Abstract
The t(8;21)(q22;q22) translocation, occurring in 40% of patients with acute myeloid leukemia (AML) of the FAB-M2 subtype (AML with maturation), results in expression of the RUNX1-CBF2T1 [AML1-ETO (AE)] fusion oncogene. AML/ETO may contribute to leukemogenesis by interacting with nuclear corepressor complexes that include histone deacetylases, which mediate the repression of target genes. However, expression of AE is not sufficient to transform primary hematopoietic cells or cause disease in animals, suggesting that additional mutations are required. Activating mutations in receptor tyrosine kinases (RTK) are present in at least 30% of patients with AML. To test the hypothesis that activating RTK mutations cooperate with AE to cause leukemia, we transplanted retrovirally transduced murine bone marrow coexpressing TEL-PDGFRB and AE into lethally irradiated syngeneic mice. These mice (19/19, 100%) developed AML resembling M2-AML that was transplantable in secondary recipients. In contrast, control mice coexpressing with TEL-PDGFRB and a DNA-binding-mutant of AE developed a nontransplantable myeloproliferative disease identical to that induced by TEL-PDGFRB alone. We used this unique model of AML to test the efficacy of pharmacological inhibition of histone deacetylase activity by using trichostatin A and suberoylanilide hydroxamic acid alone or in combination with the tyrosine kinase inhibitor, imatinib mesylate. We found that although imatinib prolonged the survival of treated mice, histone deacetylase inhibitors provided no additional survival benefit. These data demonstrate that an activated RTK can cooperate with AE to cause AML in mice, and that this system can be used to evaluate novel therapeutic strategies.
The t(8;21)(q22;q22) translocation, which fuses the RUNX1 (AML1/PEBPα/CBFA2) gene on chromosome 21 with the ETO (MTG8) gene on chromosome 8, is a common mutation associated with cases of acute myeloid leukemia (AML) of the FAB-M2 subtype (AML with maturation) (1). Expression of the resulting AML1-ETO (AE) fusion gene is detected in 40% of M2-AML patients and 12% of all newly diagnosed cases of AML (2, 3). The correlation between AE expression and the leukemic phenotype strongly suggests a causative role for AE in transformation. AE transcripts have been detected in nonneoplastic progenitors from AML patients in remission, suggesting that the translocation is an early event in the leukemogenic process (4). Furthermore, t(8;21) translocation and AE expression can be detected in neonatal Guthrie blood spots, implying an in utero origin of the translocation preceding development of AML in children by as much as 10 years (5, 6).
Several murine models have demonstrated that AE alone is not sufficient to induce leukemia. Mice expressing an inducible AE transgene in bone marrow cells remained disease-free for a normal life span of 24 mo (7). When expression of AE was targeted to the myeloid lineage by using the human MRP8 promoter, again the mice had no discernable phenotype (8). However, when additional random mutations were introduced by using the powerful mutagen N-ethyl-N-nitrosourea (ENU), roughly one-half of the hMRP8-AE transgenic mice developed an AML-like phenotype (8). Mice in which a Runx1-Eto fusion was generated by an in vivo translocation by using Cre recombinase also failed to develop disease (9). In another model, a conditional knock-in was generated by inserting an inducible AE cDNA within a wild-type Runx1 allele (10). Although this approach produced adult mice haploinsufficient at the Runx1 locus that were expressing AE in bone marrow, these mice did not develop AML unless treated with ENU (10). However, myeloid progenitors from these mice did appear to have increased survival over wild-type progenitors when cultured in the presence of cytokines (10). Retroviral-mediated expression of AE alone in bone marrow also fails to induce leukemia in wild-type mice (11) but contributes to leukemic transformation in interferon consensus sequence-binding protein-deficient mice (12). Taken together, these models suggest that, although AE may provide progenitors with a survival advantage, additional mutations are required to produce an AML phenotype.
Recent data have demonstrated that activating mutations in platelet-derived growth factor (PDGFR) family (type III) receptor tyrosine kinases, including FLT3 and c-KIT, are common in AML patients (13, 14). In addition, an individual diagnosed with t(5;12)-positive chronic myelomonocytic leukemia (CMML) subsequently developed AML with the t(8;21) (15). The t(5;12) translocation associated with CMML fuses the TEL (ETV6) gene to the PDGFβ receptor (PDGFβR/PDGFRB) gene, generating a fusion protein with constitutive tyrosine kinase activity. Thus, the development of an M2-AML phenotype in this patient correlated with coexpression of TEL-PDGFRB (TP) and AE.
We wanted to directly determine whether TP could cooperate with AE to induce AML. Here, we demonstrate that mice transplanted with bone marrow cells expressing both of these fusion oncogenes developed many features of human M2-AML. Malignant blasts from these mice were easily transplanted into secondary recipients. Previous studies have suggested that AML1/ETO may promote leukemogenesis by repressing target gene expression through the recruitment of nuclear corepressors, including histone deacetylases (HDAC) (16–20). However, HDAC inhibitors, trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA), do not ameliorate disease progression in our model.
Materials and Methods
Mice. Balb/c mice and B6129 F1 mice 6–8 weeks of age were purchased from Taconic Farms. Runx1 (Cbfa2) heterozygous mice were generously provided by Nancy Speck (Dartmouth Medical School). All mice were maintained in an accredited animal facility according to proper institutional guidelines.
Cell Culture. 293T cells, NIH 3T3 cells, and TP-tag 32D cells were grown in RPMI medium 1640, 10% FCS, and 1× Pen/Strep in a humidified incubator at 37°C, 5% CO2. Primary murine bone marrow mononuclear cells were plated in transplant medium consisting of DMEM, 20% FCS, and 1× Pen/Strep with 10 ng/ml stem cell factor, 6 ng/ml IL-3, 10 ng/ml thrombopoietin, and 50 ng/ml Flt-3.
Plasmids and Constructs. The pMSCV-TP-AE (MSCV, murine stem cell virus) and pMSCV-TP-148 were generated as follows. The TP and AE cDNAs were first subcloned into the EcoRI and XbaI sites of pIRES (IRES, internal ribosome entry site), respectively (CLONTECH). This construct was digested with NheI and subsequently incubated with Klenow fragment (Promega) to generate a blunt end. The TP-IRES-AE cassette was then purified from this construct following digestion with SalI. TP-IRES-AE was ligated into HpaI/SalI-digested pMSCV-neoEB (CLONTECH). pMSCV-AE-eGFP (eGFP, enhanced GFP) was produced by cloning the AE cDNA as a blunt fragment into the HpaI of pMSCV-IRES-eGFP. The TP cDNA was subcloned into the EcoRI site of MSCV-IRES-eGFP to yield pMSCV-TP-eGFP. The AE and AE(148) cDNAs were generous gifts from S. W. Heibert (Vanderbilt University, Nashville, TN).
Retrovirus Production. Retrovirus was produced with the constructs above by individually cotransfecting them with Ecopac (Cell Genesys, Foster City, CA) into 293T cells by using either Superfect reagent (Qiagen, Chatsworth, CA) or calcium phosphate precipitation. Retrovirus was subsequently collected in the conditioned medium. Viral titer was estimated by quantitative PCR by using DNA purified from transduced 3T3 cells.
Retroviral Transduction/Bone Marrow Transplantation. The method of transducing bone marrow cells with retrovirus has been described (21). Briefly, bone marrow mononuclear cells were isolated from the femurs and tibias of donor mice pretreated with 200 mg/kg 5-fluorouracil. Bone marrow cells were cultured in transplant medium for 48 h. The cells were then transduced by two rounds of centrifugation with retroviral stock. Syngeneic recipient mice conditioned with 1,000 cGy were transplanted intravenously with ≈1 × 106 unfractionated transduced bone marrow cells.
Preparation of Tissue from Normal and Diseased Mice. Moribund mice were killed, and necropsies were performed. Peripheral blood was isolated to prepare smears and obtain complete blood counts by using a Hemavet (CDC Technologies, Oxford, CT). Spleen weight was recorded. Various internal organs, neoplastic tissue, and tibias were placed in 10% buffered formalin and processed to obtain paraffin sections for histological staining with hematoxylin/eosin. Splenocytes, prepared by homogenizing a portion of the spleen followed by red cell lysis, were frozen in FCS/10% DMSO for subsequent analysis. Differentials were obtained from cytospin preparations of total bone marrow cells stained with Wright–Giemsa. Single-cell suspensions of total bone marrow were also stored in freezing medium for further analysis.
Detection of TEL/PDGFβR and AML1/ETO Protein. Whole cell protein extracts from bone marrow cells of moribund mice were prepared by boiling directly in 1× Laemmli sample buffer. The extracts were analyzed by Western blotting with PDGFRB-specific antibodies (BD Biosciences) and anti-ETO antibodies (generously provided by P. Erickson, Vanderbilt University). Blots were stripped and reprobed with antibodies raised against β-actin (Sigma) to control for protein loading.
Treatment of Secondary Recipients. Secondary transplants were performed by injecting freshly isolated bone marrow or splenocytes intravenously into sublethally irradiated (500 cGy) B6129 F1 female mice. Cohorts of MSCV-TP-AE secondary recipients were treated daily with i.p. injection of vehicle (PBS, 10% DMSO), 1 mg/kg TSA (Sigma), 50 mg/kg imatinib (Gleevec; Novartis, Basel), or TSA plus imatinib. Another group of secondary recipients was treated with vehicle, 2 mg/kg TSA per day, or 5 mg/kg TSA per day. In a third secondary transplant, treatment with 5 mg/kg/day TSA was compared with 50 mg/kg SAHA per day (Aton Pharma, Tarrytown, NY). Mice were treated until disease was apparent and were analyzed as described above.
Kaplan–Meier Analysis. Kaplan–Meier plots were generated on groups of mice on the basis of cumulative survival after transplantation by using STATVIEW software (SAS Institute, Cary, NC).
Flow Cytometry. Single cell suspensions of bone marrow cells were washed once each with PBS and Flow Buffer (PBS, 0.1% BSA). Then, 5 × 105 cells in 0.1 ml of Flow Buffer were incubated either alone or with appropriate antibodies to detect the following murine antigens: CD34, Gr-1, CD11b, CD 117 (c-Kit), Sca-1, B220, CD3, and Ter-119 (BD PharMingen). After incubation on ice for 1 h, cells were washed three times with Flow Buffer and subjected to flow cytometry on a FACScan (BD Biosciences) or a MoFlo (DAKO/Cytomation). Analysis was performed on 7-aminoactinomycin D-negative cells by using either CELLQUEST (BD Biosciences) or F LOJO software (Tree Star, San Carlos, CA). Quadrants were set on the basis of isotype control antibodies conjugated to phycoerythrin, FITC, or allophycocyanin.
Southern Blotting. Genomic DNA was isolated from splenocytes of analyzed mice by using Puregene (Gentra Systems). DNA was digested with XbaI or XhoI and analyzed by Southern blotting (22). Blots were hybridized with a 0.6 kb of EcoNI probe (from the Ψ region of MSCV-IRES-GFP) that was labeled with the Rediprime II kit (Amersham Pharmacia). After washing, hybridizing bands were visualized by autoradiography.
Analysis of Histone Acetylation. Histones were purified from bone marrow of B6129 mice 2 h after treatment with vehicle, TSA, or SAHA, as described (23). Acid-extracted histones (2.5 μg) were analyzed for acetylated histone H4 and total histone H4 protein by Western blotting with the appropriate antibodies (Upstate Biotechnology, Lake Placid, NY).
Results
Retroviral-Mediated Expression of AE Alone Is Not Sufficient to Induce Leukemia in Wild-Type or Runx1+/- Mice. We initially transduced bone marrow mononuclear cells isolated from both wild-type (Runx1+/+) and Runx1+/- mice with a retroviral construct capable of coexpressing AE and an eGFP marker from a single transcript by using the IRES of the encephalomyocarditis virus (MSCV-AE-eGFP, Fig. 1A). The murine stem cell virus (MSCV) backbone was used because of its documented ability to maintain expression in murine hematopoietic stem cells as well as multiple hematopoietic lineages (24). Transduction efficiency in our laboratory has been documented to be at least 40% (data not shown). Wild-type mice were compared with Runx1+/- mice to determine whether Runx1 haploinsufficiency in the presence of high-level retroviral expression of AML1/ETO would contribute to a disease phenotype (25). Transduced cells were then transplanted into lethally irradiated syngeneic recipients and monitored for disease.
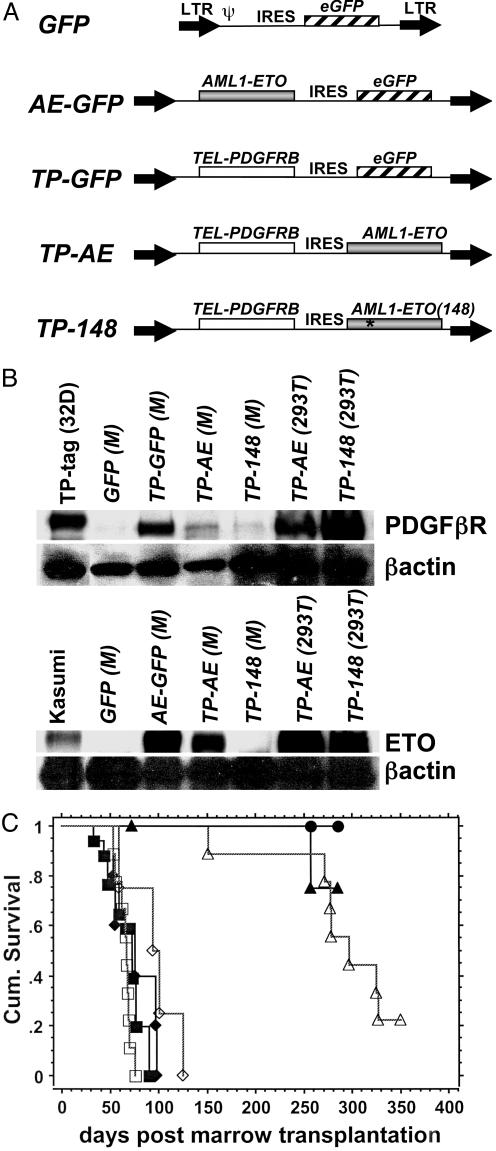
Fig. 1.
Rapid mortality in mice coexpressing TP and AE. (A) Structure of retroviral constructs derived from the MSCV vector. LTR, long terminal repeats; Ψ, packaging signal. (B) Expression of retroviral constructs. Whole cell protein extracts from bone marrow (M) of the indicated mice or transfected 293T cells were analyzed by immunoblotting with antibodies to PDGFβR, ETO, or β-actin. TP-tag (32D), 32D cell line expressing His-tagged TEL-PDFGβR; Kasumi, AML1/ETO expressing cell line from a M2-AML patient. (C) Kaplan–Meier plot showing survival of mice after transplantation with bone marrow cells transduced with the retroviral constructs. MSCV-eGFP in wild-type B6129 mice (•, n = 4); MSCV-AE-eGFP in B6129 wild-type +/+ (▴, n = 5) or Runx1+/- mice (▵, n = 8); MSCV-TP-AE in +/+ (▪, n = 10) or Runx1+/- (□, n = 9); and MSCV-TP-148, +/+ (♦, n = 10), Runx1+/- (⋄, n = 5).
At 2 months posttransplant, MSCV-AE-eGFP mice appeared healthy and displayed normal hematopoiesis in the peripheral blood and bone marrow (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org), despite expression of AML1/ETO fusion protein in the bone marrow (Fig. 1B). Percentages of MSCV-AE-eGFP bone marrow cells staining positive for the myeloid marker, Gr-1, and progenitor markers, Sca-1 and CD 117 (c-Kit), were similar to MSCV-eGFP controls (see Fig. 5). After a latency period of many months, several MSCV-AE-eGFP mice developed signs of illness including splenomegaly, thrombocytopenia, and anemia regardless of Runx1 haploinsufficiency (Fig. 1C and data not shown). These mice demonstrated increased splenic extramedullary hematopoiesis and abnormal myelopoiesis in the bone marrow but without distinct morphologic features of AML (data not shown and Fig. 5). At 9 months posttransplant, the percentage of Gr-1+/GFP+ cells in the bone marrow increased dramatically as shown by flow cytometry (see Fig. 5). These marrow cells displayed a range of Gr-1 expression, reflecting the myelodysplasia observed, and had increased expression of Sca-1 and CD 117 (see Fig. 5). These findings are consistent with those of de Guzman et al. (11), who recently reported that retroviral expression of AE failed to induce leukemia. The survival of our MSCV-AE-eGFP mice and the documentation of normal hematopoiesis before the development of disease are consistent with results from other investigators and suggest that additional mutations are required for leukemic transformation.
Coexpression of TP and AE Rapidly Generates a Transplantable AML That Requires an Intact AML1/ETO RUNX1 DNA-Binding Domain. We next used our retroviral system to test whether an additional specific mutation could cooperate with AE to induce leukemia in vivo. We chose to examine TP based on the clinical evidence that it was reported to be coexpressed with AE in a t(5;21)-positive chronic myelomonocytic leukemia patient who subsequently developed AML (15). Retroviral expression of TP alone in mouse bone marrow results in a rapidly fatal myeloproliferative disease characterized by splenomegaly, anemia, thrombocytopenia, and an excess of mature neutrophils in various organs and the peripheral blood (21). We hypothesized that AE would cooperate with TP to disrupt myeloid differentiation resulting in an accumulation of immature myeloid cells characteristic of leukemia.
Bone marrow cells isolated from either wild-type or Runx1+/- mice were transduced with a MSCV retroviral vector designed to coexpress TP and AE (MSCV-TP-AE, Fig. 1 A), which lacks eGFP. These transduced cells were transplanted into lethally irradiated syngeneic wild-type recipients. In sharp contrast to mice expressing only AE, MSCV-TP-AE-expressing mice rapidly developed lethal disease within 2 months posttransplant, irrespective of Runx1 genotype (Fig. 1 B and C). MSCV-TP-AE mice presented with clinical characteristics including splenomegaly, hepatomegaly, anemia, thrombocytopenia, and moderately elevated white blood cells counts (mean, 54; range, 6.7–131.9; n = 9). Although mice expressing TP alone have similar gross morphology, their white blood counts are extremely high (21). Analysis of bone marrow from MSCV-TP-AE mice revealed a striking predominance of blasts, ranging from 20% to 60%, having slightly basophilic staining cytoplasm, prominent nucleoli, numerous vacuoles, and abnormal granules, all characteristics of AE-positive patient blasts (Fig. 2A) (3). The normal splenic architecture in MSCV-TP-AE mice was disrupted by foci of myeloid blasts (see Fig. 6, which is published as supporting information on the PNAS web site). The neoplastic myeloid blasts also infiltrated other organs including the liver (Fig. 6). The accumulation of myeloid blasts in MSCV-TP-AE mice clearly differed from the accumulation of mature neutrophils in mice expressing TP alone (Fig. 2 A and B) (21). Mesenteric masses were observed in 7/19 MSCV-TP-AE mice due to infiltration of blasts and lymphocyte proliferation. Analysis of DNA isolated from the spleens of MSCV-TP-AE mice by Southern blotting revealed multiple integrations of the retroviral construct suggesting that the blasts were oligoclonal in nature (see Fig. 7, which is published as supporting information on the PNAS web site). Therefore, in contrast to the phenotypes observed when either oncogene is expressed alone, coexpression of AE and TP resulted in the fatal accumulation of neoplastic cells with myeloid blast morphology.
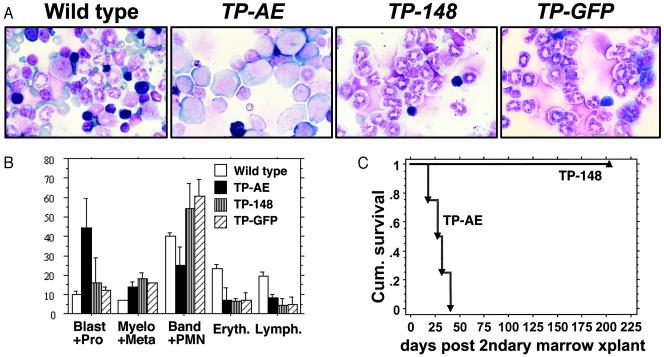
Fig. 2.
MSCV-TP-AE mice develop transplantable AML. (A) Bone marrow cells stained with May–Grunwald/Giemsa. (B) Bone marrow differential counts. Averages of the different populations based on 200 total cells are shown ±SD. Wild type, n = 3; MSCV-TP-AE, n = 5; MSCV-TP-148, n = 5, MSCV-TP-GFP, n = 3. Blast+Pro, myeloblasts and promyelocytes; Myelo+Meta, myelocytes and metamyelocytes; Band+PMN, band and mature neutrophils; Eryth., erythroid precursors; Lymph., lymphocytes. (C) Survival of MSCV-TP-AE or MSCV-TP-148 secondary recipients. MSCV-TP-AE secondary recipients (▾, n = 5) develop a rapidly fatal myeloid leukemia similar to the primary recipients demonstrating transplantability of the leukemic phenotype. However, recipients of MSCV-TP-148 splenocytes remain disease-free (▴, n = 10).
We also assessed the ability of TP to cooperate with an AE mutant, containing a point mutation (L148D) in the DNA-binding RUNX1 homology domain (MSCV-TP-148, Fig. 1 A). This mutant lacks the ability to bind DNA and fails to repress transcription in transient transfection assays (26, 27). MSCV-TP-148 mice developed a fatal myeloproliferative disorder (Fig. 1C) characterized by extreme leukocytosis (white blood count: mean, 174; range, 80.6–395.8; n = 7), splenomegaly, anemia, and thrombocytopenia (data not shown), identical to the phenotype reported in mice expressing TP alone (21). Similar to MSCV-TP-eGFP mice, MSCV-TP-148 mice were found to have an excess of mature neutrophils in the bone marrow (Fig. 2 A and B) and various organs, including the spleen and liver (see Fig. 6). Unlike the focal pattern of the immature myeloid blasts in the MSCV-TP-AE mice, the mature myeloid cells diffusely infiltrated the organs of the MSCV-TP-148 mice (see Fig. 6). Mesenteric masses that contained mature neutrophils and proliferating lymphocytes were also found in 3/8 MSCV-TP-148 mice. Thus, an intact RUNX1 DNA-binding domain is critical for AE to induce acute leukemia in this model system.
TP-induced myeloproliferative disease (MPD) is not transplantable in secondary recipients (21). We used transplantation into secondary recipients as an in vivo functional assay to further characterize cooperation between AE and TP. Freshly isolated splenocytes from moribund MSCV-TP-AE or MSCV-TP-148 mice were injected into sublethally irradiated (500-cGy) secondary recipients. These mice were then monitored for the development of disease. Secondary recipients of MSCV-TP-AE splenocytes rapidly developed a leukemic phenotype identical to that seen in primary mice (Fig. 2C). Tertiary recipients of MSCV-TP-AE splenocytes also developed AML (data not shown). In contrast to MSCV-TP-AE-induced leukemia, the MPD in MSCV-TP-148 mice was not transplantable in secondary recipients (Fig. 2C). These results demonstrate that TP and AE cooperate to induce a transplantable AML in mice.
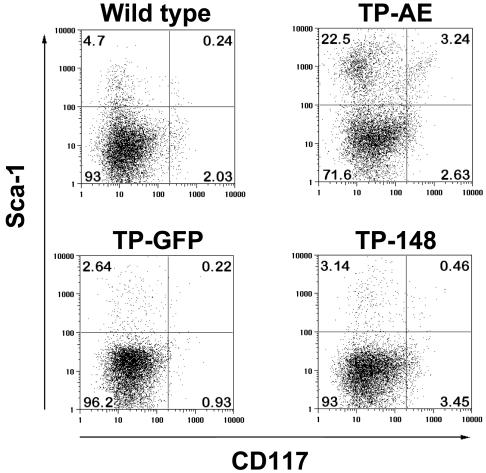
Leukemic Blasts in TP-AE Mice Have an Immature Myeloid Phenotype. We next performed flow cytometry to further characterize the blasts in our transplanted mice. The MSCV-TP-AE construct does not contain a f luorescent marker; thus, viable (7-aminoactinomycin D-negative) cells were gated by forward and side scatter. When compared with wild-type mice, bone marrow cells from the MSCV-TP-AE mice had an increased percentage of Mac-1-expressing cells (see Fig. 8, which is published as supporting information on the PNAS web site). Sorting of MSCV-TP-AE bone marrow revealed that the cells having a blast-like morphology were predominantly in the Gr-1lo/Mac-1+ and Gr-1-/Mac-1+ populations, suggestive of an immature myeloid morphology (data not shown). In contrast, the majority of cells in the bone marrow of MSCV-TP-148 mice were Gr-1hi/Mac-1+ (74.8%), consistent with the accumulation of mature neutrophils (see Fig. 8). MSCV-TP-AE bone marrow cells also had increased CD34 expression, characteristic of progenitor cells, which was not observed in MSCV-TP-148 mice (see Fig. 8). Most striking was the increase in the percentage of Sca-1+/CD 117- and Sca-1+/CD 117+ cells in the TP-AE mice (Fig. 3). These data support the morphologic findings that cells with an immature myeloid phenotype are accumulating in MSCV-TP-AE mice.
Fig. 3.
Neoplastic cells from MSCV-TP-AE mice have an immature myeloid immunophenotype. Bone marrow cells isolated from MSCV-TP-AE, MSCV-TP-148, MSCV-TP-eGFP, and wild-type mice were analyzed by flow cytometry with antibodies to detect expression of the progenitor cell markers, CD 117 (c-Kit) and Sca-1.
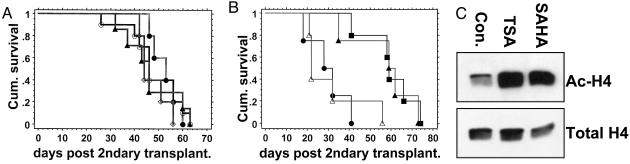
MSCV-TP-AE Secondary Recipients Provide a Useful Preclinical Model of M2-AML. We next used our model system to test the effects of pharmacological doses of HDAC inhibitors on the development of MSCV-TP-AE-induced leukemia. We hypothesized that the HDAC inhibitors TSA and SAHA would prevent the proposed chromatin modifications mediated by the AML1/ETO–HDAC association to effectively delay or prevent disease progression. TSA (1 mg/kg per day) has been previously shown to block tumor-induced angiogenesis and tumor growth in mice (28, 29). SAHA has been reported to delay disease progression in a mouse model of acute promyelocytic leukemia (30). Doses of TSA at 2 mg/kg per day and 5 mg/kg per day had no effect on disease progression in MSCV-TP-AE secondary recipients (Fig. 4A). Treatment of secondary recipients with SAHA (50 mg/kg per day) was likewise ineffective in preventing disease progression (Fig. 4A). Thus, MSCV-TP-AE secondary recipient mice rapidly developed leukemia that was not influenced by pharmacological doses of HDAC inhibitors.
Fig. 4.
Survival of MSCV-TP-AE secondary recipients treated with HDAC inhibitors and/or imatinib mesylate. (A) B6129 F1 secondary recipient mice were transplanted with splenocytes from a primary MSCV-TP-AE mouse and then treated daily with vehicle (DMSO, ▴), 2mg/kg TSA (•), 5mg/kg TSA (○), or 50 mg/kg SAHA (⋄) per day. (B) B6129 F1 secondary recipient mice were transplanted with splenocytes from a primary MSCV-TP-AE mouse and then treated daily with vehicle (10% DMSO/PBS) (•), 1mg/kg TSA per day (▵), 50 mg/kg imatinib per day (▪), or TSA plus imatinib (▴)(n = 5 for each arm). (C) Acetylation of histone H4 in the bone marrow of mice 2 h after treatment with vehicle, 5 mg/kg TSA per day, or 50 mg/kg SAHA per day.
We then hypothesized that leukemia generated in our model by two distinct mutations may respond to combination therapy with an HDAC inhibitor and imatinib mesylate, a tyrosine kinase inhibitor. Imatinib has previously been shown to improve the survival of mice with TP-induced malignancies (31). MSCV-TP-AE secondary recipients were divided into cohorts that received daily doses of vehicle, TSA (1 mg/kg) alone, imatinib (50 mg/kg) alone, or a combination of imatinib and TSA. Mice treated with TSA alone had no increase in survival when compared with controls (Fig. 4B). Mice treated with imatinib alone or imatinib plus TSA had significantly increased survival over mice treated with TSA alone or controls (Fig. 4B). Yet, the combination of imatinib and TSA was no more effective than imatinib alone (Fig. 4B). To verify that doses of TSA and SAHA were acting in vivo to inhibit HDAC and to promote histone acetylation, we analyzed acetylation of histone H4 in bone marrow of wild-type B6129 mice treated with vehicle, TSA, or SAHA. Hyperacetylation of H4 was detected in strain-matched wild-type mice in response to TSA and SAHA treatments, demonstrating effective in vivo concentrations of these drugs (Fig. 4C).
Discussion
Here, we present the previously undescribed findings that coexpression of an activated receptor tyrosine kinase, TEL/PDGFβR, with AML1/ETO in primary hematopoietic cells induces AML. In contrast, retroviral-mediated overexpression of AE alone in bone marrow cells resulted in a disease most similar to a myelodysplastic syndrome with refractory anemia that developed only after a long latency and was not influenced by Runx1 haploinsufficiency. A similar syndrome, termed oligoblastic leukemia or t(8;21)-positive myelodysplastic syndrome, has been described in patients with the t(8;21) that do not have frank leukemia (32–34). Mice expressing both AE and TP (MSCV-TP-AE) rapidly developed a transplantable leukemia characterized by an infiltration of myeloid blasts in the bone marrow and other organs. This leukemic phenotype differed significantly from the nontransplantable myeloproliferative disease generated by coexpression of TP with an AE DNA binding mutant (MSCV-TP-148) that was identical to mice expressing TP alone (21).
The rapid onset and mortality of the leukemia induced by TP and AE suggest these two mutations are sufficient to generate AML. However, our data do not exclude the possibility of other spontaneous mutations occurring within this short time frame. In addition, insertional mutagenesis mediated by integration of the retroviral construct could potentially constitute an oncogenic event, although MSCV is presumed to integrate randomly. The competitive emergence of oligoclonal leukemic blasts in the presence of nontransduced progenitors provides further support for sufficiency.
The MSCV-TP-AE myeloid blasts displayed morphologic features that have been reported in patients with AE-positive M2-AML (3). Flow cytometric data also suggested an immature myeloid phenotype of a significant percentage of cells in the bone marrow of MSCV-TP-AE mice. However, the immunophenotyping also demonstrated some heterogeneity within the leukemic population. For example, of the 25% of MSCV-TP-AE bone marrow cells that were Sca-1+, only 3.2% were also CD117+. This may reflect a subpopulation of leukemic stem cells that give rise to slightly more differentiated leukemic blasts as hypothesized by Bonnet and Dick (35).
Tissue culture models have suggested AML1/ETO may exert its dominant negative effects by interacting with nuclear corepressors, including HDACs (16–20). Corepressor interaction domains were found to be critical for AML1/ETO to block the differentiation of hematopoietic progenitors into mature granulocytes in vitro (36). A frequently cited implication of this model is that pharmacological inhibition of HDAC activity would be anticipated to inhibit AE-associated leukemia. However, administration of TSA into mice secondarily transplanted with mononuclear cells from MSCV-TP-AE mice did not result in clinical improvement at doses up to 5 mg/kg/day. Likewise, treatment of secondary recipients with SAHA was ineffective in preventing disease progression. Perhaps the chromatin modifications initiated by AML1/ETO represent only an initial step in a series of chromatin modifications in the promoters of target genes. For example, AML1/ETO-mediated histone deacetylation through HDAC recruitment may be followed by additional silencing chromatin modification, such as DNA methylation, that may not respond to HDAC inhibition alone. In the case of another myeloid leukemia-associated fusion protein, PML-RARα, HDAC recruitment does appear to be followed by recruitment of DNA methyltransferases (37). Another explanation may be that the unique contributions of the cooperating TP mutation induced additional chromatin modifications that rendered these HDAC inhibitors ineffective. MSCV-TP-AE mice provide a tool to dissect the molecular mechanisms through which activated receptor tyrosine kinases and AE cooperate to induce AML and to evaluate new therapeutic strategies for M2-AML.
Supplementary Material
Acknowledgments
We thank S. W. Hiebert for the AE and AE(148) cDNAs. We are also grateful for the Runx1+/- mice provided by N. Speck. Dr. Marie LaRegina provided expert pathological analysis of tissue specimens from affected mice. This work was supported by National Institutes of Health Grants NIH NHLBI T32 HL07088 (to J.L.G.) and CA81197-04 (to M.H.T.) and a grant from the Leukemia and Lymphoma Society of America (to M.H.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TP, TEL-PDGFRB; AE, AML1-ETO; AML, acute myeloid leukemia; MSCV, murine stem cell virus; HDAC, histone deacetylase; TSA, trichostatin A; SAHA, suberoylanilide hydroxamic acid; PDGFR, platelet-derived growth factor receptor; IRES, internal ribosome entry site; eGFP, enhanced GFP.
References
- 1.Miyoshi, H., Shimizu, K., Kozu, T., Maseki, N., Kaneko, Y. & Ohki, M. (1991) Proc. Natl. Acad. Sci. USA 88, 10431-10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Langabeer, S. E., Walker, H., Rogers, J. R., Burnett, A. K., Wheatley, K., Swirsky, D., Goldstone, A. H. & Linch, D. C. (1997) Br. J. Haematol. 99, 925-928. [DOI] [PubMed] [Google Scholar]
- 3.Nucifora, G., Dickstein, J. I., Torbenson, V., Roulston, D., Rowley, J. D. & Vardiman, J. W. (1994) Leukemia 8, 1533-1538. [PubMed] [Google Scholar]
- 4.Miyamoto, T., Weissman, I. L. & Akashi, K. (2000) Proc. Natl. Acad. Sci. USA 97, 7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mori, H., Colman, S. M., Xiao, Z., Ford, A. M., Healy, L. E., Donaldson, C., Hows, J. M., Navarrete, C. & Greaves, M. (2002) Proc. Natl. Acad. Sci. USA 99, 8242-8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiemels, J. L., Xiao, Z., Buffler, P. A., Maia, A. T., Ma, X., Dicks, B. M., Smith, M. T., Zhang, L., Feusner, J., Wiencke, J., et al. (2002) Blood 99, 3801-3805. [DOI] [PubMed] [Google Scholar]
- 7.Rhoades, K. L., Hetherington, C. J., Harakawa, N., Yergeau, D. A., Zhou, L., Liu, L. Q., Little, M. T., Tenen, D. G. & Zhang, D. E. (2000) Blood 96, 2108-2115. [PubMed] [Google Scholar]
- 8.Yuan, Y., Zhou, L., Miyamoto, T., Iwasaki, H., Harakawa, N., Hetherington, C. J., Burel, S. A., Lagasse, E., Weissman, I. L., Akashi, K., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz, F., Refaeli, Y., Trumpp, A. & Bishop, J. M. (2000) EMBO Rep. 1, 133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi, M., O'Brien, D., Kumaravelu, P., Lenny, N., Yeoh, E. J. & Downing, J. R. (2002) Cancer Cells 1, 63-74. [DOI] [PubMed] [Google Scholar]
- 11.de Guzman, C. G., Warren, A. J., Zhang, Z., Gartland, L., Erickson, P., Drabkin, H., Hiebert, S. W. & Klug, C. A. (2002) Mol. Cell. Biol. 22, 5506-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwieger, M., Lohler, J., Friel, J., Scheller, M., Horak, I. & Stocking, C. (2002) J. Exp. Med. 196, 1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilliland, D. G. & Griffin, J. D. (2002) Curr. Opin. Hematol. 9, 274-281. [DOI] [PubMed] [Google Scholar]
- 14.Beghini, A., Peterlongo, P., Ripamonti, C. B., Larizza, L., Cairoli, R., Morra, E. & Mecucci, C. (2000) Blood 95, 726-727. [PubMed] [Google Scholar]
- 15.Golub, T. R., Barker, G. F., Lovett, M. & Gilliland, D. G. (1994) Cell 77, 307-316. [DOI] [PubMed] [Google Scholar]
- 16.Amann, J. M., Nip, J., Strom, D. K., Lutterbach, B., Harada, H., Lenny, N., Downing, J. R., Meyers, S. & Hiebert, S. W. (2001) Mol. Cell. Biol. 21, 6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelmetti, V., Zhang, J., Fanelli, M., Minucci, S., Pelicci, P. G. & Lazar, M. A. (1998) Mol. Cell. Biol. 18, 7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiebert, S. W., Lutterbach, B. & Amann, J. (2001) Curr. Opin. Hematol. 8, 197-200. [DOI] [PubMed] [Google Scholar]
- 19.Wang, J., Hoshino, T., Redner, R. L., Kajigaya, S. & Liu, J. M. (1998) Proc. Natl. Acad. Sci. USA 95, 10860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang, J., Hug, B. A., Huang, E. Y., Chen, C. W., Gelmetti, V., Maccarana, M., Minucci, S., Pelicci, P. G. & Lazar, M. A. (2001) Mol. Cell. Biol. 21, 156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomasson, M. H., Sternberg, D. W., Williams, I. R., Carroll, M., Cain, D., Aster, J. C., Ilaria, R. L., Jr., Van Etten, R. A. & Gilliland, D. G. (2000) J. Clin. Invest. 105, 423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Southern, E. M. (1992) Biotechnology 24, 122-139. [PubMed] [Google Scholar]
- 23.Yoshida, M., Kijima, M., Akita, M. & Beppu, T. (1990) J. Biol. Chem. 265, 17174-17179. [PubMed] [Google Scholar]
- 24.Hawley, R. G., Fong, A. Z., Burns, B. F. & Hawley, T. S. (1992) J. Exp. Med. 176, 1149-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song, W. J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I. C., Haworth, C., Hock, R., et al. (1999) Nat. Genet. 23, 166-175. [DOI] [PubMed] [Google Scholar]
- 26.Lenny, N., Meyers, S. & Hiebert, S. W. (1995) Oncogene 11, 1761-1769. [PubMed] [Google Scholar]
- 27.Linggi, B., Muller-Tidow, C., van de Locht, L., Hu, M., Nip, J., Serve, H., Berdel, W. E., van der Reijden, B., Quelle, D. E., Rowley, J. D., et al. (2002) Nat. Med. 8, 743-750. [DOI] [PubMed] [Google Scholar]
- 28.Kim, M. S., Kwon, H. J., Lee, Y. M., Baek, J. H., Jang, J. E., Lee, S. W., Moon, E. J., Kim, H. S., Lee, S. K., Chung, H. Y., et al.. (2001) Nat. Med. 7, 437-443. [DOI] [PubMed] [Google Scholar]
- 29.Vigushin, D. M., Ali, S., Pace, P. E., Mirsaidi, N., Ito, K., Adcock, I. & Coombes, R. C. (2001) Clin. Cancer Res. 7, 971-976. [PubMed] [Google Scholar]
- 30.He, L. Z., Tolentino, T., Grayson, P., Zhong, S., Warrell, R. P., Jr., Rifkind, R. A., Marks, P. A., Richon, V. M. & Pandolfi, P. P. (2001) J. Clin. Invest. 108, 1321-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasson, M. H., Williams, I. R., Hasserjian, R., Udomsakdi, C., McGrath, S. M., Schwaller, J., Druker, B. & Gilliland, D. G. (1999) Blood 93, 1707-1714. [PubMed] [Google Scholar]
- 32.Xue, Y., Yu, F., Zhou, Z., Guo, Y., Xie, X. & Lin, B. (1994) Leuk. Res. 18, 761-765. [DOI] [PubMed] [Google Scholar]
- 33.Taj, A. S., Ross, F. M., Vickers, M., Choudhury, D. N., Harvey, J. F., Barber, J. C., Barton, C. & Smith, A. G. (1995) Br. J. Haematol. 89, 890-892. [DOI] [PubMed] [Google Scholar]
- 34.Kojima, K., Omoto, E., Hara, M., Sasaki, K., Katayama, Y., Nawa, Y., Kimura, Y., Azuma, T., Takimoto, H. & Harada, M. (1998) Ann. Hematol. 76, 279-282. [DOI] [PubMed] [Google Scholar]
- 35.Bonnet, D. & Dick, J. E. (1997) Nat. Med. 3, 730-737. [DOI] [PubMed] [Google Scholar]
- 36.Hug, B. A., Lee, S. Y., Kinsler, E. L., Zhang, J. & Lazar, M. A. (2002) Cancer Res. 62, 2906-2912. [PubMed] [Google Scholar]
- 37.Di Croce, L., Raker, V. A., Corsaro, M., Fazi, F., Fanelli, M., Faretta, M., Fuks, F., Lo Coco, F., Kouzarides, T., Nervi, C., et al. (2002) Science 295, 1079-1082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.