Abstract
Previous studies have demonstrated that the specificity of Src homology 2 (SH2) and phosphotyrosine-binding domain interactions are mediated by phosphorylated tyrosines and their neighboring amino acids. Two of the first phosphotyrosine-based binding sites were found on middle T antigen of polyoma virus. Tyr-250 acts as a binding site for ShcA, whereas Tyr-315 forms a binding site for the SH2 domain of the p85 subunit of phosphatidylinositol 3-kinase. However, genetic analysis of a given phosphotyrosine's role in signaling can be complicated when it serves as a binding site for multiple proteins. The situation is particularly difficult when the phosphotyrosine serves as a secondary binding site for a protein with primary binding determinates elsewhere. Mutation of a tyrosine residue to phenylalanine blocks association of all bound proteins. Here we show that the mutation of the amino acids following the phosphorylated tyrosine to alanine can reveal phosphotyrosine function as a secondary binding site, while abrogating the phosphotyrosine motif's role as a primary binding site for SH2 domains. We tested this methodology by using middle T antigen. Our results suggest that Tyr-250 is a secondary binding site for phosphatidylinositol 3-kinase, whereas Tyr-315 is a secondary binding site for a yet-to-be-identified protein, which is critical for transformation.
Middle tumor antigen (MT) is the principal oncoprotein of murine polyomavirus, a member of the Papovaviridae family. As its name indicates, polyomavirus is capable of inducing transformation in a wide variety of cell types. MT induces transformation by associating with and modulating the activities of numerous host cell proteins involved in cell cycle control and proliferation. MT contains a membrane anchoring domain at its C terminus that is essential for transformation. It forms a complex with host cell signaling molecules in a sequential manner (1, 2). Its initial association with protein phosphatase 2A (3, 4) allows for the recruitment of Src family tyrosine kinases (5–11), which in turn phosphorylate several tyrosine residues on MT (2). This leads to its association with and activation of signaling molecules such as ShcA, phosphatidylinositol 3-kinase (PI3-kinase), and phospholipase C-γ, which bind regions surrounding phosphotyrosine residues 250, 315, and 322, respectively (12–15). In this way, MT acts much like a constitutively activated tyrosine kinase receptor. More than two decades of research on MT have shed light on a number of important concepts in growth factor-mediated signal transduction. In fact, the first tyrosine kinase assays were done on MT immunoprecipitates (16–18), and PI3-kinase was first identified as a MT binding partner essential for its transforming ability (19–23).
The phosphorylated tyrosine residue 315 (Y315) of MT and the three carboxyl amino acid residues function as a binding site for the Src homology 2 (SH2) domain of the p85 subunit of PI3-kinase (24). This region of MT contains the preferred PI3-kinase binding motif, YMXM (25). It has been previously shown that substituting tyrosine with phenylalanine (Y315F) in MT abrogates p85 association and PI3-kinase activity, resulting in a significant loss of transformation ability (14, 19, 26, 27). Indeed the mutation of Y315 to phenylalanine was the first reported point mutation in any molecule to block PI3-kinase binding, paving the way for the elucidation of the exact nature of the PI3-kinase binding site. However, it should be noted that the Y315F mutant retains a degree of residual transforming activity that appears to vary with cell type (28–30).
Although the association with and activation of PI3-kinase has been shown to be necessary, it is not a sufficient condition for MT-induced transformation. MT mutants that activate PI3-kinase but are defective for ShcA association are also transformation defective (31, 32). ShcA, via its phosphotyrosine-binding domain, binds MT at its tyrosine residue 250 (Y250) region, which contains an NPXY motif (12, 13). Either a mutation in Y250 or Pro-248 completely abolishes ShcA association with MT (13, 27, 32).
Our view of the interactions of MT with cellular proteins has been limited by the genetics available to probe such interactions. Systematic mutations have identified tyrosines that are important for transformation. For each tyrosine pointed to by the genetics, a single binding protein has been identified. Here we have widened our genetic study of the phosphotyrosine-based binding motifs to include the amino acids surrounding the key phosphotyrosines. We found that both Y250 and Y315 play complex roles in their interactions with cellular proteins. Y250 serves as an important component of the PI3-kinase binding site on MT in addition to its reported role in ShcA binding. Furthermore, we provide genetic and biochemical evidence that Y315 plays a key function in addition to its known role as a PI3-kinase binding site. When the PI3-kinase binding site at Y315 is mutated from YMPM to YAAA, MT loses PI3-kinase binding but retains significant transforming activity. By adding selected phosphotyrosine motifs to WT MT and the Y315F mutant, we show that Y315 appears to serve as a binding site for two phosphotyrosine binding proteins, just as Y250 serves as a binding site for both ShcA and PI3-kinase. Although the identity of the second protein is not yet known, it appears to be important in transformation.
Materials and Methods
Detailed protocols for all of the methods used are in Supporting Materials and Methods, which are published as supporting information on the PNAS web site, www.pnas.org.
Plasmid Construction. To create the pLJ-YAAA and pLJ-FAAA mutants, double-stranded oligos were subcloned into a pGMT vector and then into a pLJ vector. Site-directed mutagenesis was performed (Stratagene) to create a point mutation for M253A. For the “add-back” mutants, double-stranded oligos were subcloned into a unique restriction site, EspI (32). Insertion into the EspI site resulted in an additional serine at the beginning and a leucine at the end of the insertion, respectively.
Cells and Tissue Culture. The Bosc 23 viral packaging cell line (33) was grown in DMEM (Invitrogen) with 10% FBS in 10% CO2. BALB/c3T3 fibroblasts (clone A31) were grown in DMEM with 10% calf serum (CS) in 10% CO2.
Retroviral Infections and Establishment of BALB/c3T3 Clones. Bosc 23 cells were transfected by using the N,N-bis[2-hydroxyethyl]2-aminoethane sulfonic acid method as described (34). Infections with viral supernatants, drug selection, and isolation of G418-resistant colonies were carried out as described (35).
Antibodies, Immunoprecipitations, and Immunoblotting. Anti-ShcA antibody (Upstate Biotechnology, Lake Placid, NY) and anti-MT antibody 18-8 (36) were used for immunoprecipitations. Anti-ShcA antibody (BD Transduction Laboratories, San Diego), anti-MT antibody F4 (36), anti-phospho-Akt antibody (Cell Signaling Technologies, Beverly, MA), and anti-Akt antibody (Cell Signaling Technologies) were used for immunoblotting. Anti-p85 antibody R32 (K. Auger and T.M.R., unpublished work) was used for immunoprecipitations and immunoblotting. Cells were incubated in DMEM with 0.1% CS overnight. Then they were rinsed and lysed in 1% Nonidet P-40 lysis buffer. The cell lysates were used in Western blots or further used for immunoprecipitation.
Lipid Kinase Assays. Lipid kinase assays were performed as described (37).
Focus Formation Assays. Cells were infected with titered retroviral supernatants. Infected cells were split into three plates the next day. For focus formation, the medium was changed every 3–4 days with DMEM containing 7% CS for ≈3 weeks without drug selection; for colony formation, the cells were selected in 500 μg/ml G418, and the medium was changed every 3–4 days. The focus formation assay and G418 drug selected plates were fixed and stained.
Soft Agar Assays. Cells stably expressing MT and its mutants were mixed with 0.3% agarose in DMEM with 10% CS and plated into six-well dishes containing 0.8% agarose in DMEM with 10% CS. Cells were fed every 5–7 days with 1 ml of DMEM with 10% CS without agarose and allowed to grow for 3–5 weeks before being scored for growth.
Results
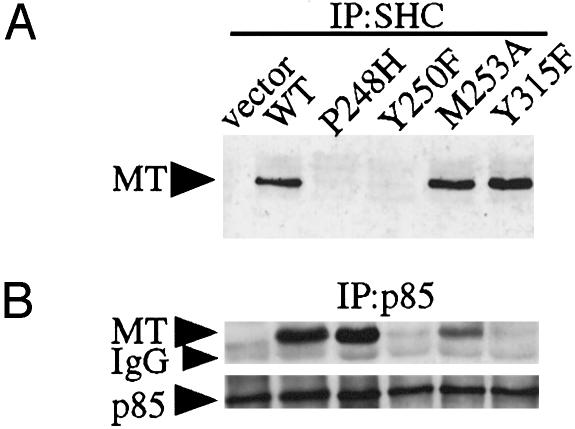
Y250 Is Embedded in a YXXM Motif and Is Necessary for Efficient p85 Binding to MT. It has previously been reported that both Y250F and P248H mutations in MT (Fig. 1) are transformation defective (27, 32) and lack ShcA association (13) (Fig. 2A). However, the effects of these mutations on MT's association with PI3-kinase activity have never been compared. As shown in Fig. 2B, the P248H mutant was comparable to WT in its p85 binding and MT-associated PI3-kinase activity (data not shown). However, the Y250F mutant showed low levels of associated PI3-kinase activity and was in fact similar to Y315F, a known MT mutant defective for binding p85. Thus, Y250 but not ShcA binding per se is essential to PI3-kinase binding. Both P248H and Y250F mutants failed to induce transformation (Table 1) (27, 31, 32).
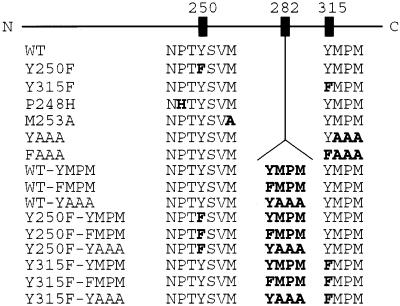
Fig. 1.
Schematic overview of the WT MT and its mutants. Each mutant is characterized by the status of the amino acids in the ShcA and PI3-kinase binding sites and/or by the presence of added sequences at amino acid 282. N, N terminal; C, C terminal.
Fig. 2.
Characterization of the Y250F, P248H, and M253A mutants. (A) Y250F and P248H lack ShcA association, whereas M253A mutant retains ShcA association. Cell lysates were immunoprecipitated (IP) with an anti-ShcA antibody. Samples were analyzed by Western blotting with an anti-MT antibody. (B) p85 association with MT requires an intact tyrosine residue at 250. Cell lysates were immunoprecipitated (IP) with an anti-p85 antibody and immunoblotted with an anti-MT antibody (Upper) and an anti-p85 antibody (Lower).
Table 1. Focus formation by MT mutants.
| Virus | % | Virus | % |
|---|---|---|---|
| Control | 0 | WT-YMPM | 98 ± 3 |
| WT | 100 | WT-FMPM | 85 ± 2 |
| Y250F | 1 ± 2 | WT-YAAA | >100 |
| Y315F | 6 ± 1 | Y250F-YMPM | 3 ± 3 |
| P248H | 0 | Y250F-FMPM | 0 |
| M253A | 11 ± 7 | Y250F-YAAA | 0 |
| YAAA | 73 ± 5 | Y315F-YMPM | 62 ± 0.7 |
| FAAA | 4 ± 4 | Y315F-FMPM | 12 ± 9 |
| Y315F-YAAA | 17 ± 15 |
BALB/c3T3 cells were infected with viral supernatants and cultured with DMEM containing 7% CS for ≈3 weeks. Viral titers were measured in each experiment, and results are normalized for titer. The number of foci formed by WT MT was defined as 100% and used to normalize between experiments. The numbers shown represent the average with SD of at least three independent experiments.
The YMPM sequence at residues 315–318 in MT has been suggested to be required for p85 binding (25). The sequence surrounding Y250 of MT is NPTYSVM. In addition to the NPTY motif required for ShcA binding, this sequence contains a YXXM motif, a potential p85 binding site (25). It seemed plausible that MT might use two YXXM sites to bind p85, as is the case with the platelet-derived growth factor (PDGF) receptor (38). To test whether the YXXM motif at Y250 functions in p85 binding, we created a mutant in which alanine was substituted for methionine at residue 253 (M253A) (Fig. 1). Biochemical analysis revealed that M253A had an intermediate phenotype: it bound more p85 (Fig. 2B) and had more PI3-kinase activity (data not shown) than a Y315F or Y250F mutant, but it had less activity than WT MT. To assess the biological consequences of the mutation, a focus formation assay was carried out. The M253A mutant was not able to transform cells as efficiently as WT MT but was more potent than a Y315F mutation, consistent with a previous study in which isoleucine was substituted for methionine at residue 253 (Table 1) (27). This finding suggests that Y250 is the major determinant for p85 binding with M253 contributing a relatively smaller amount of the binding energy. Overall, our genetic and biochemical data are consistent with the idea that Y250 provides a second binding site for PI3-kinase.
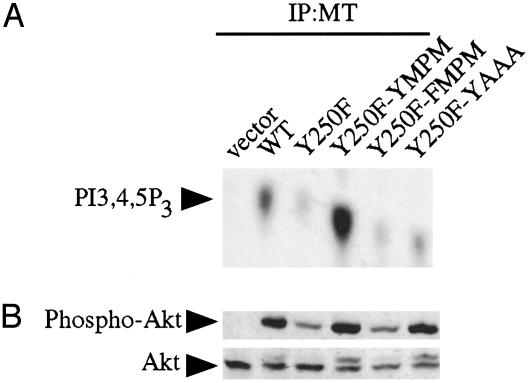
PI3-Kinase Binding Can Be Restored to Y250F by Adding a YXXM Motif at Another Site. To test this hypothesis, we examined whether restoring a PI3-kinase binding site in a Y250F background would restore PI3-kinase binding. Previously it was shown that it is possible to restore ShcA binding and transformation in a P248H background by inserting an NPXY sequence between amino acids 282 and 283 (32). Using this insertion site, we created a MT allele that contained the sequence EEEEEYMPM inserted after L282 in a Y250F background. This insert corresponds to the sequence of amino acids surrounding Y315 in WT MT. The glutamic acid residues were included to ensure efficient phosphorylation of the new YMPM motif by pp60src (39, 40). This allele of MT was designated Y250F-YMPM (Fig. 1). As a control, we also created a second add-back by using FMPM in the place of YMPM, which was designated Y250F-FMPM. Interestingly, as shown in Fig. 3A, Y250F-YMPM showed an increased amount of associated PI3-kinase activity compared with WT MT, whereas Y250F-FMPM showed levels of PI3-kinase association comparable to Y250F. Cells expressing the Y250F-YMPM allele also showed increased Akt phosphorylation compared with cells expressing WT MT (Fig. 3B). These data suggest that in WT MT PI3-kinase may compete with ShcA for Y250, whereas in Y250F-YMPM, PI3-kinase can engage its second binding site without competition. Notably, the Y250F-YMPM allele fails to transform BALB/c3T3 cells presumably because it is not capable of binding ShcA (Table 1).
Fig. 3.
Characterization of the Y250F-YMPM and Y250F-YAAA mutants. (A) In vitro lipid kinase assay. Anti-MT immunoprecipitates (IP) were analyzed for their ability to phosphorylate phosphatidylinositol-4,5-bisphosphate. PI3-kinase product phosphatidylinositol-3,4,5-trisphosphate (PI3,4,5P3) is indicated. Y250F-YMPM showed more MT-associated PI3-kinase activity than WT MT. Y250F-YAAA restored a significant amount of PI3-kinase activity compared with Y250F. (B) Y250F-YMPM and Y250F-YAAA alleles activate Akt. Equal amounts of cell lysates were analyzed by Western blotting with an anti-phospho-Akt Ser-473 antibody (Upper) and an anti-Akt antibody (Lower).
Y250 Forms a Secondary Binding Site for PI3-Kinase. We wanted to test further the relative contributions of Y250 and the remainder of the YXXM motif to this second binding site. To this end, we made an additional add-back by using the same site as before, Y250F-YAAA, which supplies a phosphorylatable tyrosine but no additional binding determinants for the SH2 domains of PI3-kinase. Consistent with the data for the M253A mutation, the Y250F-YAAA add-back significantly restored MT-associated PI3-kinase activity (Fig. 3A) and Akt activation (Fig. 3B). Conventionally, the difference between primary and secondary binding sites is defined functionally. The secondary binding site depends critically on the phosphotyrosine but depends less on the amino acids making up the rest of the motif. The observation that the interaction of p85 with Y250 is relatively insensitive to mutation in the carboxyl amino acids suggests a model in which Y250 forms a secondary binding site for PI3-kinase with the YMPM motif at 315–318 acting as the primary site. As we will show next, mutation of the amino acids carboxyl to Y315 has a much more dramatic effect on PI3-kinase binding.
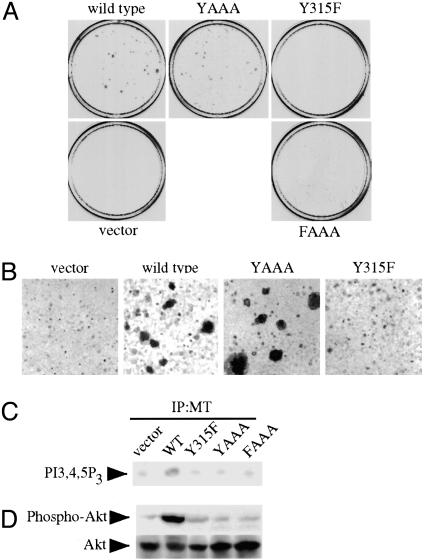
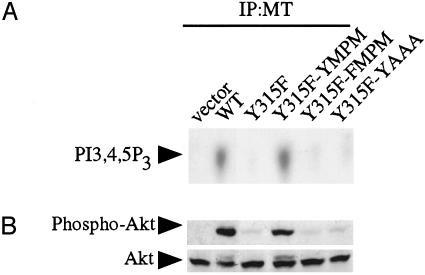
Y315 May Play Another Role Other than PI3-Kinase Binding. To investigate the role of the Y315 region in effector binding, we replaced the MPM residues C-terminal to the Y315 with AAA (henceforth referred to as YAAA) (Fig. 1). As a control, we also made an FAAA mutant. As expected, the level of p85 association with the YAAA mutant, like the Y315F mutant, is nearly undetectable (data not shown). Likewise, the MT-associated PI3-kinase activity of YAAA does not exceed that of the Y315F mutant (Fig. 4C). To our surprise, however, the YAAA mutant was capable of inducing transformation in a focus assay (Table 1 and Fig. 4A) and in a soft agar assay (Fig. 4B) to a degree close to WT MT. YAAA transformation was much more efficient than that of the Y315F mutant, which is defective for p85 association.
Fig. 4.
The YAAA mutant transforms cells much better than Y315F but fails to bind PI3-kinase. (A) Focus formation assay of the 315 region MT mutants. YAAA can induce transformation at levels approaching those of WT MT. BALB/c3T3 cells were infected with virus supernatants. Infected cells were pooled and split equally. One set of cells was grown for 3 weeks in 7% CS to test for focus formation. The other was selected in media containing 500 μg of G418 media to test for colony formation and viral titers (data not shown). Viral titers of all viruses were similar (data not shown). (B) YAAA is able to form large colonies in soft agar. Cells were grown in suspension for 4–5 weeks. (C) In vitro lipid kinase assay. Anti-MT immunoprecipitates (IP) were analyzed for their ability to phosphorylate phosphatidylinositol-4,5-bisphosphate. PI3-kinase product phosphatidylinositol-3,4,5-trisphosphate (PI3,4,5P3) is indicated. (D) The YAAA allele does not activate Akt. Equal amounts of cell lysates were analyzed by Western blotting with an anti-phospho-Akt Ser-473 antibody (Upper) and then an anti-Akt antibody (Lower).
Because activation of PI3-kinase is believed to be necessary for MT transformation, we tested for Akt activation in these mutants to see whether PI3-kinase might be activated via another pathway. YAAA, like Y315F, only poorly activated Akt (Fig. 4D). This ruled out the possibility that YAAA-induced transformation was caused simply by association with PI3-kinase and strongly suggested that Y315 is part of a complex binding site, like Y250. In addition, the inability of the YAAA mutant to bind PI3-kinase is consistent with a model in which Y315 is the primary binding site for PI3-kinase. Phosphopeptide mapping demonstrated that Y315 in YAAA is efficiently phosphorylated (data not shown). Thus the failure of the YAAA mutant to bind PI3-kinase is not because of the lack of phosphorylation of Y315 in the context of YAAA.
Construction of a MT Allele That Binds and Activates PI3-Kinase but Fails to Transform as Efficiently as WT MT. To test our hypothesis that Y315 plays a role in transformation in addition to PI3-kinase activation, we wanted to see whether restoring PI3-kinase binding in a Y315F background would fully restore transformation. We again turned to the strategy of making add-back mutants. In this case we added a YMPM, FMPM, or YAAA motif to Y315F. These add-back alleles of MT were designated Y315F-YMPM, Y315F-FMPM, and Y315F-YAAA, respectively (Fig. 1).
We first analyzed the biochemical and biological properties of the Y315F add-back mutants. The Y315F-YMPM mutant restored MT associated PI3-kinase activity to levels comparable to those seen for WT MT (Fig. 5A). Both Y315F-FMPM and Y315F-YAAA failed to restore PI3-kinase association. Although it was clearly transforming, the Y315F-YMPM failed to restore transformation activity completely to the level of WT MT (Table 1). It remained possible that the failure of the YMPM add-back to rescue transformation was caused by a failure of the add-back allele to activate PI3-kinase. To test this, we examined the phosphorylation of Akt in cells expressing the various add-back alleles. Notably, Akt phosphorylation in cells expressing Y315F-YMPM was comparable to that seen in cells expressing WT MT (Fig. 5B). This was true both for phosphorylation of Ser-473 (Fig. 5B) and Thr-308 of Akt (data not shown). Therefore, it appears that PI3-kinase association can be maintained when two PI3-kinase binding sites are available for binding. However, as seen in the case of Y315F-YMPM, PI3-kinase binding is not sufficient for WT levels of transformation. Something intrinsic about Y315 itself, aside from PI3-kinase binding, must contribute to MT's transforming ability. Taken together, these data strongly suggest that Y315 is critical for binding a second protein in addition to PI3-kinase and that this “protein X” is unable to efficiently bind to the add-back alleles, possibly because of steric constraints.
Fig. 5.
Y315F-YMPM restores PI3-kinase association with MT but fails to transform BALB/c3T3 cells to the level of WT MT. (A) In vitro lipid kinase assay. Anti-MT immunoprecipitates (IP) were analyzed for their ability to phosphorylate phosphatidylinositol-4,5-bisphosphate. PI3-kinase product phosphatidylinositol-3,4,5-trisphosphate (PI3,4,5P3) is indicated. (B) Y315-YMPM allele activates Akt to the level of WT MT. Equal amounts of cell lysates were analyzed by Western blotting with an anti-phospho-Akt Ser-473 antibody (Upper) and then an anti-Akt antibody (Lower).
A Model for MT Transformation. In our tentative model, PI3-kinase binds to Y315 as its primary binding site and Y250 as a secondary site. We hypothesize that protein X binds to Y315. Furthermore, Y315 appears to be its secondary binding site, with its primary binding site, and its identity, currently unknown. This sharing would lead to competition between PI3-kinase and protein X for binding at Y315. The YAAA mutant presumably transforms relatively efficiently, because although it fails to bind PI3-kinase as efficiently as WT MT, it binds protein X with higher affinity than WT MT. This model makes an exciting and testable prediction: an add-back mutant in a WT background might produce a MT allele that could bind both PI3-kinase and protein X optimally. Thus we might expect that a YAAA add-back in a WT background would transform even better than WT MT.
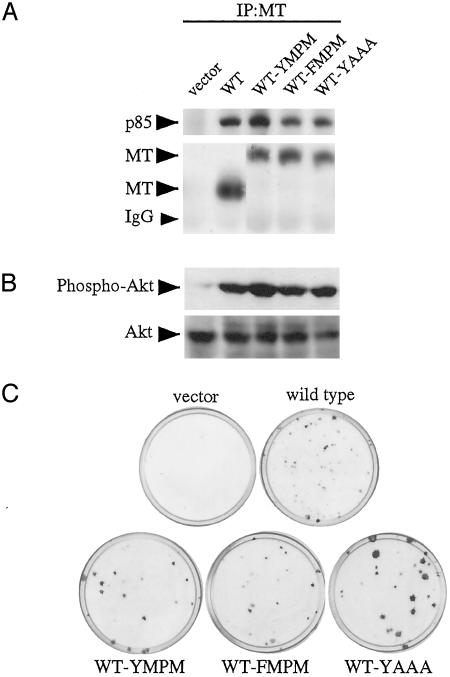
To test our model, we constructed three additional add-back alleles in a WT background, which we designated WT-YMPM, WT-YAAA, and WT-FMPM (Fig. 1). As shown in Fig. 6A, WT-YMPM displayed higher levels of p85 binding and MT-associated PI3-kinase activity (data not shown) than WT MT. Both FMPM and YAAA add-backs failed to increase p85 and PI3-kinase association above WT MT. WT-YMPM displayed higher levels of Akt phosphorylation than did WT MT (Fig. 6B). WT-YAAA and WT-FMPM displayed levels of Akt phosphorylation similar to WT MT. This was true both for phosphorylation of Akt on Ser-473 (Fig. 6B) and Thr-308 (data not shown). In focus formation assays we found that all three add-backs transformed (Fig. 6C and Table 1). Strikingly, the WT-YAAA add-back made foci considerably larger than WT MT. These data are consistent with a model in which PI3-kinase and protein X compete for binding with Y315. Hence, adding back a second YMPM motif increases PI3-kinase binding, whereas adding back a YAAA motif enhances transformation, presumably via enhanced binding of protein X.
Fig. 6.
WT-YMPM shows enhanced PI3-kinase association but similar transformation activity to WT, whereas WT-YAAA shows enhanced transformation activity without affecting PI3-kinase association with MT. (A) WT-YMPM shows enhanced p85 binding to MT. Cell lysates were immunoprecipitated (IP) with an anti-MT antibody. Samples were analyzed by Western blotting with an anti-p85 antibody (Upper) and an anti-MT antibody (Lower). (B) Enhanced transformation by WT-YAAA is independent of Akt activation. Equal amounts of cell lysates were analyzed by Western blotting with an anti-phospho-Akt Ser-473 antibody (Upper) and then an anti-Akt antibody (Lower). (C) Focus formation assay of Y315F add-back mutants. WT-YMPM does not increase BALB/c3T3 cells transformation, whereas WT-YAAA showed enhanced transformation. BALB/c3T3 cells were infected with viral supernatants and cultured with DMEM containing 7% CS for ≈3 weeks. Viral titers of all viruses were similar (data not shown).
Discussion
More than two decades of research on MT have shed light on important concepts in growth factor signal transduction (2). Studies on MT have helped to define YMXM and NPTY as PI3-kinase (14, 25) and ShcA (13) binding consensus sequences, respectively. In the current study we have refined our understanding of each of these binding sites, finding evidence that each site appears to be complex, with two or more cellular proteins potentially competing for binding to a single phosphotyrosine.
Our data suggest a model for PI3-kinase binding to MT. In addition to its traditional binding site, Y315, Y250 may play a role as a direct binding site for PI3-kinase. Previous work has also implicated Y250 in PI3-kinase binding but suggested that it played an indirect role, via ShcA binding. Specifically, it has been demonstrated that ShcA, via its interaction with Grb2 and Gab1, can provide a binding site for p85, at least in some cell types (30). Here we show that in BALB/c3T3 cells Y250 may play a much more direct role in binding PI3-kinase. In these cells, a P248H mutant, which blocks ShcA binding, has little effect on PI3-kinase binding and activation of Akt (data not shown), whereas mutation of Y250 blocks both PI3-kinase binding and ShcA binding. Moreover, PI3-kinase binding may be added back to a Y250F mutant by adding a second YMPM motif. Interestingly, Y250 is also part of a YXXM motif, which means MT has two YXXM motifs similar to the PDGF receptor (38). In the case of the PDGF receptor, the tyrosines Y740, which is part of a YMDM motif, and Y751, a YVPM motif, both are required for recruiting PI3-kinase. Thus, in our study MT is behaving like the PDGF receptor in its requirements for PI3-kinase binding. We do not think that our data contradict the previous data showing Gab1 involvement in PI3-kinase binding (30). In the previous work, Gab1 binding appeared to be relevant only under certain defined circumstances. Specifically, Gab1 appeared to play a role under conditions of relatively high MT expression. Notably, our retroviral vectors express MT relatively weakly.
In our model, Y250 is a secondary binding site for p85. We picture p85 binding to phosphorylated Y315 tightly via an interaction with one of its SH2 domains (24) and the consensus YMXM motif at Y315 (25). The second SH2 domain of p85 appears to interact with Y250 with much of the binding energy coming from the phosphotyrosine contact. This idea is supported by two pieces of data presented here. First, the methionine at 253 is of relatively little importance, as M253A has significantly higher PI3-kinase activity than Y250F or Y315F. Furthermore, Y250F-YAAA, but not Y315F-YAAA, restored PI3-kinase activity and was capable of activating Akt. It would be interesting to know whether this kind of binding site hierarchy also exists in growth factor receptors.
PI3-kinase and ShcA may be competing for binding to Y250. Notably, both the P248H and the Y250F-YMPM add-back mutant bind more PI3-kinase than WT MT, presumably because these mutants have a good secondary binding site for PI3-kinase, which is not shared with ShcA. We have also observed a reciprocal situation for ShcA. Mutants that feature decreased binding of PI3-kinase, e.g., Y315F, etc., show elevated levels of MT in ShcA immunoprecipitates (Fig. 2 A and data not shown).
Our data also suggest that Y315 plays a role in transformation that is separate from its role in PI3-kinase binding. Three facts support this argument. First, the YAAA mutant transforms cells markedly better than the Y315F mutant, although YAAA activates the PI3-kinase pathway no better than Y315F. Second, the Y315F-YMPM add-back restores binding of PI3-kinase activity to the level of WT MT but transforms BALB/c3T3 cells less well than WT MT. Finally, the WT-YAAA add-back transforms cells better than WT MT even though it binds no more PI3-kinase than WT MT.
It is possible that our YAAA mutant and YAAA add-back mutants are actually binding some protein that is not normally bound by WT MT. However, we think this is unlikely because the amino acid sequences YAAA, or EEEEEYAAA in our addback mutants, are not known to be a specific binding site for any protein. In fact, other YXXX mutants are also able to rescue MT-induced transformation to varying degrees without PI3-kinase pathway activation (data not shown). We feel our data are most easily reconciled with a model postulating the existence of protein X. We hypothesize that protein X binds to the phosphorylated form of Y315 in competition with PI3-kinase, but has its primary binding site elsewhere on MT. In the YAAA mutant, there is no competition between protein X and PI3-kinase, which allows increased protein X binding, at least partially compensating for the loss of PI3-kinase binding in transformation. This finding suggests that protein X, like PI3-kinase, plays an important role in transformation. This hypothesis is supported by the WT-YAAA add-back, which presumably also decreases competition between p85 and protein X and clearly transforms better than WT MT (without binding PI3-kinase better than WT MT). Interestingly, the behavior of MT with respect to PI3-kinase and protein X binding is similar to the behavior of PDGF receptor binding to PI3-kinase and Nck, where both proteins share Y751 on the PDGF receptor (41). However, Nck is believed to use Y751 as its primary binding site, whereas our data suggest Y315 is a secondary binding site for protein X, because EEEEEEYAAA is not a primary binding site for any known SH2 or phosphotyrosine-binding domain. We have tested the limits of protein X's flexibility in binding Y315 by changing amino acids 316–318 to the very bulky WPW. The YWPW mutant was transformtion-defective with a phenotype similar to Y315F (data not shown).
The identity of protein X remains unclear. We believe that most SH2 domains might bind with considerable affinity to the YAAA motif if it were a secondary binding site. The best documented example of this type of behavior is found in the SH2 domain of pp60c-src. The isolated SH2 domain of pp60c-src has been shown to bind to a pYAAA peptide efficiently, although nonspecifically (42). In the context of the pp60c-src molecule, it has been shown that down-regulation of tyrosine kinase activity depends on an intramolecular interaction of the SH2 domain with Y527 in its phosphorylated state (43). This intramolecular SH2 interaction is quite similar to the secondary binding site concept that we are proposing for YAAA and protein X. Notably, the intramolecular SH2 interaction in pp60c-src does not depend on the makeup of the amino acids COOH-terminal to Y527. Indeed, all amino acids following Y527 can be deleted without any activation of pp60c-src. It seems likely that the same type of behavior would be found for most, if not all, other SH2 domains. Several tests were done to see whether protein X might be a known binding partner of MT. We have been unable to detect any changes in pp60c-src binding or activity in MT immunopreciptiates (data not shown). In the case of the PDGF receptor, loss of PI3-kinase binding can be compensated by increased phospholipase C-γ binding (44). However, we have not been able to observe enhanced phospholipase C-γ binding (data not shown). ShcA is unlikely to be protein X, based on our observation that YAAA transforms much better than Y315F, whereas both have a similar profile in terms of ShcA and PI3-kinase binding. Furthermore, WT-YAAA transforms significantly better than WT without changing ShcA and PI3-kinase association. We have also examined Nck binding without seeing any differences (data not shown). Whatever the identity of protein X, our data suggest that it plays an important role in MT transformation. It is possible that other functionally important proteins like protein X will be discovered if this type of mutational analysis is applied to phosphotyrosine binding sites in other signaling proteins.
Supplementary Material
Acknowledgments
We thank Drs. Joanne Chan, Ole Gjoerup, Helen McNamee, Romesh Subramanian, and Lei Zhang for critical reading of the manuscript and Meacie Fairfax for the manuscript preparation. This work was supported by National Institutes of Health Grants RO1 CA30002-21 and PO1 CA89021-02 (to T.M.R.) and CA34722 and CA50661 (to B.S.).
Abbreviations: MT, middle tumor antigen; PI3-kinase, phosphatidylinositol 3-kinase; Y315, tyrosine residue 315; Y250, tyrosine residue 250; PDGF, platelet-derived growth factor; SH2, Src homology 2; CS, calf serum.
References
- 1.Ichaso, N. & Dilworth, S. M. (2001) Oncogene 20, 7908–7916. [DOI] [PubMed] [Google Scholar]
- 2.Dilworth, S. M. (2002) Nat. Rev. Cancer 2, 951–956. [DOI] [PubMed] [Google Scholar]
- 3.Pallas, D. C., Shahrik, L. K., Martin, B. L., Jaspers, S., Miller, T. B., Brautigan, D. L. & Roberts, T. M. (1990) Cell 60, 167–176. [DOI] [PubMed] [Google Scholar]
- 4.Walter, G., Ruediger, R., Slaughter, C. & Mumby, M. (1990) Proc. Natl. Acad. Sci. USA 87, 2521–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glover, H. R., Brewster, C. E. & Dilworth, S. M. (1999) Oncogene 18, 4364–4370. [DOI] [PubMed] [Google Scholar]
- 6.Courtneidge, S. A. & Smith, A. E. (1983) Nature 303, 435–439. [DOI] [PubMed] [Google Scholar]
- 7.Courtneidge, S. A. & Smith, A. E. (1984) EMBO J. 3, 585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornbluth, S., Sudol, M. & Hanafusa, H. (1987) Nature 325, 171–173. [DOI] [PubMed] [Google Scholar]
- 9.Kypta, R. M., Hemming, A. & Courtneidge, S. A. (1988) EMBO J. 7, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, S. H., Harvey, R., Espino, P. C., Semba, K., Yamamoto, T., Toyoshima, K. & Smith, A. E. (1988) EMBO J. 7, 3845–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horak, I. D., Kawakami, T., Gregory, F., Robbins, K. C. & Bolen, J. B. (1989) J. Virol. 63, 2343–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dilworth, S. M., Brewster, C. E., Jones, M. D., Lanfrancone, L., Pelicci, G. & Pelicci, P. G. (1994) Nature 367, 87–90. [DOI] [PubMed] [Google Scholar]
- 13.Campbell, K. S., Ogris, E., Burke, B., Su, W., Auger, K. R., Druker, B. J., Schaffhausen, B. S., Roberts, T. M. & Pallas, D. C. (1994) Proc. Natl. Acad. Sci. USA 91, 6344–6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talmage, D. A., Freund, R., Young, A. T., Dahl, J., Dawe, C. J. & Benjamin, T. L. (1989) Cell 59, 55–65. [DOI] [PubMed] [Google Scholar]
- 15.Su, W., Liu, W., Schaffhausen, B. S. & Roberts, T. M. (1995) J. Biol. Chem. 270, 12331–12334. [DOI] [PubMed] [Google Scholar]
- 16.Eckhart, W., Hutchinson, M. A. & Hunter, T. (1979) Cell 18, 925–933. [DOI] [PubMed] [Google Scholar]
- 17.Smith, A. E., Smith, R., Griffin, B. & Fried, M. (1979) Cell 18, 915–924. [DOI] [PubMed] [Google Scholar]
- 18.Schaffhausen, B. S. & Benjamin, T. L. (1979) Cell 18, 935–946. [DOI] [PubMed] [Google Scholar]
- 19.Kaplan, D. R., Whitman, M., Schaffhausen, B., Raptis, L., Garcea, R. L., Pallas, D., Roberts, T. M. & Cantley, L. (1986) Proc. Natl. Acad. Sci. USA 83, 3624–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitman, M., Kaplan, D. R., Schaffhausen, B., Cantley, L. & Roberts, T. M. (1985) Nature 315, 239–242. [DOI] [PubMed] [Google Scholar]
- 21.Kaplan, D. R., Whitman, M., Schaffhausen, B., Pallas, D. C., White, M., Cantley, L. & Roberts, T. M. (1987) Cell 50, 1021–1029. [DOI] [PubMed] [Google Scholar]
- 22.Courtneidge, S. A. & Heber, A. (1987) Cell 50, 1031–1037. [DOI] [PubMed] [Google Scholar]
- 23.Whitman, M., Downes, C. P., Keeler, M., Keller, T. & Cantley, L. (1988) Nature 332, 644–646. [DOI] [PubMed] [Google Scholar]
- 24.Yoakim, M., Hou, W., Liu, Y., Carpenter, C. L., Kapeller, R. & Schaffhausen, B. S. (1992) J. Virol. 66, 5485–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songyang, Z., Shoelson, S. E., Chaudhuri, M., Gish, G., Pawson, T., Haser, W. G., King, F., Roberts, T., Ratnofsky, S., Lechleider, R. J., et al. (1993) Cell 72, 767–778. [DOI] [PubMed] [Google Scholar]
- 26.Carmichael, G., Schaffhausen, B. S., Mandel, G., Liang, T. J. & Benjamin, T. L. (1984) Proc. Natl. Acad. Sci. USA 81, 679–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markland, W., Oostra, B. A., Harvey, R., Markham, A. F., Colledge, W. H. & Smith, A. E. (1986) J. Virol. 59, 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oostra, B. A., Harvey, R., Ely, B. K., Markham, A. F. & Smith, A. E. (1983) Nature 304, 456–459. [DOI] [PubMed] [Google Scholar]
- 29.Markland, W. & Smith, A. E. (1987) Biochim. Biophys. Acta 907, 299–321. [DOI] [PubMed] [Google Scholar]
- 30.Ong, S. H., Dilworth, S., Hauck-Schmalenberger, I., Pawson, T. & Kiefer, F. (2001) EMBO J. 20, 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Druker, B. J., Ling, L. E., Cohen, B., Roberts, T. M. & Schaffhausen, B. S. (1990) J. Virol. 64, 4454–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Druker, B. J., Sibert, L. & Roberts, T. M. (1992) J. Virol. 66, 5770–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pear, W. S., Nolan, G. P., Scott, M. L. & Baltimore, D. (1993) Proc. Natl. Acad. Sci. USA 90, 8392–8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, C. A. & Okayama, H. (1988) BioTechniques 6, 632–638. [PubMed] [Google Scholar]
- 35.Cherington, V., Morgan, B., Spiegelman, B. M. & Roberts, T. M. (1986) Proc. Natl. Acad. Sci. USA 83, 4307–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pallas, D. C., Schley, C., Mahoney, M., Harlow, E., Schaffhausen, B. S. & Roberts, T. M. (1986) J. Virol. 60, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auger, K. R., Wang, J., Narsimhan, R. P., Holcombe, T. & Roberts, T. M. (2000) Biochem. Biophys. Res. Commun. 272, 822–829. [DOI] [PubMed] [Google Scholar]
- 38.Kashishian, A., Kazlauskas, A. & Cooper, J. A. (1992) EMBO J. 11, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smart, J. E., Oppermann, H., Czernilofsky, A. P., Purchio, A. F., Erikson, R. L. & Bishop, J. M. (1981) Proc. Natl. Acad. Sci. USA 78, 6013–6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patschinsky, T., Hunter, T., Esch, F. S., Cooper, J. A. & Sefton, B. M. (1982) Proc. Natl. Acad. Sci. USA 79, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishimura, R., Li, W., Kashishian, A., Mondino, A., Zhou, M., Cooper, J. & Schlessinger, J. (1993) Mol. Cell. Biol. 13, 6889–6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bradshaw, J. M. & Waksman, G. (1999) Biochemistry 38, 5147–5154. [DOI] [PubMed] [Google Scholar]
- 43.Xu, W., Harrison, S. C. & Eck, M. J. (1997) Nature 385, 595–602. [DOI] [PubMed] [Google Scholar]
- 44.DeMali, K. A., Whiteford, C. C., Ulug, E. T. & Kazlauskas, A. (1997) J. Biol. Chem. 272, 9011–9018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.