2c). Of the remaining eight areas, four appear to contain multiple GAF binding sites because they display either bimodal (three areas) or multipeak profiles (one area) (Fig. 2d); the other four exhibit profiles that appear as half of a monotonic curve with signal precipitously dropping off. In two of these latter four, we observed two half monotonic binding profiles arranged as near mirror images of one another (Fig. 2e). We interpret these profiles as either direct binding of GAF to the ends of transposons elsewhere in the genome, or GAF binding nearby the ends of transposons elsewhere in the genome, because the sharp decrease of binding outside the ends of transposon is what one would expect in the case of cross-hybridization of the entire transposon. The average number of GATC sites in these cases does not differ on either side of the GAF binding site, so this mirror image “half-site” profile cannot be due to scarcity of methylation targets on only one side of binding sites. As in the Adh–cactus region, the binding profiles in the 82F region also fall into three categories: half monotonic curve, monotonic curve, or multipeak patterns (Fig. 2b). The 20 areas of low GAF binding displayed a similar range of binding profiles as the 26 areas of high GAF binding (Table 4).
Fig. 2.
GAF binding profiles. (a) GAF binding profiles across the entire 2.9-Mb Adh–cactus region. (b) GAF binding profiles in the 82F region. (c) Example of monotonic GAF binding profile in the CG11860 gene. Peak signal is seen in the first exon and intron of CG11860. Three GAGAG/CTCTC consensus binding sites are in the 5′ UTR of CG11860. (d) Example of a multimodal GAF binding profile, showing multiple binding sites of GAF throughout the Imaginal disk growth factor (Idgf) gene region. (e) Example of a pattern likely due to cross-hybridization. Two half monotonic curves are arranged as a near mirror image of one another with sharply decreasing signal at the ends of a blood transposable element at 2L:15329890 to 15339590. Black diamonds, GAGAG/CTCTC consensus binding sites; solid bars, exons; lines connecting solid bars, introns; gray bars, transposons. Standard errors are shown.
Most of the areas in the Adh–cactus region associated with high GAF binding ratios are within the vicinity of sequences that contain annotated genes, with 15 that are <3 kb from the nearest start codon, and 18 that are within 10 kb of the nearest start codon (Table 4). Although high GAF binding ratios were commonly associated with putative regulatory sequences 5′ or 3′ of transcription units (nine instances), 5 of the 23 GAF binding sites are contained within 5′ or 3′ UTRs and 9 occurred within introns (Table 4). None occurred within coding regions. There was a single instance where no annotation features were identified in a 10-kb vicinity of GAF binding (the closest gene was >25 kb away). This may be caused by regulatory sequences acting from a distance, it may be caused by functionally irrelevant GAF binding, or it may be caused by the existence of genes not yet annotated. Considering the Adh–cactus and 82F regions as representative samples from the genome, and extrapolating from these results, we expect that there are likely >1,000 sites with high GAF binding genome-wide, and >750 more sites with low but detectable GAF binding by using the DamID assay.
GAF Binding Motif Analyses. In vitro, GAF binds to the sequence GAGAG (28). By using three independent motif-finding methods that all use genomic sequence data and GAF binding data from the tiling path microarrays, we were able to successfully identify the correct consensus GAGAG binding motif for GAF. Table 1 shows the results of analyses based on Regulatory Element Detection Using Correlation of Expression (REDUCE) (23), the Keles et al. method (24), and the Motif Discovery scan (MDscan) (25). The first two methods are similar; they were developed to perform motif selection based on a least-squares fit of a linear predictive model for expression log-ratios, but can be used without modification to analyze binding log-ratios. The MDscan method compares the probability that one motif occurs in the top ranking sequences based on binding ratios and its occurrence in the background sequences. The success of all three of these algorithms in identifying the correct binding site indicates that DNA tiling path microarrays combined with DamID mapping of binding sites will provide a robust source of data for cis-regulatory motif-finding algorithms.
Table 1. Results of using motif-finding algorithms with GAF binding site microarray data.
|
REDUCE
|
Keles, et al.
|
MDscan
|
|||
|---|---|---|---|---|---|
| 5-mer | 6-mer | 5-mer | 6-mer | 5-mer | 6-mer |
| GAGAG (K1,M1) | AGAGAG (K1,M3-5,M7-9) | GAGAG (R1,M1) | AGAGAG (R1,M3-5,M7-9) | GAGAG (R1,K1) | CACACA |
| GAGCG (K3) | GAGAGA (K9,M6,M10) | CAGGA (R3) | AGAGCG (R3) | ATCAA | ACACAC |
| CAGGA (K2) | AGAGCG (K2) | GAGCG (R2) | TACATA (R5) | ACCAA | AGAGAG (R1,K1) |
| AGAGA (K10,M6) | GAGAGC | AAGGA (R6) | CAGCTG | AACAA | AGAGAG (R1,K1) |
| AGGAC | TACATA (K3) | ATGGC | CAGGAC (R8) | AGCAA | AGAGAG (R1,K1) |
REDUCE, Keles et al., and MDscan algorithms were used with the same data set. For each method, the top five ranked 5-mer and 6-mer motifs are shown. Rankings of these motifs based on the other two methods are indicated in parentheses. The REDUCE method is denoted by R, the method developed by Keles et al. is denoted by K, and the MDscan method is denoted by M. GAGAG was the top motif identified by each of the three methods.
Scanning of genome sequence revealed that GAGAG/CTCTC motifs were contained in almost all DNA fragments showing peak levels of signal in the 46 areas we identified, but also in 2,115 DNA fragments without appreciable binding signal. All of the areas with high levels of binding contained at least one GAGAG/CTCTC site in the DNA fragments that showed peak signal on the microarrays, allowing the precise coordinates of GAF binding to be predicted. The average number of such sites in DNA fragments with peak signal was 4.3, whereas the median was 3 sites. In the 20 areas with low levels of binding, often more than one adjacent fragment showed indistinguishable levels of peak signal. The average number of GAGAG/CTCTC sites in these DNA fragments was 2.2, whereas the median was 1 site. Thus, we find an overall correlation between signal strength and the number of potential binding sites for GAF. For one case, no GAGAG/CTCTC sites were identified even though the binding patterns observed were monotonic and in nonrepetitive DNA sequence. This case could be caused by weak binding site(s) that do not match the exact consensus, or perhaps this is a false positive.
Independent Verification of GAF Binding Sites Using ChIP. We verified several candidate GAF binding sites by using ChIP from both Kc167 cell chromatin extracts and in embryonic chromatin extracts (26). We tested five fragments shown by DamID to bind GAF and seven fragments that were not positive in the GAF–DamID assay. Among the five fragments that were positive for DamGAF binding, four were from the high-level GAF-binding fragment list and one was from the low-level GAF-binding fragment list. All of the fragments tested, both those postive and negative for GAF binding in the DamID assay, contained at least one copy of the GAF-binding motif (GAGAG/CTCTC) (28).
All five of the GAF DamID-positive DNA fragments also were positive for binding in the GAF ChIP assays from both Kc167 cells and embryos (Table 2). No difference between DamID- and ChIP-positive GAF binding sites was noted in Kc167 cells, and only one of the seven DamID-negative fragments was ChIP-positive in the embryonic chromatin extracts. These results indicate that the ChIP and DamID assays both accurately reflect bona fide GAF binding sites in vivo. Although the correspondence between DamID and GAF assays was striking, there was only a moderate correlation between the quantitative values of the DamID and GAF positive data for Kc167 cells (0.54), and thus the quantitative results from the two techniques are complementary. Finally, these results also indicate that GAF distribution in embryos and in embryonically derived Kc167 cells is largely overlapping, but qualitatively and perhaps quantitatively different.
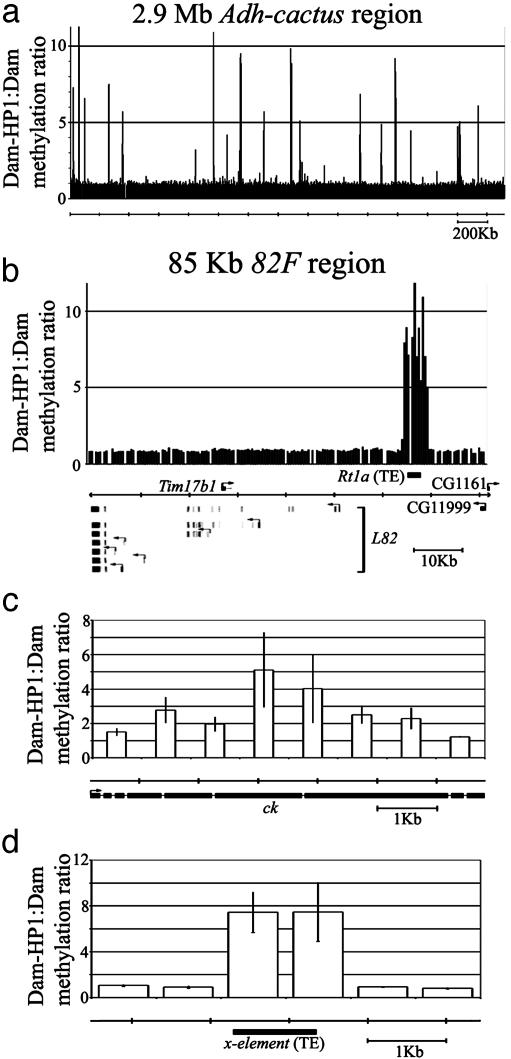
HP1 Binding Profile. We identified 17 areas in the 2.9 Mb Adh–cactus region, and one area in the 150 kb 82F region, that were associated with significant Dam-HP1:Dam ratios (Fig. 3 a and b and Table 5, which is published as supporting information on the PNAS web site). Fifteen of the seventeen areas yielded high HP1 binding ratios. All but one of these areas contain transposons or other repeat elements (Table 5), in agreement with previous studies showing that signal from HP1 fusion protein experiments is associated with transposable elements (8). To distinguish between cross-hybridization and direct association of HP1, we examined the local pattern of signal intensities for each area that contained repetitive DNA or transposable elements. Only 6 of the 17 areas in the Adh–cactus region showed distributions that strongly indicated direct association of HP1, with four showing monotonic, and four showing bimodal or complex patterns of signal distribution (Fig. 3c). The single area of HP1 binding in the 82F region also displayed a multimodal signal distribution, indicating multiple binding sites. All other areas we identified showed profiles that indicated cross hybridization (Fig. 3d). Thus, the use of tiling path microarrays allowed us in at least some cases to distinguish between bona fide association of HP1 with chromosomes and cross-hybridization. Microarrays with more sparsely spaced DNA fragments do not allow this distinction to be made.
Fig. 3.
HP1 binding profiles. Profiles displayed as in Fig. 2. (a) HP1 binding profiles across the entire 2.9 Mb Adh–cactus region. (b) HP1 binding profiles in the 82F region. (c) Binding of HP1 in the crinkled gene. (d) An example of clear cross-hybridization from remote sequences associated with HP1 binding.
We found patterns indicating real HP1 binding within the coding sequence of only one gene, crinkled (ck), which encodes a non-muscle myosin involved in bristle formation (Fig. 3c) (15, 29). Whether ck has any role in Kc167 cells is unknown, but based on microarray analyses, ck is expressed at moderate levels in these cells (L.V.S. and K.P.W., unpublished results). Thus, HP1 binding within ck does not prevent its expression. Interestingly, HP1 appears to bind to the transcribed region of the ck locus rather than to the promoter region. No repetitive elements are present in the region of strong HP1 binding in ck (the nearest repeat element/transposon is >30 kb away), indicating that HP1 is recruited to the ck gene through a mechanism distinct from its strong association with repetitive DNA.
Finally, we compared HP1 and GAF binding sites and found that they rarely overlapped (Fig. 4a, which is published as supporting information on the PNAS web site). In only one case did we observe GAF and HP1 binding profiles very near one another (Fig. 4b). These results indicate that, on a local level, GAF and HP1 binding sites are largely independent of one another in the Adh–cactus region.
Summary. Our results demonstrate the feasibility of protein–DNA interaction mapping with tiling path DNA microarrays that cover large tracts of a complex genome. We found that data from genomic tiling path arrays allowed the sites of chromosomal association to be readily discerned for both a site-specific transcription factor and a general heterochromatin-associated protein. Because all GAF-binding fragments identified with DamID were verified with ChIP, either approach is capable of yielding accurate and high-resolution binding site mapping for chromatin-associated proteins. ChIP can be complementary to DamID, and when suitable antibodies are available for a DNA-associated protein, ChIP can be used either with candidate targets or with microarrays to cross-validate binding sites. Additional studies will be required to determine the biological relevance of the dozens of GAF and HP1 binding sites we observed. Nevertheless, these results indicate that genomic tiling path microarrays will be valuable for mapping the binding sites of a wide range of regulatory proteins in Drosophila. These methods should be applied equally well for mapping DNA–protein interactions in cells isolated from animals, and will aid in the comprehensive delineation of genome-wide regulatory networks that control gene expression and development.
Supplementary Material
Acknowledgments
We thank Scott A. Rifkin for computational assistance and Harmen Bussemaker for helpful discussions and support. This work is supported by a Human Frontiers in Science Program grant (to B.V.S. and K.P.W.), a Centre National de la Recherche Scientifique grant (to G.C.), a National Science Foundation grant (to H.Z.), and a National Human Genome Research Institute grant (to K.P.W.).
Abbreviations: ChIP, chromatin immunoprecipitation; Dam, DNA adenine methyltransferase.
References
- 1.Reinke, V. & White, K. P. (2002) Annu. Rev. Genomics Hum. Genet. 3, 153–178. [DOI] [PubMed] [Google Scholar]
- 2.White, K. P., Rifkin, S. A., Hurban, P. & Hogness, D. S. (1999) Science 286, 2179–2184. [DOI] [PubMed] [Google Scholar]
- 3.Reinke, V., Smith, H. E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S. J., Davis, E. B., Scherer, S., Ward, S. & Kim, S. K. (2000) Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- 4.Hill, A. A., Hunter, C. P., Tsung, B. T., Tucker-Kellogg, G. & Brown, E. L. (2000) Science 290, 809–812. [DOI] [PubMed] [Google Scholar]
- 5.Jiang, M., Ryu, J., Kiraly, M., Duke, K., Reinke, V. & Kim, S. K. (2001) Proc. Natl. Acad. Sci. USA 98, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2199–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbeitman, M. N., Furlong, E. E., Imam, F., Johnson, E., Null, B. H., Baker, B. S., Krasnow, M. A., Scott, M. P., Davis, R. W. & White, K. P. (2002) Science 297, 2270–2275. [DOI] [PubMed] [Google Scholar]
- 8.van Steensel, B., Delrow, J. & Henikoff, S. (2001) Nat. Genet. 27, 304–308. [DOI] [PubMed] [Google Scholar]
- 9.Iyer, V. R., Horak, C. E., Scafe, C. S., Botstein, D., Snyder, M. & Brown, P. O. (2001) Nature 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 10.Ren, B., Robert, F., Wyrick, J. J., Aparicio, O., Jennings, E. G., Simon, I., Zeitlinger, J., Schreiber, J., Hannett, N., Kanin, E., et al. (2000) Science 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 11.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 12.Horak, C. E., Mahajan, M. C., Luscombe, N. M., Gerstein, M., Weissman, S. M. & Snyder, M. (2002) Proc. Natl. Acad. Sci. USA 99, 2924–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren, B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R. A. & Dynlacht, B. D. (2002) Genes Dev. 16, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinmann, A. S., Yan, P. S., Oberley, M. J., Huang, T. H. & Farnham, P. J. (2002) Genes Dev. 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Misra, S., Crosby, M. A., Mungall, C. J., Matthews, B. B., Campbell, K. S., Hradecky, P., Huang, Y., Kaminker, J. S., Millburn, G. H., Prochnik, S. E., et al. (2002) Genome Biol. 3, RESEARCH0083-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner, M., Misra, S., Roote, J., Lewis, S. E., Blazej, R., Davis, T., Doyle, C., Galle, R., George, R., Harris, N., et al. (1999) Genetics 153, 179–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reese, M. G., Hartzell, G., Harris, N. L., Ohler, U., Abril, J. F. & Lewis, S. E. (2000) Genome Res. 10, 483–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stowers, R. S., Russell, S. & Garza, D. (1999) Dev. Biol. 213, 116–130. [DOI] [PubMed] [Google Scholar]
- 19.van Steensel, B. & Henikoff, S. (2000) Nat. Biotechnol. 18, 424–428. [DOI] [PubMed] [Google Scholar]
- 20.Biggin, M. D. & Tjian, R. (1988) Cell 53, 699–711. [DOI] [PubMed] [Google Scholar]
- 21.James, T. C., Eissenberg, J. C., Craig, C., Dietrich, V., Hobson, A. & Elgin, S. C. (1989) Eur. J. Cell Biol. 50, 170–180. [PubMed] [Google Scholar]
- 22.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 23.Bussemaker, H. J., Li, H. & Siggia, E. D. (2001) Nat. Genet. 27, 167–171. [DOI] [PubMed] [Google Scholar]
- 24.Keles, S., Van Der Laan, M. & Eisen, M. B. (2002) Bioinformatics 18, 1167–1175. [DOI] [PubMed] [Google Scholar]
- 25.Liu, X. S., Brutlag, D. L. & Liu, J. S. (2002) Nat. Biotechnol. 20, 835–839. [DOI] [PubMed] [Google Scholar]
- 26.Cavalli, G., Orlando, V. & Paro, R. (1999) in Chromosome Structural Analysis: A Practical Approach, ed. Bickmore, W. A. (Oxford Univ. Press, Oxford), pp. 20–37.
- 27.Cleveland, W. S., Grosse, E. & Shyu, W. M. (1992) in Statistical Models, eds. Chambers, J. M. & Hastie, T. J. (Brooks/Cole, Belmont, CA), pp. 309–373.
- 28.Omichinski, J. G., Pedone, P. V., Felsenfeld, G., Gronenborn, A. M. & Clore, G. M. (1997) Nat. Struct. Biol. 4, 122–132. [DOI] [PubMed] [Google Scholar]
- 29.Gubb, D., Shelton, M., Roote, J., McGill, S. & Ashburner, M. (1984) Chromosoma 91, 54–64. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.