Abstract
Iron is limiting in the human host, and bacterial pathogens respond to this environment by activating genes required for bacterial virulence. Transcriptional regulation in response to iron in Gram-negative bacteria is largely mediated by the ferric uptake regulator protein Fur, which in the presence of iron binds to a specific sequence in the promoter regions of genes under its control and acts as a repressor. Here we describe DNA microarray, computational and in vitro studies to define the Fur regulon in the human pathogen Neisseria meningitidis group B (strain MC58). After iron addition to an iron-depleted bacterial culture, 153 genes were up-regulated and 80 were down-regulated. Only 50% of the iron-regulated genes were found to contain Fur-binding consensus sequences in their promoter regions. Forty-two promoter regions were amplified and 32 of these were shown to bind Fur by gel-shift analysis. Among these genes, many of which had never been described before to be Fur-regulated, 10 were up-regulated on iron addition, demonstrating that Fur can also act as a transcriptional activator. Sequence alignment of the Fur-binding regions revealed that the N. meningitidis Fur-box encompasses the highly conserved (NATWAT)3 motif. Cluster analysis was effective in predicting Fur-regulated genes even if computer prediction failed to identify Fur-box-like sequences in their promoter regions. Microarray-generated gene expression profiling appears to be a very effective approach to define new regulons and regulatory pathways in pathogenic bacteria.
Iron starvation is used by many pathogens as a signal that they are in a host environment resulting in the expression of virulence factors that are transcriptionally regulated by iron through the ferric uptake regulator protein Fur. Fur forms a dimer together with ferrous iron and binds to a consensus sequence (Fur-box) that overlaps the promoters of iron-regulated genes, resulting in the inhibition of transcription. Fur homologues have been identified in many Gram-negative and -positive bacteria, including important human pathogens (1–5). Recent studies indicate that Fur functions as a regulator of genes involved in a variety of cellular processes such as acid shock response, chemotaxis, metabolic pathways, bioluminescence, and the production of toxins and virulence factors (6–11). In some organisms Fur may also act as a positive regulator in controlling gene expression (12–15), although interactions between the Fur protein and the operator region of the iron-activated genes have not been studied in detail.
The pathogenic neisseriae also appear to regulate the expression of several genes in response to iron, including specific virulence factors (16). However, studies on the Fur-dependent iron regulation of Neisseria genes have been hampered because of the inability to construct a fur null mutant. A missense mutant of Neisseria gonorrhoeae fur was shown to be altered in the iron regulation of a broad range of genes supporting the existence of a large set of genes that may be under Fur control (17). Our more recent studies in N. gonorrhoeae have established that the gonococcal Fur protein binds to the promoter regions of several genes involved in diverse metabolic pathways (18). These initial reports led us to postulate that pathogenic neisseriae possess a broad array of genes whose expression is under the control of the transcriptional regulatory protein Fur. To further define the gene repertoire that is regulated by iron and Fur in neisseriae, in the present study we used DNA microarray technology to monitor the kinetics of expression of the entire gene repertoire of Neisseria meningitidis MC58 (19, 20) in response to iron.
Experimental Procedures
Growth Conditions. N. meningitidis MC58 cultures were grown in chemically defined medium with 12.5 μM desferal (iron-depleted) for 3 h. After this adaptation to iron starvation, half of the culture was supplemented with 100 μM ferric nitrate, and growth continued for a 5-h period. Aliquots of the two cultures were removed at different time points, and bacterial RNA was prepared as described below.
Microarray Procedures, Hybridization, and Analysis. DNA microarray analysis was performed as described (19). The hybridization probe was constituted by a mixture of the differently labeled cDNA derived from bacterial cultures grown under iron-replete or -depleted conditions. For each image, the signal value of each spot was determined by subtracting the mean pixel intensity of the background value and normalizing to the median of all spot signals. The spots, which gave a negative value after background subtraction, were arbitrarily assigned the standard deviation value of negative controls. The data resulting from direct and reverse labeling were averaged for each spot. Expression ratios were obtained at each time point by the direct comparison of RNA obtained from bacterial cultures grown in the presence of iron with RNA obtained from bacterial cultures grown in the presence of desferal. The data of each time point represent the average of at least three independent experiments. The accuracy and statistical significance of the expression ratios were determined by applying the analysis provided by the CYBER-T program (http://visitor.ics.uci.edu/genex/cybert). To process our data with CYBER-T, for each spot we calculated log 2 ratios and a paired expression value to estimate the mean expression level within the experimental and control data sets. Genes whose expression ratios changed >2-fold and had P values <0.01 were considered up- or down-regulated. Genes with a P value >0.01 were not considered as regulated, regardless of their differential expression level. The P value obtained by CYBER-T calculations represents the probability of a specific gene to be differentially expressed under the two experimental conditions.
Clustering Analysis. The data sets generated by microarray analysis over the 5-h growth time were subjected to clustering analysis by ARRAYSCOUT software (LION bioscience, Heidelberg). Hierarchical clustering was used to analyze the expression patterns of up- and down-regulated genes. The noncentroid UPGMA (unweighted pair group method with arithmetic mean; ref. 21) algorithm was applied to generate a set of hierarchical clusters on the expression profiles of the 235 regulated genes.
RT-PCR. Total RNA was isolated from N. meningitidis MC58 by using the RNeasy kit (Qiagen, Valencia, CA), and RT-PCR was performed by using the SuperScript RT-PCR kit (Invitrogen) as recommended by the manufacturer. Five-microliter samples were collected at five PCR cycle intervals and electrophoresed on a 1% agarose gel. The densities of the bands were measured by using Bio-Rad QUANTITY ONE 4.0 quantitation software.
Fur Binding Studies. The ability of Fur to bind to gene promoterproximal regions was determined by electrophoretic mobility-shift assay (EMSA) on PCR-amplified fragments as described (18).
Results
Global Analysis of N. meningitidis Gene Expression During Growth Under Iron-Replete and -Depleted Conditions. To examine the simultaneous expression of the entire gene repertoire of N. meningitidis MC58 in response to iron, RNA profiles of bacteria grown in the presence and absence of iron were compared over a 5-h period by using whole-genome microarrays. Growth of N. meningitidis MC58 in the presence of iron altered the expression of 235 genes (11% of the N. meningitidis MC58 genome) by >2-fold in three independent experiments (Table 1 and Fig. 4, which are published as supporting information on the PNAS web site, www.pnas.org). A high percentage of genes were up-regulated (153 genes), whereas 80 were down-regulated and 2 were either up- or down-regulated depending on the time of growth. Functional classification of the genes with an altered expression profile revealed that the most abundant group of genes was the family of hypothetical genes (84 genes), of which 49 were up-regulated and 34 were down-regulated (Fig. 4). Three other well represented families that were primarily up-regulated on iron addition included genes involved in energy metabolism, protein synthesis, and cell envelope assembly. In addition to the genes belonging to the hypothetical family, the most abundant families of down-regulated genes were those encoding proteins involved in cellular and transport processes, including genes that are directly responsible for iron uptake and transport.
Computational Analysis of the Fur-Binding Sequence in the Promoter/Operator Regions of Iron-Regulated Genes. In an attempt to predict which of the N. meningitidis iron-regulated genes are under the control of the transcriptional regulatory protein Fur, we first grouped all iron-regulated N. meningitidis genes into putative transcriptional units (defined as a cluster of adjacent genes having intergenic regions <50 bp in length). From this analysis, the 235 iron-regulated genes were combined into 203 putative transcriptional units. We then performed a computational analysis on the promoter-proximal regions of all iron-regulated transcriptional units in search for potential Fur-binding sequences. The homology search method was performed by using the FINDPATTERN program of the WISCONSIN PACKAGE (Accelrys, San Diego), under default parameters allowing a maximum of 37% mismatches. The 400-bp upstream regions of each transcriptional unit were scanned for the presence of (i) the Escherichia coli Fur consensus sequence (at least 12 of 19 bp identity) (22), (ii) the Neisseria Fur consensus sequence (at least 13 of 21 bp identity) (23), (iii) the (NATWAT)3 sequence (12 of 18 bp matching) recently proposed as the E. coli Fur-box (6, 24), and (iv) the Fur-binding sequences of the Neisseria gonorrhoeae fur and fbpA genes (at least 12 of 19 and 13 of 33 bp matching, respectively) recently mapped by DNase footprint experiments (23, 25). This analysis revealed that ≈50% of all N. meningitidis iron-regulated genes (including both up- and down-regulated genes) could be under the direct control of Fur (Table 2, which is published as supporting information on the PNAS web site). In particular, 104 regions had homology to at least one of the five Fur-binding sequences used for the computational analysis. Of these regions, 70 were found to carry a DNA sequence with sufficient homology to the Neisseria or E. coli Fur consensus binding sequence within 250-bp upstream sequences. A subset of these genes were also found to contain sequences with homology to the Fur consensus binding sequence composed of the three repeats of the sequence NATWAT. In some instances, distinct Fur-boxes were found more than once on the same intergenic region. Typically, the Fur-binding sequences were located within 150 bp of the transcriptional start site of the down-regulated genes, whereas the Fur-binding sequences of the iron-activated genes had a more scattered distribution (data not shown).
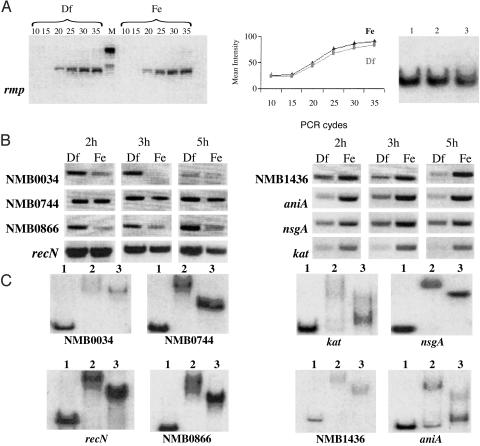
Biochemical Confirmation of Fur-Binding Activity. To support the reliability of both microarray data and Fur-box computer predictions, we selected four iron-repressed genes (recN, NMB0034, NMB0744, and NMB0866) and four up-regulated genes (pan1, kat, nspA, and NMB1436) carrying highly conserved Fur-boxes and subjected them to two types of analysis. First, we measured their transcription levels under iron-replete and -depleted conditions by RT-PCR, starting from the same RNA samples used in microarray experiments. Secondly, the upstream promoter regions of these genes were PCR-amplified and used in gel-shift analysis with E. coli and/or Neisseria Fur. These two analyses confirmed the microarray and computer prediction data, respectively (Table 2, Fig. 1).
Fig. 1.
RT-PCR validation of microarray expression ratios, and EMSA of promoter regions of up- and down-regulated genes. (A Left and Center) RT-PCR and relative quantification on rmp mRNA, used as negative control, obtained from a 2-h culture grown under iron-replete (Fe) and -depleted (Df) conditions. PCR cycles at which transcripts were analyzed are indicated above each lane. M, molecular weight. (A Right) Fur binding to the rmp promoter region by EMSA analysis. Lanes: 1, no protein; 2, E. coli Fur; 3, Neisseria Fur. (B) RT-PCR analysis of representative down- and up-regulated genes obtained from bacterial cultures sampled at 2, 3, and 5 h of growth in iron-replete (Fe) and -depleted (Df) conditions. (C) EMSA analysis of the promoter-proximal regions of the iron-regulated genes represented in B. Lanes: 1, no protein; 2, E. coli Fur; 3, Neisseria Fur.
In addition, 34 regulated transcriptional units were randomly selected and their upstream promoter regions were amplified and subjected to gel-shift analysis. The selection included 20 transcriptional units with recognizable Fur-box sequences and 14 genes for which both computer analysis and manual inspection failed to identify a sufficiently conserved Fur consensus sequence in their upstream regions (Table 2). Overall, 32 of 42 promoter-proximal regions subjected to gel-shift analysis belonged to the class of down-regulated transcriptional units; the remaining 10 regions were from up-regulated genes (Table 2). Both the E. coli and Neisseria Fur proteins were found to bind to the promoter/operator regions of 24 of 32 down-regulated transcriptional units and 8 of 10 up-regulated transcriptional units.
The availability of experimental data for Fur-binding activity on 42 promoter-proximal regions offered the opportunity to define the reliability of the in silico prediction of the Neisseria Fur-box. We found that 27 of 28 promoter regions carrying recognizable Fur-boxes were capable of binding both E. coli and Neisseria Fur (Table 2). In the case of the 14 amplified fragments with no recognizable Fur-box sequences, 9 were not retarded by Fur in the gel-shift experiments; however, the remaining 5 fragments showed an affinity for both the E. coli and Neisseria Fur proteins (Table 2).
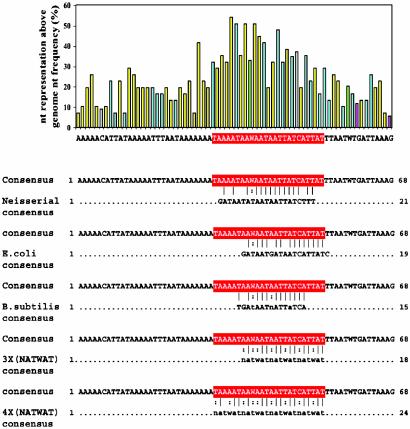
Analysis of the N. meningitidis Fur Consensus Binding Sequence. The gel-shift analysis described above allowed us to identify 32 DNA fragments capable of interacting with both Neisseria and E. coli Fur, 27 of which had a predictable Fur-box. We therefore hypothesized that sequence alignment of these fragments, together with the alignment of the promoter-proximal regions of previously characterized Neisseria Fur-dependent transcriptional units (17, 18, 26), should define a sufficiently reliable N. meningitidis Fur consensus sequence. To facilitate this alignment, we first scanned each fragment for the presence of a E. coli-like Fur sequence, and then launched the homology search on a limited portion of each of these fragments of ≈70 nt with the E. coli-like Fur-binding sequence. In a few cases, more than one Fur-box was predicted on the same fragment, bringing to 32 the total number of regions aligned for Fur-box consensus sequence determination. The analysis defined a conserved region of ≈40 nucleotides centered around a highly conserved sequence constituted by a 4-fold repetition of the NATWAT hexamer recently proposed by Escolar et al. (24) to be the E. coli Fur-binding sequence (Fig. 2). Based on our present analysis, the N. meningitidis Fur-box includes sequences homologous to the proposed E. coli and N. gonorrhoeae Fur-boxes (thus fully justifying the ability of E. coli Fur to bind the N. meningitidis iron-regulated promoter-proximal regions) but extends over a longer nucleotide stretch in which AT nucleotides are particularly abundant.
Fig. 2.
Multiple sequence alignment of upstream regions of EMSA-positive genes. (Upper) Graph reporting the representation of each nucleotide of the derived consensus sequence above the MC58 genomic nucleotide frequency. (Lower) Pairwise alignment of the derived consensus with the previously proposed Fur-boxes. In red is highlighted the most conserved region of the derived consensus, matching with the previously proposed Fur-binding consensus sequence (16–18).
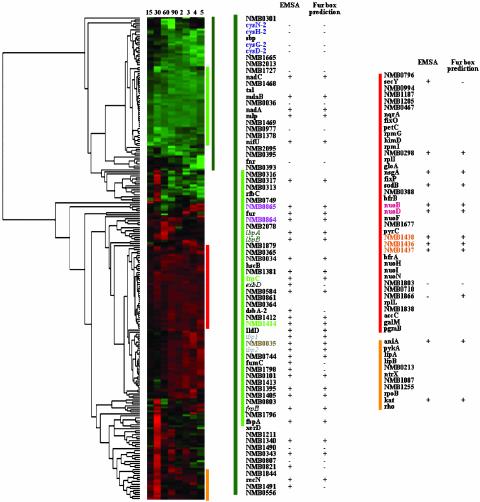
Cluster Analysis of the Neisseria Iron-Regulated Genes. The availability of RNA expression profiles at eight different time points of the growth cycle allowed us to perform a cluster analysis of all of the Neisseria iron-regulated genes. When unsupervised hierarchical clustering was performed, a cluster of 39 down-regulated genes could be defined, at the eighth level of hierarchy, that was remarkably enriched with genes experimentally proven to bind Fur (Fig. 3). This cluster included 26 Fur-regulated genes, subjected either to gel-shift analysis in this study, or to gel-shift and/or footprint analyses in N. gonorrhoeae (17, 18, 26). Our cluster analysis indicated that a few genes demonstrating a positive shift in the gel-shift analysis grouped in clusters with genes that were negative in the gel-shift assay, indicating that not all of the genes found to bind to the Fur protein had a similar RNA expression profile. In addition, 9 of 11 up-regulated genes whose promoter regions were shown to bind Fur also grouped in a common cluster, characterized by constantly up-regulated genes (Fig. 3). Taken together, cluster analysis clearly indicates that all genes showing a constant down-regulation under iron-replete conditions are regulated by Fur. Genes having different transcription profiles cannot be excluded a priori from the Fur regulon and likely represent genes with a more complex regulatory mechanism in which Fur may not be the exclusive regulatory protein.
Fig. 3.
Unsupervised hierarchical clustering of N. meningitidis iron-regulated genes. Genes are grouped according to the similarity of their transcriptional profile, over the growth period. Gene expression ratios are represented by bars; green and red indicate down- and up-regulated genes, respectively. Vertical lines indicate the clusters containing the genes analyzed by EMSA, the names of which are reported nearby, together with the results of EMSA analysis and the computational search of Fur-boxes. The dark-green vertical bars indicate the down-regulated gene clusters, and the light-green vertical bars indicate the cluster of highly down-regulated, Fur-dependent genes. The names of genes organized as single transcriptional units are visualized in the same color.
Discussion
Extensive physiological studies in the pathogenic neisseriae have revealed relatively few examples of the regulation of gene expression by DNA-binding proteins. Among these, the regulatory protein Fur has been proposed to function in the control of several putative virulence factors required for survival within the human host. However, a detailed analysis of the role of Fur in Neisseria gene regulation has not previously been performed. In this study we have used microarray technology together with computational analysis and in vitro binding studies to identify targets of the Fur transcriptional regulatory protein in N. meningitidis. Our studies demonstrate that iron can regulate a broad array of N. meningitidis genes through both Fur-dependent and -independent pathways.
Our microarray analysis indicates that under iron-depleted conditions several virulence-associated genes are overexpressed. These include NMB0393, NMB0977, and NMB1857, which are homologous to E. coli genes involved in toxin production and multidrug resistance (27), NMB0103, which encodes a putative bacteriocin resistance protein, and genes encoding surface proteins potentially involved in cell adhesion: the adhesion complex protein NMB2095, the type IV pilin-related protein NMB2016, the lipoprotein NMB1898, the FrpC protein (NMB1415), and the frpA/C-related genes NMB0364, NMB0584, NMB1402, NMB1405, NMB1412, and NMB1414. The observations that most of these genes group in the same cluster family and that the promoter-proximal region of several of these genes bind Fur in the gel-shift analysis strongly suggest that the regulation of these genes is directly mediated by Fur. DNA recombination and repair mechanisms are additional critical processes for Neisseria pathogenesis and may support bacterial adaptation to the host environment. Iron starvation has been reported to induce pilin antigenic variation by increasing the frequency of recombination events (28). In agreement with these studies, our results indicate that the recN gene is under the control of Fur and suggest that in N. meningitidis Fur may be directly involved in DNA recombination and repair processes.
An interesting finding in this study was the observation that growth of N. meningitidis under iron-replete conditions resulted in an increased expression of a broad array of genes, a remarkable proportion of which carry potential Fur-binding sequences in their promoter-proximal regions. In particular, we have shown that the expression of secY and sodB, previously demonstrated to be components of the gonococcal Fur regulon (18), was also increased under iron-replete conditions in N. meningitidis, and that Fur was capable of binding the promoter region of these genes. Other interesting genes that were up-regulated during growth with iron were nspA and aniA. The NspA protein is known to elicit a protective antibody response against N. meningitidis serogroups A, B, and C (29, 30) and was recently proposed as a vaccine candidate for the prevention of meningococcal disease. The aniA gene, which encodes a nitrite oxide reductase, is one of the key enzymes involved in anaerobic nitrate/nitrite respiration and nitrogen assimilation, and was shown to be a major antigen in patients with gonococcal disease (31). Adjacent to aniA and transcribed in the opposite direction is the norB gene, which encodes a nitric oxide reductase, a second enzyme of the nitrogen assimilation pathway. This gene is also constantly up-regulated during growth with iron and appears to have a well conserved Fur-box sequence in its upstream promoter region. Our results suggest that aniA and norB may undergo at least two levels of positive regulation; one mediated by Fur, and another involving the positive regulator Fnr, the gene of which is known to be activated under strict anaerobic conditions (32, 33). The fine-tuned regulation of these two genes may have important biological implications. Although this concerted mechanism of regulation awaits experimental support, a possible role for fnr in N. meningitidis virulence could be envisaged, whereby Fnr, in concert with Fur, regulates a number of genes that become relevant for optimal bacterial survival and growth in the host.
Several of the iron-regulated genes identified in this study belong to the hypothetical gene family, highlighting how much remains to be understood about the biology and biochemistry of iron regulation in Neisseria. Particularly interesting is a three-gene operon (NMB 1436–38) that is up-regulated on iron addition. Computational analysis indicates that one of these genes encodes a protein carrying an iron–sulfur cluster, a motif often encountered in enzymes involved in the detoxification of oxygen radicals. We have recently constructed a N. meningitidis mutant carrying the complete deletion of this operon and found that the mutant is highly sensitive to hydrogen peroxide (R.G. and G.G., unpublished studies). Thus, it is reasonable to speculate that the function of this operon is to scavenge the highly toxic oxygen radicals produced in the cell by the iron-catalyzed Fenton reaction.
Whereas the repressive mechanism of Fur has been thoroughly investigated (6), the mechanism of positive regulation by this protein has not been well elucidated (13, 14). A recent study (15) suggests that part of the activating properties of Fur may be defined at the posttranscriptional level. In E. coli, a small RNA (RyhB) that is negatively regulated by Fur has been shown to down-regulate a set of iron-storage proteins when iron is limiting (15). Although our search of the N. meningitidis MC58 genome did not identify a ryhB homolog, we cannot rule out the existence of such a mechanism in N. meningitidis. Fur may also exploit different mechanisms of DNA binding that are operator-dependent, resulting in differential regulation. Delany et al. (12) have demonstrated that in Helicobacter pylori the Fur protein binds to the iron-activated pfr promoter and that Fe2+ decreases the efficiency of binding. Furthermore, these investigators have reported that structural differences are apparent between the iron-activated pfr promoter and an iron-repressed promoter.
The data presented here indicate that iron can also regulate gene expression in a Fur-independent manner. Approximately 50% of the up- and down-regulated genes do not present a canonical Fur consensus sequence in their upstream promoter regions. Likewise, we did not observe binding of the Fur protein to the promoter regions of several of these genes. The existence of iron-dependent, Fur-independent regulation is attractive and deserves further experimental analysis. In this context, it has recently been reported that growth of Pseudomonas aeruginosa and Pasteurella multocida in the presence of iron also results in the increased expression of a high proportion of genes (34, 35). However, the Fur-box was observed in only a subset of the P. aeruginosa-regulated genes, suggesting an involvement of additional regulatory proteins in regulation of iron-regulated genes.
The binding sequence of the Fur protein has been continuously revisited by several investigators and has been highly debated. de Lorenzo et al. (36) have proposed a 19-bp consensus, based on DNaseI analysis. Furthermore, an analysis of 33 gene promoters identified to be iron and Fur regulated by a Fur titration assay revealed a similar 19-bp consensus (10). A 21-bp Neisseria consensus that shares ≈80% homology to the E. coli consensus has been derived and proposed to be the binding sequence of the Neisseria Fur protein (23). Although the 19- and 21-bp consensuses serve as good indicators to identify a Furregulated gene, the Fur-binding sequence identified in the promoters of several Fur-regulated genes differed from the classical Fur-binding sequence (37–42). In light of these results, the Fur-binding sequence has been reinterpreted to be a combination of three repeats of 6-bp 5′-NAT(A/T)AT-3′ rather than a 19-bp palindrome (24). The presence of hexameric repeats with varying degree of homology may allow for binding of Fur with varying affinity, thus allowing a wide degree of regulation for Fur-dependent genes (6, 12, 24). Recent studies of Fur-regulated genes in B. subtilis have proposed that an overlapping 7-1-7 heptamer motif is the minimal recognition unit for high-affinity Fur binding (43, 44). Our alignment of the promoter-proximal regions of the N. meningitidis iron and Fur-regulated genes identified in this study suggests that the hexameric repeat 5′NATW(A/T)AT 3′ is the Fur-binding unit and is in agreement with the proposed requirement of three minimal repeat units for efficient Fur binding (24).
Although computer predictions and EMSA analysis allow for the identification of Fur-binding regions, caution should be taken in assigning a priori the biological significance to these results. Indeed, neither analysis addresses the affinity of Fur for specific promoter/operator regions or, ultimately, the relevant function of these interactions in vivo. In this context, it is interesting to point out that when the upstream regions of 600 iron-insensitive genes were subjected to computer analysis using the consensus Fur-box defined in this study, 31% of the genes appeared to carry a putative Fur-binding site. Although this percentage is statistically lower than the 51% frequency found in the iron-regulated genes (see Results), it is still remarkably high. The high percentage of Fur-boxes in proximity of genes insensitive to iron regulation can be explained if one assumes a low affinity of Fur for these regions.
By using hierarchical cluster analysis we were able to group the Neisseria iron-regulated genes into gene clusters having similar transcription activities throughout the growth cycle. We then examined whether there were clusters containing only genes with Fur-binding sequences. Indeed, a gene cluster was generated solely composed of Fur-binding genes. The genes of this group are characterized by an expression profile of constant down-regulation, which for some genes was as low as 10-fold and never <1.5-fold with respect to expression under iron-replete conditions. Considering that 32 of the 68 down-regulated transcriptional units were subjected to gel-shift analysis and that previous studies had demonstrated the Fur-binding activity of 5 additional down-regulated transcriptional units (17, 18), this analysis appears to be a powerful tool to predict Fur-regulated genes (gel-shift data available for 46 genes, corresponding to 58% of all down-regulated genes). Considering that in the promoter region of some of these genes a canonical Fur-box sequence cannot be recognized, cluster analysis, rather than Fur-box prediction, appears to be the most reliable approach for predicting Fur-binding activity. Our gel-shift and cluster analyses have also shown that genes exist that bind Fur but belong to different cluster families. One possible interpretation of these results is that under the experimental conditions used, transcription of these genes is affected by, in addition to Fur, other regulatory factors. Should this be experimentally demonstrated, cluster analysis would become particularly useful to elucidate new mechanisms of iron gene regulation involving more than one regulatory protein.
Supplementary Material
Acknowledgments
We thank Sarika Agarwal, Borys Szmigielski, Sergio Balloni, Claudio Donati, V. Masignani, and M. Scarselli for assistance with computer analysis, Isabel Delany for helpful discussions, Filippo Randazzo and Joel Berger for microarray spotting and designing, and A. Maiorino for expert secretarial assistance. This study was supported by Public Health Service Grants U19AI 38515 and AI48611 (to C.A.G.).
Abbreviation: EMSA, electrophoretic mobility-shift assay.
References
- 1.Berish, S. A., Subbarao, S., Chen, C. Y., Trees, D. L. & Morse, S. A. (1993) Infect. Immun. 61, 4599-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bsat, N. & Helmann, J. D. (1999) J. Bacteriol. 181, 4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill, P. J., Cockayne, A., Landers, P., Morrissey, J. A., Sims, C. M. & Williams, P. (1998) Infect. Immun. 66, 4123-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Staggs, T. M. & Perry, R. D. (1992) Mol. Microbiol. 6, 2507-2516. [DOI] [PubMed] [Google Scholar]
- 5.Thomas, C. E. & Sparling, P. F. (1994) Mol. Microbiol. 11, 725-737. [DOI] [PubMed] [Google Scholar]
- 6.Escolar, L., Perez-Martin, J. & de Lorenzo, V. (1999) J. Bacteriol. 181, 6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karjalainen, T. K., Evans, D. G., Evans, D. J., Jr., Graham, D. Y. & Lee, C. H. (1991) Microb. Pathog. 11, 317-323. [DOI] [PubMed] [Google Scholar]
- 8.Litwin, C. M. & Calderwood, S. B. (1993) Clin. Microbiol. Rev. 6, 137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prince, R. W., Cox, C. D. & Vasil, M. L. (1993) J. Bacteriol. 175, 2589-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stojiljkovic, I., Baumer, A. J. & Hantke, K. (1994) J. Mol. Biol. 236, 531-548. [DOI] [PubMed] [Google Scholar]
- 11.Vasil, M. L., Ochsner, U. A., Johnson, Z., Colmer, J. A. & Hamood, A. N. (1998) J. Bacteriol. 180, 6784-6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delany, I., Spohn, G., Rappuoli, R. & Scarlato, V. (2001) Mol. Microbiol. 42, 1297-1309. [DOI] [PubMed] [Google Scholar]
- 13.Dubrac, S. & Touati, D. (2000) J. Bacteriol. 182, 3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubrac, S. & Touati, D. (2002) Microbiology 148, 147-156. [DOI] [PubMed] [Google Scholar]
- 15.Masse, E. & Gottesman, S. (2002) Proc. Natl. Acad. Sci. USA 99, 4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelissen, C. N., Kelley, M., Hobbs, M. M., Anderson, J. E., Cannon, J. G., Cohen, M. S. & Sparling, P. F. (1998) Mol. Microbiol. 27, 611-616. [DOI] [PubMed] [Google Scholar]
- 17.Thomas, C. E. & Sparling, P. F. (1996) J. Bacteriol. 178, 4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sebastian, S., Agarwal, S., Murphy, J. R. & Genco, C. A. (2002) J. Bacteriol. 184, 3965-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grifantini, R., Bartolini, E., Muzzi, A., Draghi, M., Frigimelica, E., Berger, J., Ratti, G., Petracca, R., Galli, G., Agnusdei, M., et al. (2002) Nat. Biotechnol. 20, 914-921. [DOI] [PubMed] [Google Scholar]
- 20.Tettelin, H., Saunders, N. J., Heidelberg, J., Jeffries, A. C., Nelson, K. E., Eisen, J. A., Ketchum, K. A., Hood, D. W., Peden, J. F., Dodson, R. J., et al. (2000) Science 287, 1809-1815. [DOI] [PubMed] [Google Scholar]
- 21.Kaufman, L. & Rousseeuw, P. J. (1990) Finding Groups in Data: An Introduction to Cluster Analysis (Wiley, New York).
- 22.Stojiljkovic, I. & Srinivasan, N. (1997) J. Bacteriol. 179, 805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genco, C. A. & Desai, P. J. (1996) Trends Microbiol. 4, 179-184. [DOI] [PubMed] [Google Scholar]
- 24.Escolar, L., Perez-Martin, J. & de Lorenzo, V. (1998) J. Mol. Biol. 283, 537-547. [DOI] [PubMed] [Google Scholar]
- 25.Desai, P. J., Angerer, A. & Genco, C. A. (1996) J Bacteriol. 78, 5020-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schryvers, A. B. & Stojiljkovic, I. (1999) Mol. Microbiol 32, 1117. [DOI] [PubMed] [Google Scholar]
- 27.Chatterjee, P. K. & Sternberg, N. L. (1995) Proc. Natl. Acad. Sci. USA 92, 8950-8954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Serkin, C. D. & Seifert, S. (2000) Mol. Microbiol. 37, 1075-1086. [DOI] [PubMed] [Google Scholar]
- 29.Martin, D., Brodeur, B. R., Hamel, J., Couture, F., de Alwis, U., Lian, Z., Martin, S., Andrews, D. & Ellis, R. W. (2000) J. Biotechnol. 83, 27-31. [DOI] [PubMed] [Google Scholar]
- 30.Moe, G. R., Tan, S. & Granoff, D. M. (1999) Infect. Immun. 67, 5664-5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Householder, T. C., Belli, W. A., Lissenden, S., Cole, J. A. & Clark, V. L. (1999) J. Bacteriol. 181, 541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anjum, M. F., Stevanin, T. M., Read, R. C. & Moir, James W. B. (2002) J. Bacteriol. 184, 2987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bagg, A. & Neilands, J. B. (1987) Biochemistry 26, 5471-5477. [DOI] [PubMed] [Google Scholar]
- 34.Paustian, M. L., May, B. J. & Kapur, V. (2001) Infect. Immun. 69, 4109-4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochsner, U. A., Wilderman, P. J., Vasil, A. I. & Vasil, M. L. (2002) Mol. Microbiol. 45, 1277-1287. [DOI] [PubMed] [Google Scholar]
- 36.de Lorenzo, V., Giovannini, F., Herrero, M. & Neilands, J. B. (1987) J. Bacteriol. 169, 2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chai, S., Welch, T. J. & Crosa, J. J. (1998) J. Biol. Chem. 273, 33841-33847. [DOI] [PubMed] [Google Scholar]
- 38.Griggs, D. W. & Konisky, J. (1989) J. Bacteriol. 171, 1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunt, M. D. & McIntosh, M. A. (1994) J. Bacteriol. 176, 3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochsner, U. A., Vasil, A. I. & Vasil, M. L. (1995) J. Bacteriol. 177, 7194-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tardat, B. & Touati, D. (1993) Mol. Microbiol. 9, 53-63. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, D. K., Beliaev, A. S., Giometti, C. S., Tollaksen, S. L., Khare, T., Lies, D. P., Nealson, K. H., Lim, H., Yates, J., III, Brandt, C. C., et al. (2002) Appl. Environ. Microbiol. 68, 881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baichoo, N. & Helmann, J. D. (2002) J. Bacteriol. 184, 5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baichoo, N., Wang, T., Ye, R. & Helmann, J. D. (2002) Mol. Microbiol. 45, 1613-1629. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.